Abstract
The extent of enthalpy–entropy compensation in protein–ligand interactions has long been disputed because negatively correlated enthalpy (ΔH) and entropy (TΔS) changes can arise from constraints imposed by experimental and analytical procedures as well as through a physical compensation mechanism. To distinguish these possibilities, we have created quantitative models of the effects of experimental constraints on isothermal titration calorimetry (ITC) measurements. These constraints are found to obscure any compensation that may be present in common data representations and regression analyses (e.g., in ΔH vs. –TΔS plots). However, transforming the thermodynamic data into ΔΔ-plots of the differences between all pairs of ligands that bind each protein diminishes the influence of experimental constraints and representational bias. Statistical analysis of data from 32 diverse proteins shows a significant and widespread tendency to compensation. ΔΔH versus ΔΔG plots reveal a wide variation in the extent of compensation for different ligand modifications. While strong compensation (ΔΔH and −TΔΔS opposed and differing by < 20% in magnitude) is observed for 22% of modifications (twice that expected without compensation), 15% of modifications result in reinforcement (ΔΔH and −TΔΔS of the same sign). Because both enthalpy and entropy changes arise from changes to the distribution of energy states on binding, there is a general theoretical expectation of compensated behavior. However, prior theoretical studies have focussed on explaining a stronger tendency to compensation than actually found here. These results, showing strong but imperfect compensation, will act as a benchmark for future theoretical models of the thermodynamic consequences of ligand modification.
Keywords: enthalpy–entropy compensation, free energy, binding thermodynamics, protein–ligand interactions, protein complexes
Introduction
The interactions between proteins and their ligands can be characterized by the free energy, enthalpy, and entropy changes associated with the binding reaction. The Gibb's free energy (ΔG) for a reaction carried out at a temperature T is related to the change in enthalpy (ΔH) and the change in entropy (ΔS) of that reaction by
| (1) |
It is often observed that the range of ΔG values for groups of related reactions is much smaller than the ranges of their associated changes in ΔH and TΔS. This has led to the idea that the differences in the enthalpic and entropic contributions are negatively correlated or “compensated” as a result of some shared features of the physical reaction mechanism.1 However, it is also known that negative correlations of enthalpy and entropy changes arise from experiment design, random measurement errors, or from the methods of analysis of measurements.1–3 Claims for enthalpy–entropy compensation in protein–ligand interactions often fail to examine other possible source of correlation and in cases where statistical analyses have been performed, the observability of compensation has remained in doubt.4–6 In addition to being an issue of fundamental interest, it remains important to establish the presence (and extent if present) of compensation in experimental studies of protein–ligand binding because such data would impinge on the assessment of models of molecular interaction and may affect how thermodynamic information is used in rational drug design.7 Consequently, here we revaluate the evidence for enthalpy–entropy compensation in protein–ligand interactions using the large quantity of isothermal titration calorimetry (ITC) data that has been produced in recent years.
Much of the historical difficulty in investigating, and consequent controversy with respect to the existence/extent of, compensation in protein reactions has its origin in the dominant early role played by van't Hoff (and Arrhenius) analyses. Until relatively recently, the simplest way to find the enthalpy and entropy changes of a reaction was via a plot of the logarithm of the equilibrium constant K (= e−ΔG/RT) against the reciprocal of the temperature. Rearranging (1) gives the van't Hoff equation describing such a plot.
| (2) |
The slope of the line is −ΔH and the intercept is ΔS (divided by the gas constant). However, this approach introduces relatively large errors in ΔH compared to the magnitude of ΔG. Because errors in the slope are correlated with errors in the intercept, errors alone can produce highly correlated changes in ΔH and ΔS for a series of reactions.2,3 Statistical tests have been proposed to discriminate cases of compensation from these artefactual correlations.4,8,9 Using such tests, it was found that many reported instances of high correlation between ΔH and ΔS for a variety of chemical reactions are indistinguishable from experimental artefacts,8 including several examples of the interactions of individual proteins with series of ligands.4–6
ITC measures the ΔH of a binding reaction directly through the heat output or input associated with a titrated reaction at constant temperature and ΔG is found from a nonlinear regression analysis of the titration curve.10 Unlike a van't Hoff analysis, these measurements are essentially independent and usually precise (e.g., mean reported errors for ΔH and ΔG are 1.5 and 0.5 kJ mol−1, respectively, in the SCORPIO database11 of ITC data, and 1.7 and 0.4 kJ mol−1 in a recent systematic analysis of replicated experiments on many protein–ligand systems12). Consequently, enthalpy–entropy correlation arising from measurement errors, which in the case of ITC results from the use of Eq. (1) to determine TΔS, is much smaller than that for a van't Hoff analysis. Indeed, the precision of ITC measurements is such that it has again become common to assume that statistical testing is unnecessary and that a high-degree correlation in a ΔH versus TΔS plot alone is sufficient evidence for compensation.13–15
Unfortunately, there are several sources of potential correlation in ITC data, which must be eliminated or accounted for in any analysis. In addition to the small correlation due to measurement errors, Cooper et al.16 have pointed out that the range of ΔG values that are accurately measurable using the most common direct ITC method is limited by the necessity to obtain an analyzable sigmoidal titration curve within the constraints of protein solubility and instrument sensitivity. This “affinity window” is narrower for direct ITC measurements than that for many other methods for monitoring binding and thus poses a particular problem for rigorous analysis of compensation. In addition, correlation can arise from “extra-experimental” factors, that is, biases in the nature of system that are selected for study.4 For example, interactions with cognate ligands are constrained in their affinity because they are usually required to be reversible and have a significant bound population at biological concentrations.4,16 Also, studies of protein with synthetic ligands often involve a series of similar changes being made to the ligand. These may each result in similar changes to ΔH and TΔS and introduce confounding correlations into the data.4,17 As a consequence of these issues, careful data selection and statistical analysis of the effects of errors and experimental factors are required for analysis of enthalpy–entropy relationships in ITC data.
Here, we combine ITC data from many proteins to investigate whether compensation is an observable feature of protein–ligand interactions. In selecting data from a wide range of systems, we minimize the potential for extra-experimental chemical biases affecting our conclusions. To enable statistical testing, we create models of the correlation expected to arise as a result of errors and the ITC affinity window; putting previous qualitative arguments16 about these factors on a quantitative footing. We show that these experimental sources of correlation are so large in traditional ΔH versus TΔS plots as to render them of no use in identifying compensation, reinforcing earlier analyses.6,18 However, we find that a new approach based on analysis of the distributions of the relative thermodynamic values (ΔΔH, TΔΔS, and ΔΔG) of all pairs of ligands that bind to each protein does allow the effects of enthalpy–entropy compensation to be distinguished from other sources of correlation. Our analysis shows that there is significant, widespread and strong tendency to enthalpy–entropy compensation in protein–ligand interactions. However, it is also clear that there is a range of degrees of compensation observed within protein–ligand systems. We reconsider previously suggested theoretical models for compensation and conclude that the prior theoretical emphasis on explaining perfect compensation is unwarranted by/inconsistent with the experimental data and that improved theories are needed to explain the varied extent of compensation actually observed.
Results
Overall correlation in ΔH versus −TΔS plots is high but is dominated by experimental constraints
The selected experimental thermodynamic data (see Methods section) describe 171 protein–ligand interactions involving 32 proteins. This dataset contains a variety of protein–peptide, protein–carbohydrate, and protein–nucleotide interactions in addition to natural and synthetic inhibitors. These data have a strong diagonal distribution in a ΔH versus −TΔS plot with a Pearson's correlation coefficient of 0.97 (Fig. 1). Li et al.15 in surveying PDBcal data have interpreted similar high correlation as evidence for enthalpy–entropy compensation. However, it is pertinent to ask to what extent limitations of the ITC method contribute to this appearance and degree of correlation?16 To answer this question, a quantitative model of these limitations is required.
Figure 1.
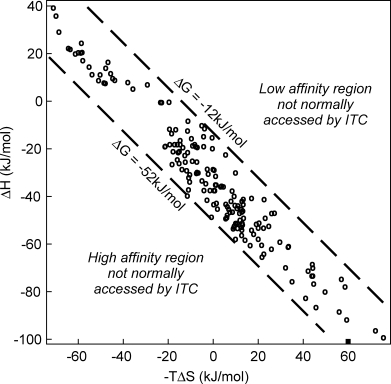
Thermodynamic parameters derived from ITC experiments for 171 protein–ligand interactions. The limitations of experimental procedures constrain measurements to the affinity window bounded by the dashed lines.
A model of the effect of the affinity window can be derived from the actual distribution of ΔG values found for all direct titration ITC experiments in the PDBcal and SCORPIO databases [Fig. 2(A)]. This distribution arises from the joint effects of extra-experimental factors and the constraints of the affinity window. Peaks in the distribution are attributable to extra-experimental selection, for example, a subset of compounds in these databases are from medicinal chemistry series, which have a more limited range of affinities.11 Underlying the distribution is a trend whereby high or low ΔG values are progressively less likely to be observed (high affinity titration curves are difficult to interpret because there is an abrupt transition to saturation; at low affinity, higher concentrations are required and few compounds are sufficiently soluble). At the centre of the window, all investigated protein–ligand systems are equally likely to yield results, and toward the extremes, the probability of an interpretable experimental result falls to zero. A probabilistic model for the effects of the affinity window Paw(ΔG) can be created [bold line in Fig. 2(A)] that approximates this underlying behavior.
Figure 2.
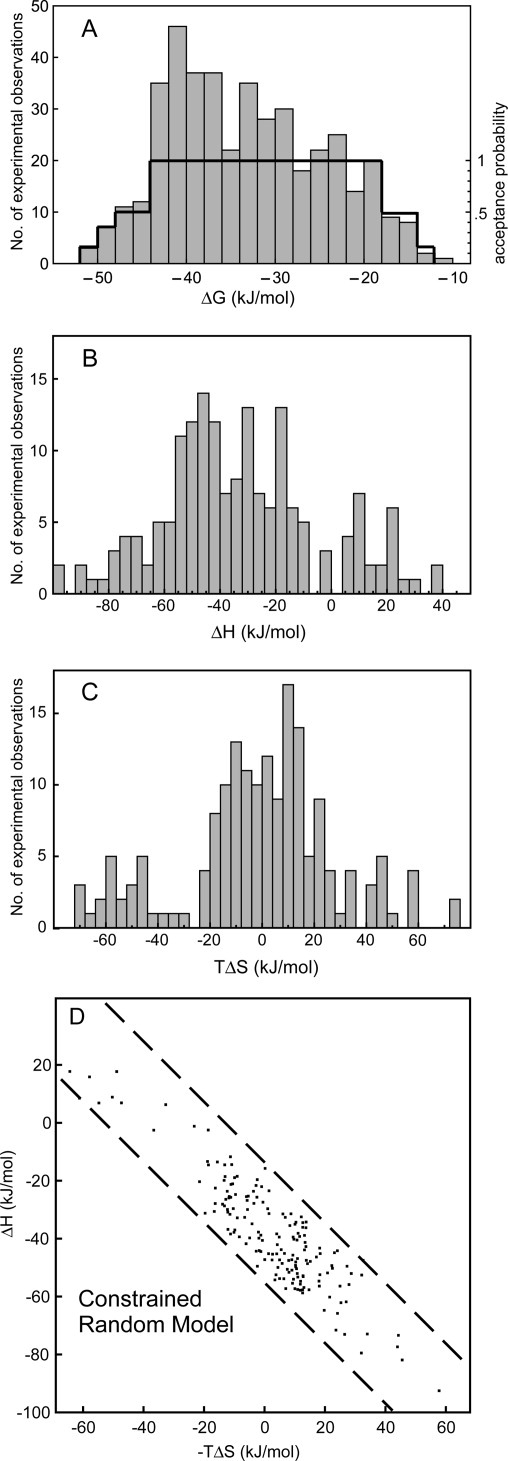
The effect of the affinity window and other experimental factors can be modeled using information from experimental measurements. (A) The distribution of all direct ITC measurements of ΔG reported in the PDBcal and SCORPIO databases is used to estimate the underlying probability of an experiment being successfully performed (bold line). This probability forms a basis for modeling the effects of the affinity window. (B) and (C) Experimental distributions of ΔH and −TΔS values found in the experimental dataset in Figure 1. Values for ΔH and −TΔS from the experimental dataset can be independently randomly sampled and then accepted with the probability defined in (A) to create a model distribution illustrating the effect of experimental constraints (D) on an otherwise random sample.
The hypothetical “constrained random” distribution of ΔH and −TΔS changes that would be observed in the presence of experimental constraints, but in the absence of any physical mechanism of compensation, can then be created by independent random resampling of the experimental values for ΔH and –TΔS [whose distributions are shown in Fig. 2(B,C)], computing the corresponding ΔG and accepting the pair of values with a probability of Paw(ΔG) (see Methods section for details). An exemplar constrained random distribution [Fig. 2(D)] appears very similar to the experiment (Fig. 1). The correlation coefficient of 20 resampled distributions ranged from 0.82 to 0.93, that is, more than 95% of the experimentally observed correlation in ΔH versus −TΔS plots of data from multiple protein systems is potentially explicable in terms of experimental constraints. What of the slightly greater correlation in the experimental data? Is this evidence of a small compensation effect? In short, no. Combining data from more than one protein in a ΔH versus −TΔS plot introduces an additional source of artefactual correlation.
Plots of the relative thermodynamics of ligands binding to each protein are more appropriate for consideration of the effects of ligand modification
The relative thermodynamics of pairs of ligands (A and B) binding to the same protein are ΔΔGA→B = ΔGB − ΔGA, ΔΔHA→B = ΔHB − ΔHA, and TΔΔSA→B = TΔSB − TΔSA. In our dataset, we find that the ranges of the ΔΔH and −TΔΔS values for such pairs of ligands are approximately half those observed for ΔH and −TΔS [compare Figs. 2(B,C) and 3(B,C)]. Consequently, we see that the appearance of a ΔH versus −TΔS plot of multiple protein–ligand systems (Fig. 1) not only reflects changes to the ligands but also contains a large contribution from interprotein variation that creates additional correlation. The overall effect of experimental constraints and interprotein variation in ΔH versus −TΔS plots is to make it statistically impracticable to identify any compensation that may be present.
Figure 3.
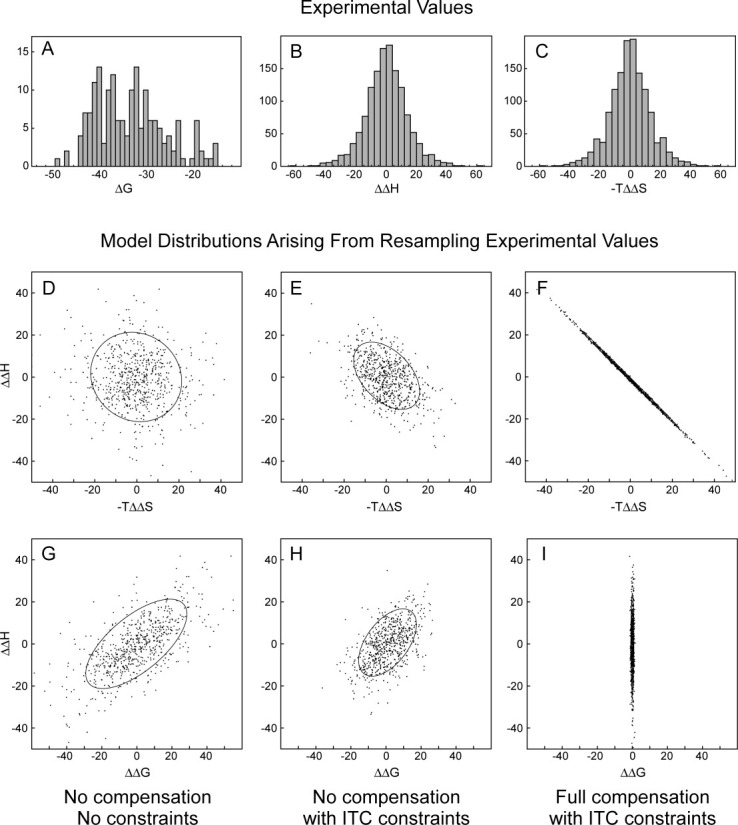
Illustrative models of the effect of experimental constraints and full/exact compensation on otherwise random distributions of the thermodynamic differences between ligands. (A), (B), and (C) Experimental distributions of ΔG for ligands and ΔΔH and −TΔΔS for pairs of ligands that bind the same protein. These can be sampled (see Methods section) to generate a model of unconstrained random changes in enthalpy and entropy (D and G), of the effect of experimental constraints of the affinity window plus correlated error (E and H), and of full compensation with measurement errors in (F and I). The ellipses surround 75% of the experimental datapoints ordered by the differences from the mean of their component coordinates.
However, if we instead plot the relative thermodynamics of all pairs of ligands that bind the same protein, we both remove the interprotein variation and are able to explicitly represent the thermodynamic consequences of all ligand modifications. Further, because the range of ΔΔH variation is smaller than ΔH, the correlation created by the affinity window is reduced and there is a greater potential for statistically distinguishing the effects of experimental constraints from any underlying physical behavior. In a similar manner to that above, models of the expected distributions of ΔΔH, −TΔΔS, and ΔΔG values under a variety of hypothetical scenarios can be created by resampling the experimental data (see Methods for details). Exemplar model distributions (Fig. 3) clearly illustrate the effects of the experimental constraints on the observed distribution and that these are distinct from those anticipated for a mechanism which creates full (exact) enthalpy–entropy compensation.
Experimental ΔΔH versus −TΔΔS and ΔΔH versus ΔΔG plots are distinct from the effects of experimental constraints
The slope of the ΔΔH versus −TΔΔS plot for data drawn from the 32 proteins is very close to unity for both the experimental data (1.01) and for the constrained random model (1.04 ± 0.08 for same number of data points) [Fig. 4(A)]. This means that there is no statistical validity in using the slope of this plot (or equivalently of ΔH vs. −TΔS for single protein) for ITC data as evidence for enthalpy–entropy compensation. However, it is also clear that experimental and constrained random model ΔΔH and −TΔΔS values do not have the same distribution. The experimental data are more narrowly distributed about the ΔΔH = −TΔΔS line (with a correlation coefficient for the model of −0.5 and for the experimental data of −0.9).
Figure 4.
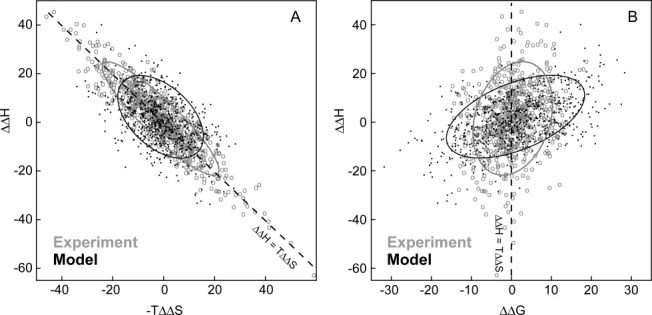
The experimental relative thermodynamics of all pairs of ligands binding each protein have features consistent with the presence of enthalpy–entropy compensation. (A) ΔΔH versus −TΔΔS plot of all experimental differences for pairs of ligands binding to each protein (open circles) superimposed on a sample drawn from a constrained random model (points). The ellipses surrounds 75% of the experimental data ordered by difference from the mean (grey = experimental data, black = model). The experimental distribution is narrower than would be expected in the absence of compensation. (B) An alternative ΔΔH versus ΔΔG plot of the same data. The experimental data are closer in average behavior to that expected for an underlying compensation mechanism, but not equivalent to the fully compensated model (cf. Fig. 3I).
It has been previously shown that ΔH versus ΔG plots are intrinsically less prone to introduce artefactual correlation that has the appearance of compensation6,18 (direct use of the experimental data avoids the correlated error between ΔH and TΔS). A plot of ΔΔH versus ΔΔG values [Fig. 4(B)] makes variation between the ligand pairs more visually apparent. Again, the experimental distribution of data appears to be different from a constrained random model. In this case, the average slopes of the distributions are different (ΔΔH/ΔΔG = 7.79 experimentally and 1.02 ± 0.03 for the constrained random model).
Can the above noted differences between experiment and model data be regarded as significant evidence for compensation being a general feature of protein–ligand interactions? An issue in determining this is that the plots (Fig. 4) are subject to potential bias due to the dominance of those systems with the most ligands (seven proteins provide 80% of the ligand pairs). To draw a robust general conclusion about enthalpy–entropy compensation, it is necessary to draw a more even sample of data from among the proteins and create a test for the statistical significance of differences from the constrained random model.
Protein–ligand interactions exhibit statistically significant greater correlation of enthalpy and entropy changes than expected by chance and constraint
Here we define two coefficients that describe the degree of compensation of the thermodynamic differences between each ligand pair. First, θ, the angle of slope of a vector joining the origin to the data point in a ΔΔH versus −TΔΔS plot (which also equals the angle of a vector joining a pair of ligands in a ΔH versus −TΔS plot) where
| (3) |
And, second, ϕ, the angle of slope of a vector joining the two ligands in a ΔΔG versus ΔΔH plot, where
| (4) |
These coefficients have advantages for statistical testing in that they vary smoothly with enthalpy and entropy changes, unlike the slopes of the vectors themselves. They are based on the two-variable version of the inverse tan function (rather than the more common single variable version) as this has values on a full circle from −π to π, thus allowing use of the robust statistical tests that exist for distributions on a circle. The nature of the distributions of θ and ϕ provided the bases for statistical examination of the effects of constraints and compensation, for example, we see from Fig. 3(F,I) that a series of reactions having perfect enthalpy–entropy compensation θ would be strongly peaked at −π/4 or 3π/4 radians and ϕ at π/2 or −π/2.
Our null hypothesis is that the experimental data are derived from a distribution in which enthalpy–entropy correlations arise solely from experimental constraints and correlated errors, that is, the constrained random model. Rejection of this null hypothesis would lead to the conclusion that there are observable effects of an unknown correlation modifying mechanism. To test this hypothesis, small samples were drawn from the experimental dataset including data from all proteins (but no more than three from each protein) and the distributions of their compensation coefficients compared to equal-sized samples from the constrained random model.
Comparison of the θ [Fig. 5(A)] and ϕ [Fig. 5(C)] distributions of the experimental samples and the constrained random model via the Kuiper statistic (see Methods section) allows us to reject the null hypothesis that the experimental samples are drawn from the constrained random model [Fig. 5(B,D)]. Further, the distribution of θ for a sample of the experimental data is more strongly peaked around the values of −π/4 or 3π/4 than the constrained random model distribution and the peaks in the experimental ϕ are displaced toward π/2 and -π/2. Protein–ligand systems overall are thus seen to exhibit a significantly greater degree of compensation that can be simply attributed to experimental factors.
Figure 5.
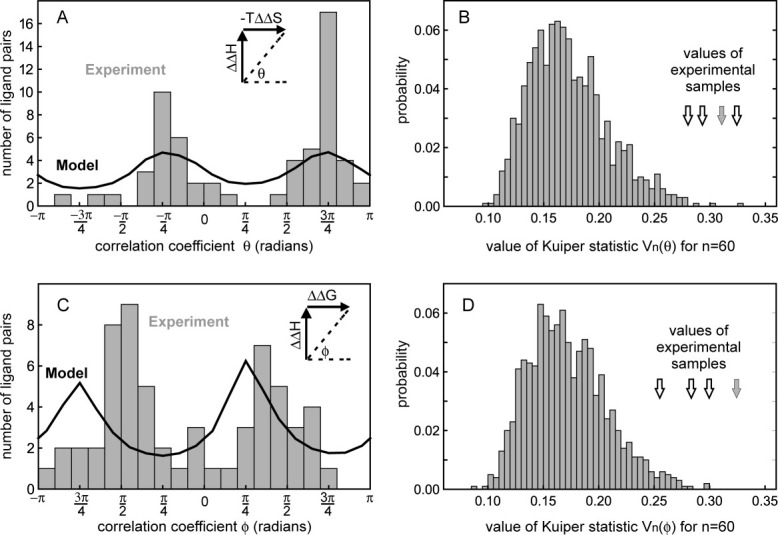
Samples of the experimental data are significantly different from an uncompensated constrained random model and show a strong tendency toward compensation. (A) The compensation coefficient θ relating ΔΔH and −TΔΔS of pairs of ligands is more strongly peaked for a sample drawn from the experimental data (histogram) around the values of −π/4 and 3π/4 radians, which correspond to exact enthalpy–entropy compensation, than the model (black line). (C) The experimental values of the compensation coefficient ϕ relating ΔΔH and ΔΔG are most likely to occur around the values of ±π/2 radians, which correspond to exact enthalpy–entropy compensation, unlike the model. The probability distribution functions of the Kuiper statistic for θ (B) and ϕ (D) generated from the constrained random model (histograms) with arrows indicating the values for four samples of the experimental data (grey arrows are the values of the samples in (A) and (C)). All experimental samples lie in the tails of the functions and are significantly different (e.g., the samples in (A) and (C) give P < 0.002 and P < 0.001, respectively).
How robust is this test of significance to assumptions about the experimental data that are inherent in the constrained random model? Because most features of the model are derived directly from the experimental distributions, the chief area of uncertainty is our assumed level of error in the experimental measurements. In particular, any greater contribution of error to the observed enthalpy variation than to the free energy variation gives rise to some additional appearance of compensation (narrowing the peaks at −π/4 or 3π/4 in the θ-plot or shifting the peaks in the ϕ-plot toward ±π/2). To account for the effects of error in all the model distributions presented here, we have included statistical errors drawn from normal distributions with widths derived from the average reported errors in the SCORPIO database (see Methods section).11 For the tests using the compensation coefficients, the standard deviations of the model ΔΔG and ΔΔH error distributions were 0.7 and 2.1 kJ/mol, respectively. These values are very similar to the variations observed in a recent single laboratory study of replicated experiments in several protein systems12 and thus appear to reasonably accurately reflect the reproducibility of ITC measurements under a single set of experimental conditions. Unfortunately, in addition to ITC measurement error, other experimental variables, for example, concentration measurements, variation in temperature, pH or ionic strength can affect the absolute accuracy or comparability of reported ΔG and ΔH values. Certainly, such systematic experimental differences do contribute to the overall variation of the experimental values (Fig. 1). However, the filters we have applied in selecting data for analysis (see Methods section) minimize the potential for variation of experimental conditions to contribute to the ΔΔG and ΔΔH values. Under a single set of experimental conditions most effects cancel or are merely proportionate to the ΔΔ-differences between ligands. Because the data points are concentrated at small values, proportionate errors affect the distributions in ΔΔ-plots only slightly. For example, a survey of the reproducibility of a single protein–ligand experiment19 showed that variations in concentration measurements between different laboratories created differences of ∼10% in the reported molar ΔH change (contributing ±4 kJ/mol to reported ΔH in the test case), whereas ΔG was only slightly affected due to its logarithmic relationship with concentration/affinity. Fortunately, such concentration errors in a series of experiments on one protein in one laboratory would normally be consistent as the “apparent” protein concentration would be standardized (and through the known stoichiometry so would that of the ligands). Consequently, concentration errors should only proportionately increase or reduce all values in a series. A hypothetical 10% error in the average ΔΔH value in our dataset amounts to only 0.8 kJ/mol, that is, smaller than the statistical error already included. Consequently, we believe that the error included in the models illustrated here (Figs. 3–5) are reasonably representative of our ΔΔ-dataset. Furthermore, additional statistical testing with an increased ΔΔH error of 5 kJ/mol (not shown) still gives significance for the Kuiper statistic at the standard P = 0.05 level, leading us to conclude that the evidence from ITC for an overall tendency to compensation across a wide variety of protein–ligand interactions is robust.
The tendency to compensation is strong but a complete range of compensation and reinforcement is observed
The experimental distributions are not equivalent to a model of full (100%) compensation within experimental errors [compare Fig. 4(A,B) with Fig. 3(F,I)]. There is also no appearance of any other simple (algebraic) relationship between enthalpy and entropy change but a broad distribution of behaviors. The variation in degree of compensation for individual interaction changes is best appreciated in the ΔΔH versus ΔΔG plot, where as a result of the interdependence of ΔΔH, −TΔΔS, and ΔΔG, compensated changes are spread over a large region of the plot [Fig. 6(A)]. The bulk of the experimental data [Fig. 6(B)] from ligand modifications (∼69%) lie in the region of greater than “10%” compensation (bounded by the lines ΔΔH = 0.1 * TΔΔS and TΔΔS = 0.1 * ΔΔH), compared to 45% in this region in the constrained random model. These include 22% of changes which are “highly” compensated with ΔΔH and TΔΔS values differing by <20% (cf. 10% for the constrained random model). For any given value of ΔΔH, the largest affinity changes occur in cases of weak compensation (largely where ΔΔH < 0.1 * TΔΔS) or reinforcement (ΔΔH and −TΔΔS have the same sign) each of which occurs for approximately 15% of modifications (cf. 20% and 35%, respectively, for the constrained random case).
Figure 6.
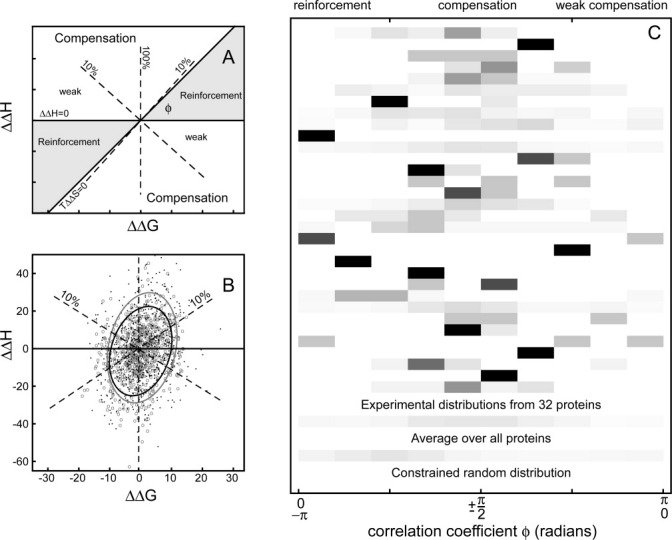
The extent of compensation in protein–ligand interactions varies substantially within protein systems. (A) A schematic relating different regions of the ΔΔH versus ΔΔG plot to different degrees of compensation (10% compensation occurs where ΔΔH = 0.1 * TΔΔS and vice versa). Reinforcement occurs when ΔΔH and −TΔΔS have the same sign. (B) The experimental data from 674 ligand modifications in 32 protein systems (grey open circles) show an overall tendency toward higher degrees of compensation (68% are compensated to better than 10%, 22% to better than 80% − more than twofold greater than random). The experimental distribution is similar to a model created assuming an intrinsic correlation = −0.91 between enthalpy and entropy changes (black points). (C) Normalized frequency distributions of the compensation coefficient ϕ for all pairs of ligands for each of the 32 proteins illustrate the strong tendency to compensation compared to the expectation of a constrained random model and also the wide variation of degree of compensation observed experimentally. Each horizontal line represents a protein (or group of proteins in the bottom two cases), where black (100%) to white (0%) shading represents the proportion of ligands in each interval.
One way to express the general properties of this variation is as an intrinsic correlation between enthalpy and entropy changes. Through trial and error, we find that a model distribution with an intrinsic correlation of ΔΔH and −TΔΔS of −0.91 subject to constraints and correlated errors has a strong resemblance to the experimental dataset [Fig. 6(B)]. This is very close to the experimental correlation for the ΔΔH and −TΔΔS plot and thus consistent with the idea that our modeling procedures are reasonable and that in practise thermodynamic changes in ligand series are sufficiently compensated that experimental constraints have only a small impact on the outcome of most experiments.
Histograms of the distributions of ϕ values for each protein show that there is a substantial variation in the degree of compensation within most protein–ligand series [Fig. 6(C)]. With the exception of some of the smaller series, whose statistical significance is doubtful, the variation between proteins seems small with a consistent central tendency to compensation.
Discussion
The analyses presented here have demonstrated that there is a significant, widespread and strong tendency for the enthalpy changes associated with chemical variation of ligands that bind to a protein to be compensated by opposite changes in entropy. Further, the accuracy of ITC measurements is such that we can see that within each protein–ligand series there are many changes to enthalpy that are well compensated by entropy change, others weakly compensated and some reinforcing. On the one hand, these observations are “intuitively” expected; the simple notion that increased strength of interaction (more favorable enthalpy) will result in reduction in freedom (reduction in entropy) implies that enthalpy and entropy changes are in opposition,20 and we know from experience that this opposition cannot be perfectly balanced in proteins, because it is possible to alter the affinity of ligands through chemical modification. On the other hand, the extent of compensation that is actually present and observable in proteins has been obscure, because common methods of measurement and presentation of the thermodynamic data contain artefacts which mimic compensation. By changing the way in which data is represented, using a large amount of precise ITC data and creating quantitative models of the artefacts, we have been able to reveal the actual extent of compensation.
We have created quantitative constrained random models of the effects of correlated error and the affinity window in ITC measurements that have been previously discussed only qualitatively.7 Studies on individual proteins have often assumed that ITC data is sufficiently accurate that a slope of a ΔH versus −TΔS plot near unity13–15 or closer to unity than a random resampled dataset4 or high correlation in multiprotein data15 can in principle provide evidence for (or is explained by) compensation. Our constrained random models show that this is not the case; in the statistical limit of large numbers of data points, the slope generated by experimental constraints is also unity with high correlation. These models reinforce previous warnings of the danger of misinterpreting features of ΔH vs. –TΔS plots as evidence of compensation.6,16,18 Exner6 has also pointed out that variations of the extent of compensation associated with individual chemical modifications of a ligand are obscured by traditional regression analysis, which, in placing a least-squares fit line through a ΔH versus −TΔS or ΔH versus ΔG plot, emphasizes the largest differences in the data.
We have overcome these problems by a new approach to analysis using plots of ΔΔH vs. –TΔΔS and ΔΔH vs. ΔΔG for all pairs of ligands that bind each protein. Plotting relative thermodynamics in this way means that there is one explicit data point corresponding to each chemical change to the ligand, and that data can be combined from different proteins while avoiding confounding effects of interprotein variation. These ΔΔ-plots also diminish any potential effect on the analysis from nonuniformity in the distribution of ΔG and ΔH measurements, such as those that arise from the difficulty of measuring small heat changes and extra-experimental influences on the affinity of investigated ligands (Fig. 2). In another difference from previous analyses, we find that it is necessary to use nonparametric statistical tests and representations of ITC data because, unlike regression analyses, these are robust to the non-normality of the experimental and model distributions that arises from the truncation imposed by the affinity window.
We introduced two “compensation coefficients”, θ and ϕ, that describe the angle of the (ΔΔH, −TΔΔS) and (ΔΔG,ΔΔH) vectors in their respective plots, which have desirable properties for quantitative and statistical comparison. Using these coefficients, we find that it is possible to detect statistically significant compensation in both ΔΔH versus −TΔΔS and ΔΔH versus ΔΔG plots of the experimental data. Although experimental error in ΔΔH tends to increase the appearance of compensation in these plots, we find that this significance is retained at more than twice reported ITC error levels, suggesting that the observation of widespread compensation is robust even if reported experimental errors are underestimated. Variation within a dataset is more clearly visualized in the ΔΔH versus ΔΔG plot or using the ϕ coefficient. Most proteins are found to have a broad distribution of ϕ with a strong central tendency to compensation (Fig. 6). The overall distributions show a more than twofold greater chance of high compensation (ΔΔH within 20% of TΔΔS) than the random case.
What then is the physical origin of this tendency to compensation? A significant strand of theory in this area is concerned to explain the almost complete compensation inferred from regression lines of unity slope through ΔH versus −TΔS plots. Generalized thermodynamic21 or statistical mechanical4 arguments, that do not consider details of molecular structure, have been formulated that predict a high degree of compensation. Expressions for the change in ΔH and −TΔS are linear and of opposite sign for both small perturbations of interactions, of populations of energy levels and of the levels themselves for a variety of hypothetical distributions of energy states. Consequently, small changes to systems tend to cancel out. Such approaches provide successful models for understanding compensation with respect to small changes in continuous variables, for example, temperature. However, perturbation approaches are not formally valid for the typical size of changes arising from ligand modifications (typically > RT). Sharp4 has suggested that “larger experimental ΔH or TΔS values presumably result from correlated perturbations,” but this idea has not been developed in detail. Further, as we have shown that there is a broad spread of behavior for protein–ligand interactions (with 31% either only weakly compensated or actually reinforcing) and that experimental error will tend to over, not under, estimate compensation, the motivation and support for these generalized theories seems weaker. In reporting a small number of observations of reinforcement, Levy and colleagues20 show that in a narrow range of circumstances the thermodynamic theory21 can give reinforcement. Sharp4 has also made an argument for such a possibility in the statistical mechanical theory, where the Gaussian distribution of states model could be “tuned” to give any desired distribution. Probably, there is room for further development of these ideas, but in making such accommodations for the experimental data these approaches lose their predictive nature and become empirically parameterized explanations, whose underlying assumptions require additional support.
An alternative view, which predicts large changes in both enthalpy and entropy as a result of chemical modification, has been propounded by Dunitz22 and Searle et al.23 This view is based on consideration of the fundamental physics of the energy of individual bonds between protein, ligand, and water molecules. The basic idea is that heat resides in the vibrational and librational motions of atoms constrained by bonds. The depth and width of the potential well describing each bond determines the energy states and thus enthalpy and entropy of the bond. In his original exposition, Dunitz22 chose a Morse potential together with parameters that illustrated the possibility of a high degree of compensation for hydrogen bonding interactions of water. However, Ford24 has pointed out that differences in the curvature and depth of the potential wells of bonds between the free and bound forms can result in a wide variety of behaviors, including reinforcement. The range of possibilities arising from variation in interactions between bound and free states for these bonding models is at least in accord with the extent of variation seen in experiment. However, at present such models are merely illustrative and not able to make any specific predictions about particular protein–ligand interactions. Many questions remain unanswered. For example, why are many changes compensated to a large extent? Is this due to the behavior of water as has been frequently suggested?1,25 A large-scale study of protein folding thermodynamics26 shows a trend in a ΔH versus ΔG plot whereby ΔHfold/ΔGfold ∼ 11. This is quite similar to the average ΔΔHbind/ΔΔGbind ∼ 8 that we see here. Is this a coincidence or a result of the process of dehydration that occurs both in folding and complex formation? Certainly, the relatively low mass of a water molecule means that variations in hydrogen bond strength are associated with relatively large changes to ΔH and ΔS. The Morse potential model also implies that larger changes to ΔH for individual bonds should be less well compensated, but there is no obvious trend for changes in the ϕ distribution with ΔΔH (data not shown). Does this suggest some weakness in the theory or simply that larger enthalpy changes are usually composites of several small changes to individual bonds?
Our discovery of an imperfect but strong tendency to enthalpy–entropy compensation in protein–ligand interactions and the development of methods of data representation and comparison that led to the discovery (particularly the use of the compensation coefficient ϕ) opens up new questions. In this regard, we follow Cornish-Bowden's maxim “that genuine but imperfect correlations are biologically more interesting than meaningless perfect ones.”5 A key question for the utility of the observations presented here is why do some changes to ligands result in compensation and others not? Are the differences seen between proteins significant, that is, are some protein's binding sites better able to compensate than others or do the differences simply reflect limitations in diversity of ligands in current experiments? Are there types of chemical changes to ligands, which are more or less likely to be compensated in interactions with proteins? Answers to these questions will have a significant impact on our attempts to rationally develop therapeutically relevant molecules.
Methods
Creation of the isothermal titration calorimetry dataset
All data on protein–ligand interactions were drawn from the SCORPIO11 and PDBcal15 databases of ITC data. Some database entries were excluded to ensure that the data form a coherent group subject to similar experimental constraints and to minimize potential extra-experimental biases in the thermodynamic values. Specifically, only data for each protein derived from direct ITC titration measurements and a single publication were selected. The heats of some binding reactions may be sensitive to changes in solution conditions (particularly ionic strength and pH) and are proportionately affected by systematic errors such as concentration measurement and instrument calibration. This restriction of data for each protein to a single publication minimizes the possible contribution of such variations to the ΔΔG and ΔΔH values in later analyses. Data for each protein were recorded at a single temperature. To isolate the effects of ligand variation, proteins were included only if data were available from more than one ligand binding at the same site. Data from studies in which a prior screen of activity or affinity was used to select candidates for ITC (i.e., almost all studies from pharmaceutical companies) were excluded as such selection biases distribution of ΔG.11 The final dataset may be found in Supporting Information Table 1.
Creation of model distributions of thermodynamic values
Model datasets were created by random resampling of the relevant experimental data and applying corrections and acceptance criteria designed to mimic experimental and other sources of correlation. The ITC affinity window is modeled by a probability of acceptance of ΔG values estimated from the observed distribution of all 422 direct ITC measurements of experimental affinities in SCORPIO11 and PDBcal15 databases [Fig. 2(A)]. This probability, Paw(ΔG), is estimated as 0.1 for ΔG = −52 to −50 kJ mol−1, 0.35 from −50 to −48 kJ mol−1, 0.5 from −48 to −44 kJ mol−1, 1 from −44 to −18 kJ mol−1, 0.5 from −18 to −14 kJ mol−1, 0.15 from −14 to −12 kJ mol−1 and otherwise zero.
The constrained random distribution for ΔH versus −TΔS [Fig. 2(D)] results from independent random sampling of these two experimental quantities, which are treated as “true” values. ΔGtrue is then computed using (1). Experimental error is mimic by addition to ΔGtrue and ΔHtrue of values drawn from normal distributions of mean zero and σ = 0.5 and 1.5, respectively. These values are the “trial” values. Computing −TΔStrial = ΔGtrial − ΔHtrial incorporates the effects of correlated experimental error. This group of trial values are then accepted as part of the model with a probability given by Paw(ΔGtrial).
A constrained random model for the relative thermodynamic quantities was created by first independently randomly selecting values of ΔG from the original experimental dataset and ΔΔH and −TΔΔS from the values for all ligand pairs that bind the same protein; these are taken as the true values. Then ΔΔGtrue = ΔΔHtrue − TΔΔStrue addition of errors (as above, but a factor of 21/2 larger) produces ΔΔGtrial and ΔΔHtrial, −TΔΔStrial = ΔΔGtrial −ΔΔHtrial and the group of data are accepted with a probability Paw(ΔGtrue + ΔΔGtrial). Approximately 281,000 groups of values for ΔΔH, −TΔΔS, and ΔΔG were accepted from 500,000 trials to construct the constrained random model dataset used in statistical testing.
An approximate model of the effect of intrinsic correlation (compensation) is achieved [Fig. 6(B)] by instead drawing ΔΔHtrue and −TΔΔStrue from a binormal distribution, that is, with a probability proportional to
| (5) |
where σ = 17.5 kJ mol−1 [the width of a normal distribution that is a reasonable approximation to the experimental distributions of both ΔΔH and –TΔΔS in Fig. 3(B,C)] and ρ is the desired correlation coefficient. The use of a normal distribution is sufficient for illustrative purposes but does narrow the distributions of ΔΔH and −TΔΔS slightly compared to experiment. In the case of a “fully compensated” distribution (Fig. 3), TΔΔStrue is set equal to the resampled value of ΔΔHtrue.
Statistical procedures
Samples of random pairs of ligands binding a single protein were drawn from the experimental dataset and compared to samples of the same size drawn from the constrained random model. To minimize potential sources of bias in the experimental sample, no protein–ligand interaction was drawn twice and no more than three pairs of ligands were included for each protein, limiting samples to a maximum of 60 pairs. The model distributions of compensation coefficients in the absence of intrinsic correlation [Fig. 5(A,C)] are computed from all data in the constrained random model.
The Kuiper statistic,27 a variant of the widely used Kolgomorov-Smirnov test for comparing two sets of unbinned data via their cumulative distribution functions, is an appropriate measure of difference between any two distributions of circular data. Importantly, the significance of a test based on the Kuiper statistic is not dependent on the choice of zero angle. Estimates of the population cumulative distributions P(θ) and P(ϕ) were generated using all of the ∼281,000 sets of values of ΔΔGA→B, ΔΔHA→B, and TΔΔSA→B in the constrained random dataset. These population cumulative distributions were then used to compute the Kuiper statistic Vn for the experimental sample by comparing its cumulative distribution, Sn(θ) or Sn(ϕ), to that appropriate for an experimental sample of size n,
| (6) |
and similarly for ϕ. The appropriate probability distribution function for the Kuiper statistic (i.e., the probability of observing a particular value of Vn by chance) was generated by resampling the constrained random distribution drawing 1000 sets of 60 datapoints and computing V60 each time. All analyses were performed using in-house Mathematica (Wolfram Research) programs.
Acknowledgments
We thank the referees for suggesting additional factors for inclusion in the statistical models.
Supplementary material
References
- 1.Lumry R, Rajender S. Enthalpy–entropy compensation phenomena in water solutions of proteins and small molecules: a ubiquitous property of water. Biopolymers. 1970;9:1125–1227. doi: 10.1002/bip.1970.360091002. [DOI] [PubMed] [Google Scholar]
- 2.Krug RR, Hunter WG, Grieger RA. Statistical interpretation of enthalpy–entropy compensation. Nature. 1976;261:566–567. [Google Scholar]
- 3.Exner O. On the enthalpy–entropy relationship. Coll Czech Chem Commun. 1964;26:1094–1113. [Google Scholar]
- 4.Sharp K. Entropy–enthalpy compensation: fact or artefact? Protein Sci. 2001;10:661–667. doi: 10.1110/ps.37801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cornish-Bowden AJ. Enthalpy–entropy compensation: a phantom phenomenon. Bioscience. 2002;27:121–126. doi: 10.1007/BF02703768. [DOI] [PubMed] [Google Scholar]
- 6.Exner O. Entropy–enthalpy compensation and anticompensation: solvation and ligand binding. Chem Commun. 2000;2000:1655–1656. [Google Scholar]
- 7.Chaires JB. Calorimetry and thermodynamics in drug design. Annu Rev Biophys. 2008;37:135–151. doi: 10.1146/annurev.biophys.36.040306.132812. [DOI] [PubMed] [Google Scholar]
- 8.Krug RR, Hunter WG, Grieger RA. Enthalpy–entropy compensation. 2. Separation of the chemical from the statistical effect. J Phys Chem. 1976;80:2341–2351. [Google Scholar]
- 9.Exner O. Statistics of the enthalpy–entropy relationship. I. The special case. Coll Czech Chem Commun. 1972;37:1425–1444. [Google Scholar]
- 10.Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- 11.Olsson TSG, Williams MA, Pitt WR, Ladbury JE. The thermodynamics of protein–ligand interaction and solvation: insights for ligand design. J Mol Biol. 2008;384:1002–1017. doi: 10.1016/j.jmb.2008.09.073. [DOI] [PubMed] [Google Scholar]
- 12.Torres FE, Recht MI, Coyle JE, Bruce RH, Williams G. Higher throughput calorimetry: opportunities, approaches and challenges. Curr Opin Struct Biol. 2010;20:1–8. doi: 10.1016/j.sbi.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krishnamurthy VM, Bohall BR, Semetey V, Whitesides GM. The paradoxical thermodynamic basis for the interaction of ethylene glycol, glycine, and sarcosine chains with bovine carbonic anhydrase II: an unexpected manifestation of enthalpy/entropy compensation. J Am Chem Soc. 2006;128:5802–5812. doi: 10.1021/ja060070r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De M, You C-C, Srivastava S, Rotello VM. Biomimetic interactions of proteins with functionalized nanoparticles: a thermodynamic study. J Am Chem Soc. 2007;129:10747–10753. doi: 10.1021/ja071642q. [DOI] [PubMed] [Google Scholar]
- 15.Li L, Dantzer JJ, Nowacki J, O'Callaghan BJ, Meroueh SO. PDBcal: a comprehensive dataset for receptor–ligand interactions with three-dimensional structures and binding thermodynamics from isothermal titration calorimetry. Chem Biol Drug Des. 2008;71:529–532. doi: 10.1111/j.1747-0285.2008.00661.x. [DOI] [PubMed] [Google Scholar]
- 16.Cooper A, Johnson CM, Lakey JH, Nöllmann M. Heat does not come in different colours: entropy–enthalpy compensation, free energy windows, quantum confinement, pressure perturbation calorimetry, solvation and multiple causes of heat capacity effects in biomolecular interactions. Biophys Chem. 2001;93:215–230. doi: 10.1016/s0301-4622(01)00222-8. [DOI] [PubMed] [Google Scholar]
- 17.Lumry R. On the interpretation of data from isothermal processes. Meth Enzymol. 1995;259:628–720. doi: 10.1016/0076-6879(95)59065-x. [DOI] [PubMed] [Google Scholar]
- 18.Beasley JR, Doyle DF, Chen L, Cohen DS, Fine BS, Pielak GJ. Searching for quantitative entropy–enthalpy compensation among protein variants. Proteins: Struct Func Genetics. 2002;49:398–402. doi: 10.1002/prot.10189. [DOI] [PubMed] [Google Scholar]
- 19.Myszka DG, Abdiche YN, Arisaka F, Byron O, Eistenstein E, Hensley P, Thomson JA, Lombardo CR, Schwarz F, Stafford W, Doyle ML. The ABRF-MIRG'02 Study: assembly state, thermodynamic, and kinetic analysis of an enzyme/inhibitor interaction. J Biomol Technol. 2003;14:247–269. [PMC free article] [PubMed] [Google Scholar]
- 20.Gallicchio E, Kubo MM, Levy RM. Enthalpy–entropy compensation in solvation and ligand binding revisited. J Am Chem Soc. 1998;120:4526–4527. [Google Scholar]
- 21.Qian H, Hopfield J. Entropy–enthalpy compensation: perturbation and relaxation in thermodynamic systems. J Phys Chem. 1996;105:9292–9299. [Google Scholar]
- 22.Dunitz JD. Win some lose some: enthalpy–entropy compensation in weak intermolecular interactions. Chem Biol. 1995;2:709–712. doi: 10.1016/1074-5521(95)90097-7. [DOI] [PubMed] [Google Scholar]
- 23.Searle MS, Westwell MS, Williams DH. Application of a generalized enthalpy–entropy relationship to binding cooperativity to weak associations in solution. J Chem Soc Perkin Trans. 1995;2:141–151. [Google Scholar]
- 24.Ford DM. Enthalpy–entropy compensation is not a general feature of weak association. J Am Chem Soc. 2005;127:16167–16170. doi: 10.1021/ja054519c. [DOI] [PubMed] [Google Scholar]
- 25.Leung DH, Bergman RG, Raymond KN. Enthalpy–entropy compensation reveals solvent reorganization as a driving force for supramolecular encapsulation in water. J Am Chem Soc. 2008;130:2798–2805. doi: 10.1021/ja075975z. [DOI] [PubMed] [Google Scholar]
- 26.Liu L, Yang C, Guo QX. A study on the enthalpy–entropy compensation in protein unfolding. Biophys Chem. 2000;84:239–251. doi: 10.1016/s0301-4622(00)00130-7. [DOI] [PubMed] [Google Scholar]
- 27.Press WH, Teukolsky SA, Vetterling WT, Flannery BP. Numerical recipes: the art of scientific computing. 3rd ed. Cambridge: Cambridge University Press; 2007. pp. 736–740. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


