SUMMARY
Insulators are multi-protein-DNA complexes thought to affect gene expression by mediating inter- and intra-chromosomal interactions. Drosophila insulators contain specific DNA binding proteins plus common components, such as CP190, that facilitate these interactions. Here we examine changes in the distribution of Drosophila insulator proteins during the heat-shock and ecdysone responses. We find that CP190 recruitment to insulator sites is the main regulatable step in controlling insulator function during heat shock. In contrast, both CP190 and DNA binding protein recruitment are regulated during the ecdysone response. CP190 is necessary to stabilize specific chromatin loops and for proper activation of transcription of genes regulated by this hormone. These findings suggest that cells may regulate recruitment of insulator proteins to the DNA in order to activate insulator activity at specific sites and create distinct patterns of nuclear organization that are necessary to achieve proper gene expression in response to different stimuli.
Keywords: Transcription, Insulator, Chromatin, Epigenetics
INTRODUCTION
Insulators mediate inter- and intra-chromosomal interactions that bring together distant regulatory elements in the genome. The specific outcome of these interactions depends on the nature of the connecting insulator sites (Phillips and Corces, 2009). For example, two insulator sites may interact to form a loop that separates an enhancer from the promoter of a gene, in which case enhancer-promoter interactions will be abolished and transcription of the gene will not take place. Alternatively, the interaction between two insulator sites may bring an enhancer in close proximity to a promoter, facilitating activation of transcription (Majumder et al., 2006; Xu et al., 2011). The CTCF insulator appears to be the main type of this class of regulatory sequences discovered so far in vertebrates. CTCF binds to specific sequences in the genome and mediates interactions among CTCF insulator sites in a process that is stabilized by cohesin (Gaszner and Felsenfeld, 2006; Wendt and Peters, 2009). In Drosophila, however, there are various DNA binding proteins that interact with distinct sites throughout the genome and show insulator activity in transgene assays. These include the Drosophila homolog of CTCF (dCTCF), Suppressor of Hairy-wing [Su(Hw)], and Boundary Element Associated Factor (BEAF) (Bushey et al., 2008; Maeda and Karch, 2007). Interestingly, all three of these DNA binding proteins are thought to recruit a common protein, Centrosomal Protein 190 (CP190), which is necessary for insulator activity (Bushey et al., 2009; Gerasimova et al., 2007; Mohan et al., 2007; Pai et al., 2004). CP190, as well as Mod(mdg4), contain BTB domains and are recruited to insulator sites by the various DNA binding proteins. CP190 and Mod(mdg4) mediate inter-insulator interactions and serve as a bridge to bring together distant insulator sites. These observations suggest that insulator activity can, in principle, be controlled by regulating the binding of Su(Hw), dCTCF, or BEAF to DNA or by modulating the interactions between these proteins and CP190 and Mod(mdg4) (Bushey et al., 2009). Additional regulatory steps may take place by recruitment of the E3 ubiquitin ligase dTopors or the RNA helicase Rm62 (Capelson and Corces, 2005; Lei and Corces, 2006).
Interactions among insulator sites throughout the genome may give rise to a specific three-dimensional organization of the chromatin that can be considered a blueprint of the transcriptional status of the cell. Since patterns of gene expression are cell lineage-specific, comparison of the genome-wide distribution of insulator proteins may give insights into the actual strategies used by cells to regulate insulator function. To begin to examine this question, we have mapped Drosophila insulator protein DNA localization genome-wide in two different cell lines and found that 5–37% of insulator protein binding sites for dCTCF, Su(Hw), BEAF, and CP190 are cell type specific (Bushey et al., 2009). Furthermore, a subset of the cell type specific CP190 sites show localization differences at sites where the amount of dCTCF, Su(Hw), or BEAF is invariant between the two cell lines. This suggests that cells may regulate insulator function by modulating the level of DNA binding protein recruitment to DNA as well as by controlling the amount of CP190 tethered to insulator sites. Therefore, cells may have insulators in which the DNA binding protein is bound to the DNA but are inactive because they lack CP190; this class of insulators maybe poised for rapid activation in response to specific stimuli. Other insulator sites appear to lack all protein components, and their activation would require the recruitment of one of the DNA binding proteins as well as CP190.
A more controlled system for studying the mechanisms by which insulator function is regulated is to perturb gene expression in a single cell type and monitor how insulator protein localization and gene expression change over time. Two widely used approaches to induce changes in gene expression in Drosophila cells are heat-shock and ecdysone treatment. Drosophila cells respond to an elevation in temperature by inducing transcription of heat-shock genes while repressing expression of the remainder of the genome, thus altering transcription on a genome-wide scale (Ashburner and Bonner, 1979). In Drosophila, postembryonic development is controlled by the steroid hormone α-ecdysone in its biologically active form, 20-hydroxyecdysone (20-HE). 20-HE binds to the ecdysone receptor complex (ECR-C), a classic nuclear receptor complex, to stimulate insect development (Riddiford et al., 2000). Treatment of cultured cells with ecdysone results in a less dramatic alteration in gene expression than heat-shock and allows analysis of expression of individual genes. These two methods of altering transcription afford the possibility of monitoring changes in insulator protein binding as well as insulator-dependent chromatin looping at specific genomic loci.
Here we assay insulator protein localization during the heat-shock and ecdysone responses. Heat-shock results in a dramatic loss of CP190 localization to DNA, while 20-HE treatment leads to changes in the pattern of CP190, Su(Hw), dCTCF and BEAF localization at a subset of sites. Specifically, in response to 20-HE, CP190 is recruited to a distinct site at the ecdysone-induced Eip75B locus, resulting in increased interactions between adjacent insulator sites and the formation of chromatin loops. The data presented here suggest that these changes in the three-dimensional architecture of the locus confine ecdysone-induced transcription to a specific set of genes in the region. Together these findings indicate that the regulation of insulator protein recruitment to DNA is an important mechanism used to establish proper chromatin organization and gene expression patterns.
RESULTS
Heat-shock causes a genome-wide reduction in CP190 localization at insulator sites
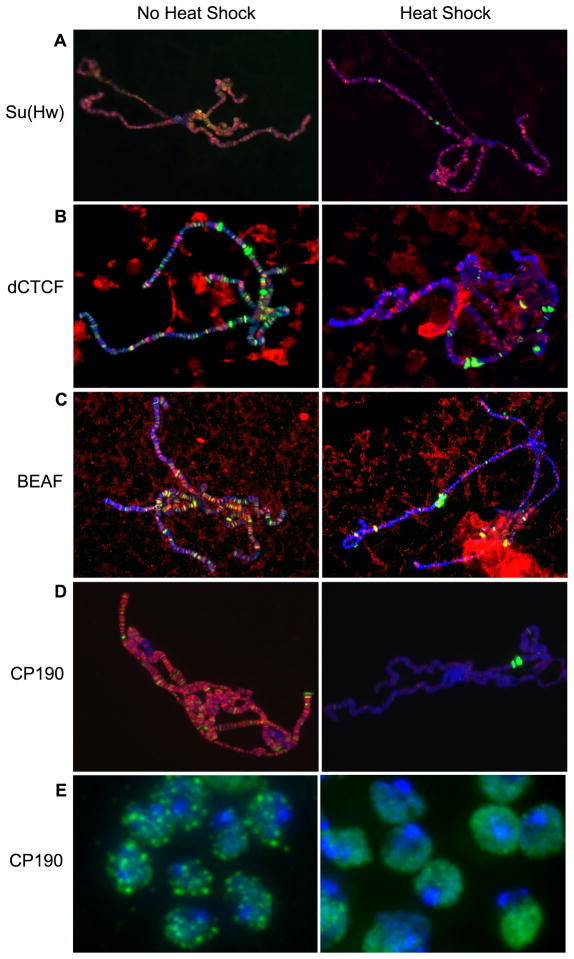
Drosophila cells undergo dramatic changes in gene expression in response to heat-shock. Transcription of most genes is turned off while expression of genes necessary for the heat-shock response is induced at high levels (Ashburner and Bonner, 1979). This system, which involves a global change in the transcriptional state of the genome, may be a powerful model to study the relationship between insulator proteins and transcription. We have previously shown that the distribution of Su(Hw) on polytene chromosomes is not significantly altered by heat-shock; nevertheless, the formation of Su(Hw) insulator bodies, which represent sites where several individual insulator sites coalesce in the nucleus, is severely disrupted (Gerasimova et al., 2000). Immunolocalization experiments using antibodies against dCTCF and BEAF indicate that the binding of these proteins to DNA is also not notably affected by the heat-shock response (Figure 1B, C). Taken together, these results suggest that changes in transcription patterns during the heat-shock response are accompanied by alterations in insulator function, but these alterations are not due to large changes in the recruitment of the DNA binding component of insulator complexes to chromatin. One possibility, suggested by the observed alterations in insulator body formation, is that regulation of insulator function during heat shock is controlled at the level of CP190 recruitment. To test this possibility we carried out immunolocalization experiments using CP190 antibodies on polytene chromosomes. The results indicate that, upon heat-shock, there is a dramatic release of CP190 from most genomic insulator sites in polytene chromosomes from third instar larvae (Figure 1D) (Oliver et al., 2010). Dismissal of CP190 from insulator sites is accompanied by a dispersion of insulator bodies: 137/160 (85.6%) of non-heat shocked cells contain CP190 insulator bodies compared to 10/160 (6.25%) of heat-shocked imaginal disc cells (Figure 1E & S1B). However, western blot analysis indicates that CP190 protein levels are unchanged upon heat-shock, suggesting that temperature elevation results in relocation of this protein from insulator bodies to a diffuse distribution throughout the nuclear space (Figure S1A). This result suggests that the global change in gene expression induced by heat-shock correlates with a decrease in insulator interactions most likely caused by regulation of CP190 recruitment to DNA.
Figure 1. Response of insulator proteins to heat-shock.
Third-instar larvae were subjected to heat-shock and polytene chromosomes were dissected and stained with antibodies recognizing (A) Su(Hw), (B) dCTCF, (C) BEAF or (D) CP190, shown in red. All polytene chromosomes (A–D) were co-stained with antibody against Pol II Ser5, shown in green, to mark the initiating form of RNA polymerase II. Upon heat-shock, Pol II Ser5 staining is reduced throughout most of the genome and is found in puffs indicating relocalization to the highly transcribed heat-shock genes. DNA is visualized with DAPI in all panels (blue). (E) Nuclei from imaginal tissue isolated from third instar larvae under heat-shock and non heat-shock conditions were stained with antibodies against CP190 (green) revealing the absence of insulator bodies after heat-shock. See also Figure S1B. The loss of CP190 insulator bodies is not due to protein degradation as shown by western blot analysis in Figure S1A.
Ecdysone treatment leads to changes in the genome-wide distribution of insulator proteins
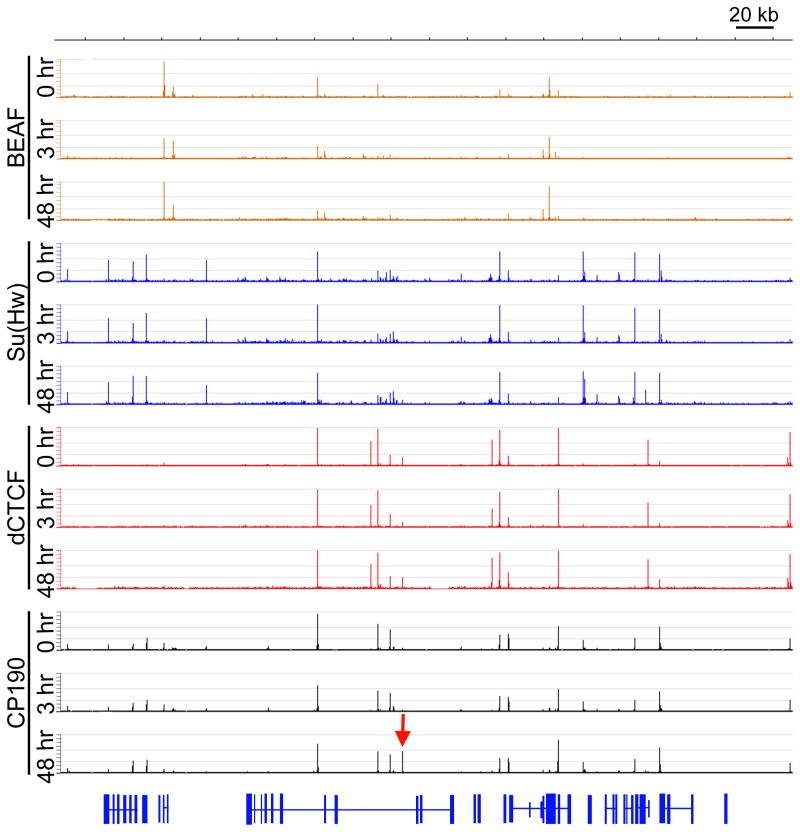
In order to determine whether a less dramatic change in gene expression than that induced by heat shock also correlates with alteration of CP190 distribution, we treated Kc cells with the hormone 20-hydroxyecdysone (20-HE). This treatment results in G2-arrest within 12–24 hr and morphological changes such as the loss of the spherical cell shape as well as appearance of cellular processes (Stevens et al., 1980). 20-HE has previously been shown to affect the expression of a few hundred Drosophila genes in Kc cells over a time course of 1–48 hr (Gauhar et al., 2009). Therefore, we used 3 hr and 48 hr time points to monitor early and late responses to the hormone. Since the changes in gene expression after 20-HE treatment are less dramatic than those observed after heat-shock, we would not expect to see a dramatic rearrangement in CP190 localization after incubation with this hormone. Consequently, insulator bodies remain unchanged in Kc cells after 3 or 48 hr of treatment with 20-HE (Figure S2). In order to more precisely determine possible changes in CP190 localization that accompany changes in gene expression after 20-HE treatment, we performed ChIP-Seq with CP190, Su(Hw), dCTCF, and BEAF at 0 hr, 3 hr, and 48 hr of incubation with this hormone (Figure 2). ChIP-seq data has been deposited in GEO under accession number GSE30740. To verify these data, we compared sites identified at 0 hr of 20-HE treatment to our previously published ChIP-chip data from Kc cells (Bushey et al., 2009). Although we identified more sites by ChIP-Seq than ChIP-chip as would be expected based on the sensitivity of the techniques, 89% of CP190 sites, 97% of Su(Hw) sites, 90% of CTCF sites, and 93% of BEAF sites identified by ChIP-chip analysis were also identified in the ChIP-Seq data sets presented here.
Figure 2. Response of insulator proteins to 20-HE treatment.
A 360 kb region of chromosome 3L including the Eip75B gene. ChIP-Seq analysis of BEAF, Su(Hw), dCTCF, and CP190 at 0, 3, and 48 hr of 20-HE treatment in Kc cells. Genes from the UCSC Genome Browser are shown in blue. The upregulated CP190 site used for 3C analysis is indicated (red arrow). Changes in insulator protein localization upon 20-HE treatment does not lead to alterations in insulator body formation as shown in Figure S2.
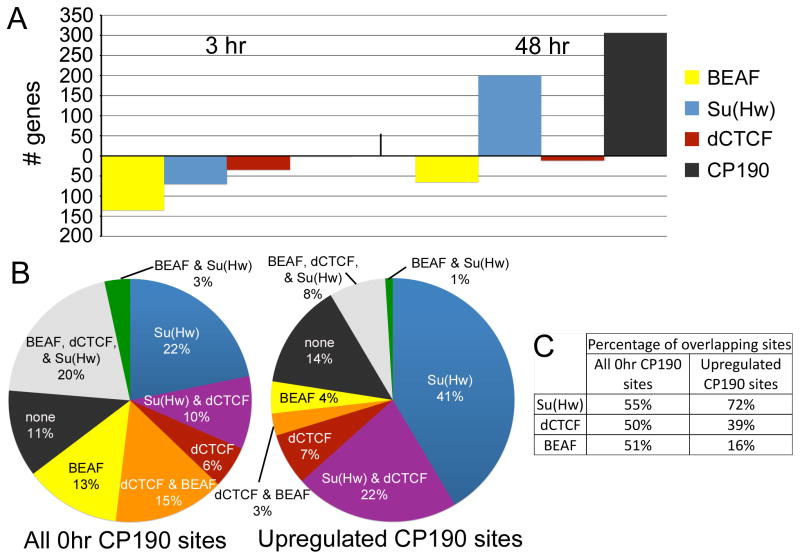
Analysis of sites that change upon 20-HE treatment indicates that, unlike the dramatic decrease in chromosome-bound CP190 observed after heat-shock, CP190 sites remain unchanged in response to 20-HE after 3 hr of treatment and there is no significant loss of CP190 binding at either time point (Figure 2 & 3A). However, 20-HE treatment results in new or increased CP190 association with DNA at 306 sites (5% of sites) after 48 hr. Comparison of the binding profiles of the three DNA binding proteins, BEAF, dCTCF, and Su(Hw), reveals similar alterations. BEAF exhibits decreased binding at 135 sites (2% of sites) at 3 hr and 65 sites (1% of sites) at 48 hr (Figure 3A), of which 58 sites are common between the two time points. dCTCF shows lost or decreased binding at 34 sites (0.6% of sites) at 3 hr followed by a decrease or loss of binding at 11 sites (0.2% of sites) at 48 hr (Figure 3A). Su(Hw) behaves in the opposite fashion, showing reduced binding at 70 sites (1% of sites) at 3 hr, and then new or increased binding at 200 sites (2% of sites) at 48 hr. (Figure 3A) The discrepancy between the responses of BEAF, dCTCF and Su(Hw) at 48 hr is not surprising since we and others have previously reported that insulators defined by these various DNA binding proteins show distinct properties (Bushey et al., 2009; Negre et al., 2010). Therefore, these observations further support the idea that BEAF, dCTCF and Su(Hw) insulators play different roles in regulating gene expression.
Figure 3. Insulator sites regulated in response to 20-HE.
(A) Graph indicates the number of insulator sites that are regulated for each protein at each time point of 20-HE treatment. Bars above the X-axis indicate upregulated genes while genes below the axis indicate downregulated genes. For a list of up- and down-regulated sites see Table S2. (B) Co-localization of CP190 sites and each of the DNA binding proteins. All CP190 sites identified at 0 hr of 20-HE treatment are represented in the pie chart on the left, while those upregulated at 48 hr of 20-HE treatment are represented in the pie chart on the right (C) The percentage of CP190 sites that overlap with each DNA binding protein is indicated. These percentages add up to over 100 % since some CP190 sites overlap with more than one DNA binding protein as indicated in B.
Analysis of insulator sites regulated in response to 20-HE treatment
Recruitment of CP190 is necessary for insulator function (Gerasimova et al., 2007; Mohan et al., 2007; Pai et al., 2004), and, therefore, we concentrated our analysis on the CP190 sites that are upregulated after 48 hr of treatment in order to examine the role of regulated insulator sites in the 20-HE response. A comparison between the 306 CP190 sites upregulated at 48 hr of 20-HE treatment and DNA binding proteins present at 48 hr of treatment reveals that upregulated CP190 sites co-localize with at least one DNA binding protein at 86% of sites (Figure 3B). This is very similar to the 89% of CP190 sites that co-localize with at least one DNA binding protein at 0 hr (Figure 3B) suggesting that these upregulated CP190 sites are authentic insulator sites. Notably, the distribution of DNA binding proteins that co-localize with these upregulated CP190 sites is very different than that for the total population of CP190 sites (Figure 3B & 3C). There is a strong enrichment for Su(Hw) sites alone or in combination with dCTCF at upregulated CP190 sites, and a depletion in BEAF sites as well as total dCTCF sites. Overall, 220 (72%) upregulated CP190 sites co-localize with a Su(Hw) site. Of these 220 CP190/Su(Hw) sites, 61 (27%) also show an increase in Su(Hw) binding. This is much greater than the percentage of Su(Hw) sites upregulated at 48 hr genome-wide (2%), indicating that upregulated CP190 sites are significantly enriched for upregulated Su(Hw) sites (p<1×10−100). These observations suggest that the recruitment of CP190 after hormone induction occurs at a specific subset of insulator sites, characterized by the presence of Su(Hw), that may represent a distinct functional class of these elements.
The data suggest that, in some cases, the formation of a new active insulator after 20-HE induction first requires recruitment of Su(Hw) followed by binding of CP190. Alternatively, at other sites, induced CP190 recruitment occurs at sites that already have one of the DNA binding proteins. This result suggests that the upregulated CP190 sites that display constant levels of DNA binding proteins may be poised insulator sites, and 20-HE induction is necessary for the recruitment of CP190 and activation of insulator function.
20-HE stimulates a CP190-dependent increase in chromatin looping at the Eip75B locus
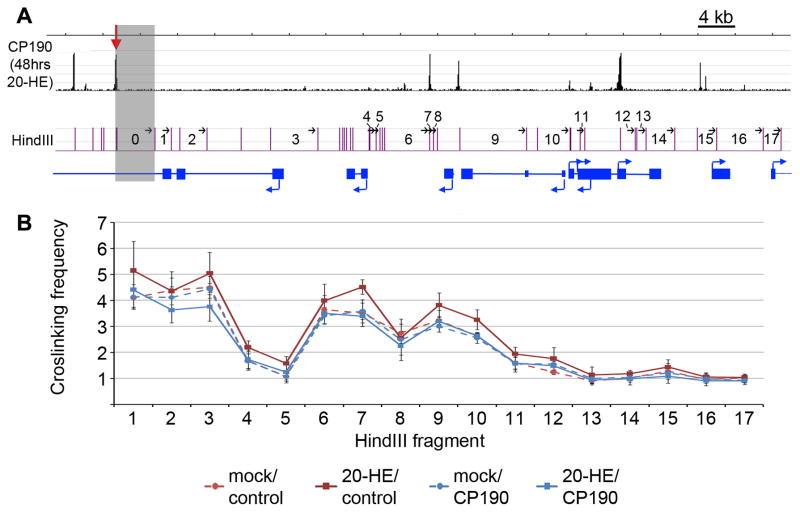
We have previously suggested that CP190 is involved in the formation of chromatin loops between insulator sites (Pai et al., 2004). The ecdysone induced protein 75B (Eip75B) locus contains a dCTCF site at which the levels of CP190 increase dramatically after 48 hr of 20-HE treatment (Figure 2). Analysis of ChIP-seq data at this locus reveals an increase in dCTCF localization as well as induced Su(Hw) localization upon 20-HE treatment, but no change in BEAF localization. These changes were validated by ChIP-qPCR analysis (Figure S2B). In order to determine the role of the increased recruitment of CP190 to this dCTCF site in the regulation of chromatin structure after hormone induction, we performed chromatin conformation capture (3C) analysis at the locus (Dekker et al., 2002).
For the 3C experiments, we used HindIII to digest genomic DNA and used fragment 0, which contains the 20-HE-induced CP190 site, as an anchor (Figure 4A shaded region). Crosslinking frequencies between the anchor and downstream fragments were quantified using a Taqman probe and quantitative PCR after 48 hr of either 20-HE or ethanol (mock) treatment. 3C analysis was also performed with cells treated with either control or CP190 RNAi. Under conditions simulating normal growth, mock hormone treatment and control RNAi, we observed weak but significant interactions between the anchor fragment and fragments 6/7 and 9/10 (Figure 4B). Although CP190 is only present within the anchor fragment at very low levels before 20-HE treatment, dCTCF is present at this site and may be sufficient to stabilize these weak interactions (Figure 2B). The strength of these two interactions increases after 20-HE treatment but not in cells treated with CP190 RNAi. Interestingly, fragments 7 and 9 contain CP190 sites before and after 20-HE induction, suggesting that the upregulated CP190 site in the anchor fragment mediates intra-chromosomal interactions with existing sites to enhance chromatin loop formation upon 20-HE treatment. Together these data suggest that de novo recruitment of CP190 to an existing dCTCF site results in alterations in the chromatin organization of the Eip75B locus upon 20-HE treatment.
Figure 4. 3C analysis of the Eip75B locus in response to 20-HE treatment.
(A) 80 kb region of chr3L containing part of the Eip75B gene. CP190 ChIP-Seq data from 48 hr of 20-HE treatment is shown in black with the upregulated CP190 site from Figure 4B indicated with a red arrow. The fragments used for 3C analysis and generated by digestion of genomic DNA with HindIII are shown in purple. Fragment 0 was used as the anchor in 3C analysis and is shaded in gray. (B) 3C analysis covering the HindIII fragments labeled as in panel A. Analysis was done with mock hormone treatment and control RNAi (dashed, red line), 20-HE treatment for 48 hr and control RNAi (solid, red line), mock hormone treatment and CP190 RNAi (dashed, blue line), and 20-HE treatment and CP190 RNAi (solid, blue line). Error bars represent the standard deviation of the mean of 4 biological replicates.
CP190-mediated chromatin organization modulates 20-HE-induced transcription
Recruitment of CP190 to a poised insulator site increases intra-chromosomal interactions and alters the three-dimensional structure of chromatin at the Eip75B locus. In order to explore the functional significance of the activation of this dCTCF insulator site by CP190 recruitment, we monitored gene expression in response to 20-HE treatment in the presence of either control or CP190 RNAi using Nimblegen microarrays that cover over 16,000 predicted genes. Expression data has been deposited in GEO under accession number GSE30686. 20-HE responsive genes were identified by comparing RNA isolated from cells at 0 hr to that at either 3 or 48 hr of 20-E induction (P < 0.1 and 1.5-fold cutoff).
In cells treated with control RNAi, we found 38 genes that were upregulated at the early, 3 hr time point and no genes that were significantly down regulated. At the 48 hr time point we found 2127 20-HE responsive genes, 1184 upregulated and 943 downregulated (Table 1 & S2). In cells treated with CP190 RNAi we found 290, 447, and 415 genes misregulated at 0 hr, 3 hr, and 48 hr of 20-HE treatment, respectively (Table 2 & S2). We observed that genes are up- and down-regulated upon CP190 knockdown at all time points, but more genes are misregulated at 3 and 48 hr of 20-HE treatment than at 0 hr. This may be a result of DNA-bound CP190 being more stable than the free pool of CP190 during RNAi treatment. As a consequence, the pre-existing CP190 sites may not be affected to the same extent as the up-regulated CP190 sites after 20-HE treatment. Alternatively, CP190 may be more important for gene regulation in response to stimuli than for steady-state growth. However, both scenarios suggest that up-regulated CP190 sites are important for proper regulation of gene expression in response to 20-HE. Further support of this conclusion comes from the finding that 24% (9 out of 38) of 3 hr 20-HE responsive genes and 11% (226 out of 2127) of 48 hr 20-HE responsive genes are misregulated upon CP190 knockdown at the respective time point suggesting that CP190 is necessary for a proper transcriptional response to 20-HE.
Table 1. Gene expression upon ecdysone treatment.
The number of genes that are up and down regulated at 3 and 48 hr of 20-HE treatment compared to 0 hr of 20-HE treatment are given. For expression levels of individual genes see Table S2.
| 20HE Treatment | Number of 20HE responsive genes | Number of genes upregulated | Number of genes downregulated |
|---|---|---|---|
| 3 hr | 38 | 38 | 0 |
| 48 hr | 2127 | 1184 | 943 |
Table 2. Gene expression in CP190 knockdown.
The number of genes that are misregulated upon CP190 RNAi treatment compared to control RNAi treatment are given. This analysis was performed at 0, 3, and 48 hours of 20-HE treatment. For expression levels of individual genes see Table S2.
| 20HE Treatment | Number of CP190 RNAi responsive genes | Number of genes upregulated | Number of genes downregulated |
|---|---|---|---|
| 0 hr | 290 | 222 | 68 |
| 3 hr | 447 | 356 | 91 |
| 48 hr | 415 | 251 | 164 |
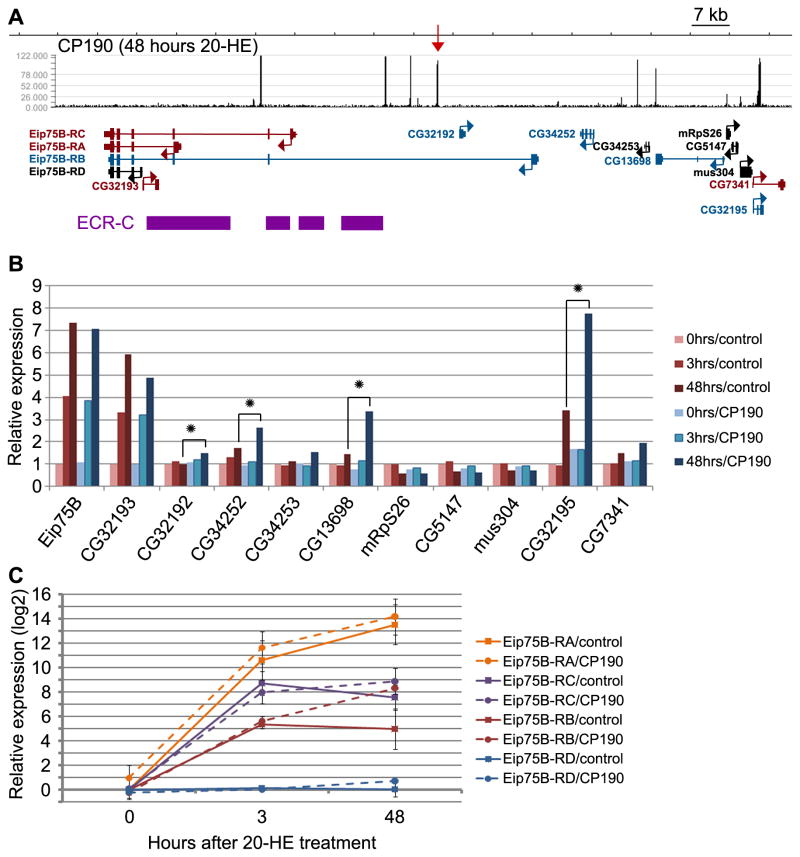
Surprisingly, the microarray data indicate that transcription of the Eip75B gene, which contains the dCTCF insulator site to which CP190 is recruited after 20-HE induction, is properly induced in response to 20-HE even in a CP190 RNAi knockdown (Figure 5B). Nevertheless, a closer look at the genes upstream of Eip75B reveals an increase in expression of four genes after 48 hr of 20-HE treatment in CP190 knockdown compared to control RNAi treatment. This observation suggests that CP190 is necessary to prevent genes surrounding Eip75B from being up-regulated upon 20-HE induction (Figure 5B).
Figure 5. Gene expression at the Eip75B locus in response to 20-HE treatment.
(A) 130 kb region of chr3L containing the Eip75B gene. CP190 ChIP-Seq data from 48 hr of 20-HE treatment is shown in black with the upregulated CP190 site indicated with a red arrow and the y-axis representing the number of tags. An expanded representation of genes including the four transcripts of Eip75B is shown; transcripts upregulated at 48 hr of 20-HE treatment in a CP190 RNAi dependent manner are shown in blue, transcripts upregulated at 48 hr of 20-HE treatment but not affected by CP190 RNAi are shown in red, and transcripts that do not change are shown in black. Binding sites for the ECR-C are shown in purple (Gauhar et al., 2009) (B) Expression level of genes within the locus in panel A pulled from our microarray analysis. Expression in control RNAi is shown in red while expression in CP190 RNAi is shown in blue. For each gene, expression levels are shown for 0, 3, and 48 hr of 20-HE treatment in light, medium and dark bars respectively. Stars indicate genes that show a change in gene expression (FDR<10%, 1.5 fold change) between control and CP190 RNAi at 48 hr of 20-HE treatment. (C) Relative expression of the four different Eip75B transcripts determined by qPCR at 0, 3, and 48 hr of 20-HE treatment. Dashed lines show expression under control RNAi and solid lines under CP190 RNAi. Error bars represent the standard deviation of the mean of 3 biological replicates. For levels of CP190 knockdown by RNAi see Figure S3.
Eip75B encodes four different transcripts expressed from four different alternative promoters (Figure 5A). However, the microarrays used for gene expression analysis only include probes for the common exons at the 3′ end of the gene. We therefore used quantitative PCR with primers that distinguish the four Eip75B transcripts to analyze the levels of each of the mRNAs. Results indicate that the four Eip75B transcripts respond differently to 20-HE treatment (Figure 5C). Transcripts Eip75B-RA and -RC are dramatically induced in response to 20-HE, whereas expression of Eip75B-RD does not change upon hormone treatment. In cells treated with control RNAi, the Eip75B-RB transcript is induced at 3 hr of 20-HE treatment, but levels begin to decrease by 48 hr. Interestingly, in cells treated with CP190 RNAi, Eip75B-RB continues to be up-regulated at 48 hr of 20-HE treatment. Therefore, the Eip75B-RB transcript behaves in a similar manner as the four misregulated genes upstream of Eip75B in cells lacking the CP190 protein. These data can be interpreted in the context of the location of binding sites for the ecdysone receptor complex (ECR-C) (Gauhar et al., 2009), which act as regulatory elements for the transcriptional activation of ecdysone-induced genes. Binding sites for ECR-C have been mapped throughout the coding region of Eip75B, such that the ecdysone-induced CP190 insulator separates ECR-C sites from the promoters of ecdysone-induced genes misregulated in cells treated with CP190 RNAi (Figure 5A).
These data suggest that activation of a poised dCTCF insulator by recruitment of CP190 is necessary to strengthen intra-chromosomal interactions with pre-existing insulator sites. These interactions create chromatin loops that may block the spread of 20-HE-induced transcription to genes surrounding Eip75B-RA. The loops formed between insulator sites could segregate the Eip75B-RB promoter as well as upstream genes from regulatory elements acting on Eip75B-RA, which continues to be upregulated at 48 hr of 20-HE treatment.
DISCUSSION
Chromatin insulators mediate inter- and intra-chromosomal interactions that establish a three-dimensional nuclear architecture and impact gene expression by controlling the juxtaposition of regulatory sequences within the nuclear space. Insulators are composed of DNA binding proteins that recruit other factors to establish or maintain these interactions. Genome-wide analyses of the distribution of these proteins in different Drosophila cell lines suggest the possibility that cells regulate insulator activity by modulating the recruitment of these two classes of proteins. To test this hypothesis, we examined changes in insulator protein localization in response to two different stimuli, heat-shock and ecdysone hormone induction, which result in changes in the transcription output of the cell. We find that insulator protein recruitment to chromatin is regulated during both the heat-shock and ecdysone responses.
Results from the analysis of polytene chromosomes during the heat shock response suggest that dismissal of CP190 from insulator sites may be a prerequisite for the broad changes in transcription that accompany temperature elevation. This implicates CP190 as a critical contributor to insulator function and suggests that its recruitment to insulator sites may play a role in regulating gene expression. Upon recovery from heat-shock, a cell must return to its original transcription profile. The observed persistence in the localization of DNA binding proteins on polytene chromosomes suggests that these proteins could act as a memory for CP190 to return to the DNA after heat-shock. Nevertheless, these findings suggest that regulatory mechanisms, perhaps in the form of covalent modifications of Su(Hw), dCTCF and BEAF, must exist to control the targeting of CP190 to a subset of sites where these proteins are present in the genome.
The steroid hormone 20-HE induces changes in gene expression that are necessary to initiate and coordinate cell proliferation, differentiation, and death that occur during molting and metamorphosis in Drosphila development (Riddiford et al., 2000). The Kc cell response to 20-HE, which includes exiting the cell cycle and altering cell morphology, resembles cell differentiation. Contrary to heat-shock, which changes the expression of all transcribed genes, Kc cells respond to treatment with 20-HE by altering transcription of a relatively small subset of genes. It is therefore not surprising that alterations in insulator protein distribution after 20-HE treatment are not as dramatic as those taking place after heat-shock. Although the majority of insulator protein binding sites remain constant upon 20-HE treatment, a subset is up and down regulated at 3 and 48 hr after hormone treatment. Interestingly, some sites upregulated at 3 hr of treatment are not maintained at 48 hr, at which time several dCTCF and BEAF sites are downregulated and Su(Hw) and CP190 sites become upregulated. This agrees with the changes in gene expression observed upon 20-HE treatment, as there are distinct sets of early and late ecdysone responsive genes, and products of the early genes repress their own transcription and induce expression of late genes (Ashburner, 1974; Burtis et al., 1990; Segraves and Hogness, 1990). Therefore, it is not surprising that many insulator sites regulated at 3 hr are distinct from those regulated at the later 48 hr time point.
We have previously shown that Su(Hw), dCTCF and BEAF show distinct distribution patterns with respect to genomic features, with BEAF found preferentially at promoters and Su(Hw) found more often at sites distant from genes (Bushey et al., 2009). Based on this distribution, we have suggested that BEAF may be involved in mediating interactions directly required for activation of transcription whereas Su(Hw) may affect gene expression more indirectly, perhaps by bringing together regions of the genome to establish different chromatin domains. In this context, it is interesting that CP190 is preferentially recruited to pre-existing Su(Hw) sites after hormone treatment. This result suggests that new CP190-mediated interactions after exposure to 20-HE may play a more general role in chromatin organization as opposed to direct regulation of the expression of specific genes.
Comparison between the genomic location of regulated insulator sites and ecdysone responsive genes does not reveal any significant correlation. This further supports the idea that insulator proteins are not typical transcription factors, and their role in regulating gene expression is more complex than protein binding to the promoter of target genes. Mechanistically, the 3C data presented here supports the idea that CP190 functions by modulating chromatin looping. Although intriguing, such a role makes it difficult to link a regulated insulator site with affected genes since insulator sites may act over long distances and a change in one insulator protein binding site can potentially regulate the expression of multiple genes.
The significance of the preferential upregulation of CP190 and Su(Hw) at 48 hr of 20-HE treatment and downregulation of dCTCF and BEAF is unclear. Since the downregulated dCTCF and BEAF sites do not localize to ecdysone responsive genes, it is possible that these are actually insulator sites that under normal growth conditions are poised for activation, but upon ecdysone treatment this poised state is lost. In the future, it will be interesting to test whether ecdysone treated cells lose the potential to respond to other environmental stimuli, thus supporting the idea that these downregulated dCTCF and BEAF sites represent a reduction in poised insulator sites.
To gain a better understanding of the functional significance of the new 20-HE-induced CP190 sites, we examined the role of a single insulator site. To this end, we selected a CP190 site within the Eip75B locus that is strongly upregulated after 48 hr of 20-HE treatment. This site contains dCTCF before and after hormone induction and may thus be poised for activation by CP190 recruitment. Interestingly, the site contains barely detectable amounts of CP190 before 20-HE treatment that are not considered statistically significant by standard peak analysis software, but that may be significant. This site can mediate interactions with adjacent insulator sites in non-treated cells, suggesting that dCTCF alone or in the presence of low levels of CP190 is sufficient to mediate interactions. Alternatively, other BTB domain-containing insulator proteins such as Mod(mdg4) may be responsible for low-level interactions in the absence of CP190. Nevertheless, interactions between this site and adjacent insulators are strengthened considerably after 48 hr of 20-HE treatment in a CP190-dependent manner. Additionally, new Su(Hw) signal appears at 48 hrs of 20-HE treatment, which is notable since the interactions identified by 3C are with sites that contain CP190, dCTCF, as well as Su(Hw). Therefore, this increase in Su(Hw) ChIP signal may be the indirect result of an increase in chromatin looping with a site directly bound by Su(Hw). Alternatively, binding of multiple insulator proteins may be necessary to achieve stable chromatin loops, and the pattern of insulator protein overlap at a given genomic location may direct interaction with other insulator sites with similar protein occupancy. Although at this point we do not understand the significance of having multiple DNA binding proteins at one genomic location, it is clear from the pie charts in Figure 3B that this is a common occurrence and therefore must have a functional output.
In addition to the 20-HE-induced Eip75B gene, the locus under study contains several additional genes whose expression is not affected by hormone treatment under normal growth conditions. Enhancer elements bound by the ecdysone receptor complex (ECR-C) responsible for 20-HE-dependent transcription of Eip75B are located within this gene (Gauhar et al., 2009). The dCTCF poised insulator site separates these regulatory sequences and the promoters of the CG32193, Eip75B-RA, Eip75B-RC and Eip75B-RD genes from the Eip75B-RB and non-ecdysone inducible genes located in the vicinity. In the absence or at 3 hr of 20-HE stimulation, this insulator is presumably not functional because it lacks CP190; this conclusion is supported by the increased transcription of Eip75B-RB after 3 hr of ecdysone treatment. Recruitment of CP190 after 48 hr of hormone induction causes activation of this insulator, explaining the drop in transcription of the Eip75B-RB RNA. This newly activated insulator may also be responsible for the absence of hormone-induced expression of genes located distal to the Eip75B-RB promoter. This hypothesis is corroborated by the activation of these genes, including Eip75B-RB, in cells lacking CP190 protein. This new CP190 site is therefore in a position that would enable it to block communication between the ECR-C and the promoters of Eip75B-RB and of other misregulated genes. Importantly this CP190 site is induced only at 48 hr when Eip75B-RB begins to be downregulated. Hence, recruitment of CP190 to a poised insulator site happens at the correct time and place for classic enhancer-blocking insulator function. Significantly, dCTCF is found at this site at 0, 3, and 48 hr of 20-HE treatment, though the insulating effect on Eip75B-RB and the other upstream genes is only observed at 48 hr, after CP190 recruitment, suggesting that dCTCF alone is not sufficient for insulator activity. This suggests that regulation of CP190 recruitment is a mechanism of regulating insulator activity and that insulator sites without CP190 may be inactive sites poised for activation.
The effect of the Eip75B 20-HE-induced insulator on the expression of adjacent genes appears to be mediated by the establishment of intra-chromosomal interactions with adjacent sites that create a specific chromatin organization. The loops formed by these interactions seem to be already in place prior to ecdysone treatment but recruitment of CP190 to the poised insulator increases the strength of the interactions or the frequency of loop formation. This suggests that, at least at this locus, there is a threshold in the plasticity of the chromatin that is critical for an insulator to affect enhancer-promoter interactions. Recruitment of CP190 regulates the level of chromatin looping within the region such that a more dynamic chromatin organization may allow for enhancer-promoter contacts, whereas a more structured and less dynamic chromatin organization may prevent these interactions. Furthermore, the promoters of transcripts affected by CP190 knockdown do not all localize within one CP190-mediated loop as suggested by classic insulator function models, again suggesting that it may be an overall decrease in chromatin mobility and not loop formation itself that causes the enhancer-blocking activity of insulator elements.
Insulator proteins have been implicated in chromatin organization and gene expression, but there have been few studies that shed light on the mechanisms by which insulator activity can be regulated in order to orchestrate the establishment of different patterns of gene expression during cell differentiation. Here we provide evidence that insulator protein recruitment is regulated in response to various stimuli and that this regulation plays a role in stabilizing chromatin loops. How CP190 is recruited to the specific site in Eip75B-RB upon 20-HE stimulation remains unclear, since dCTCF is present at the site before induction. There must be other forms of regulation, such as protein modifications, binding of additional co-factors, or release of inhibitors, that lead to CP190 recruitment to specific sites after 20-HE treatment. However, the ability to regulate CP190 recruitment to chromatin provides an important mechanism by which cells may control the organization and expression of the genome within the three-dimensional nuclear space.
EXPERIMENTAL PROCEDURES
Treatment of cells with 20-HE and dsRNA
Kc167 cells were grown in CCM3 serum free insect media (HyClone SH30065.01) at 25°C. For RNAi treatment, cells were plated at 0.5 × 106 cells/mL and incubated with 4 μg/mL of either control dsRNA targeting the β-galactosidase gene or dsRNA targeting the CP190 gene using Cellfectin II reagent (Invitrogen 10362-100). After addition of dsRNA, cells were grown for 3 days, re-plated at 0.5 × 106 cells/mL, and re-treated with 4 μg/mL of dsRNA as before. Cells were harvested after 3 additional days of growth (day 6). For ecdysone treatment, cells were plated at 0.5 × 106 cells/mL and grown overnight. Cells were treated with 20-HE (Sigma) at a final concentration of 5 × 10−7 M for 3 or 48 hr. For experiments where mock 20-HE treatment is indicated, cells were treated with EtOH. For cells treated with RNAi and 20-HE, the hormone was added on day 4 of knockdown for 48 h treatment and day 6 of knockdown for 3 h treatment; all cells were harvested on day 6. The degree of CP190 knockdown is depicted in Figure S3.
Heat-shock and immunofluorescence analysis of tissues from third instar larvae
Heat-shock treatment and immunofluorescence analysis of polytene chromosomes and imaginal tissue was done as described previously (Ivaldi et al., 2007). Cells were stained with primary antibodies in antibody dilution buffer (1xPBS, 0.1% Tween20, 1% BSA) overnight at 4°C (1:1000 rabbit α-CP190 (Pai et al., 2004), 1:100 rabbit α-Su(Hw) (Gerasimova et al., 1995), 1:50 rabbit α-BEAF (Bushey et al., 2009), 1:100 guinea pig α-dCTCF (gift from Elissa Lei), and 1:200 mouse α-Pol IIoSer5 (H14, Covance)). For immunofluorescence of imaginal discs, tissue was processed as previously described (Gerasimova et al., 2007) and slides were stained with 1:2000 rabbit α-CP190 and 1:500 mouse α-Pol IIoSer5 (H14, Covance).
ChIP-Seq analysis
Chromatin immunoprecipitation was performed as described (Bushey et al., 2009). To generate sequencing libraries ChIP DNA was prepared for adaptor ligation by end repair (‘End-It DNA End Repair Kit’ - Epicentre Cat# ER0720) and addition of ‘A’ base to 3′ ends (Klenow 3′-5′ exo- NEB Cat# M0212S). Illumina adaptors (Illumina Cat# PE-102-1001) were titrated according to prepared DNA ChIP sample concentration, and ligated with T4 ligase (NEB Cat# M0202S). Ligated ChIP samples were PCR amplified using Illumina primers and Phusion DNA polymerase (NEB Cat# F-530L) and size selected for 200–300bp by gel extraction. ChIP libraries were sequenced at the HudsonAlpha Institute for Biotechnology using an Illumina GAII sequencer. Sequences were mapped to the dm3 genome with Bowtie 0.12.3 (Langmead et al., 2009) using default settings. Peaks were then called with MACS 1.4.0alpha2 (Zhang et al., 2008) using equal numbers of unique reads for input and ChIP samples and a p-value cutoff of 1×10−10. To determine up- and down-regulated insulator sites at each time point, ChIP-Seq data for each insulator protein was normalized by total read count between the different time points. The data were then filtered by a second normalization for lane effect differences of the mean plus 1/2 standard deviation of lane specific read counts. The ChIP-timepoint-A/ChIP-timepoint-B Log2() ratio was determined and assigned for every 100 bp. Significant changes in insulator occupancy were then called as up- or down-regulated using CMARRT, a modified correlation and moving average algorithm (Kuan et al., 2008) and a stringent p-value cutoff of 1×10−10. For a list of regulated insulator sites see Table S2.
3C analysis
3C-qPCR analysis was done essentially as described (Hagege et al., 2007) with a few minor adaptations. 1 × 107 cells were cross-linked with 1% formaldehyde for 10 min at room temperature. Cell lysis was performed in 10 mM Tris pH 8, 10 mM NaCl, 0.2% NP-40, and Complete EDTA-free Mini protease inhibitors (Robbins et al.) for 15 min on ice. Following the initial phenol-chloroform/EtOH extraction, sample was resuspended in 200 μl H20. A second phenol-chloroform/EtOH extraction was then performed and the pellet was washed 3 times in 70% EtOH before final resuspension in 10 mM Tris pH 7.5. Total DNA concentration was then measured using qPCR and Sybr green detection and all samples were diluted to 50 ng/μl. Crosslinking frequency was determined using a control sample generated with BACR05B18 and a TaqMan MGB probe (Applied Biosystems) designed to the anchor fragment (6FAM-TCCTATAACAGAATTTGATCTGA-MGBNFQ). Primers used for anchor and bait fragments can be found in Table S3. qPCR was performed with TaqMan Universal PCR Master Mix (Applied Biosystems 4324018). The data presented represent an average of 4 biological replicates.
Gene expression analysis
RNA was purified from Kc cells with the RNeasy kit (Qiagen) and cDNA synthesis was performed with the High Capacity cDNA Reverse Transcription kit (Applied Biosystems). For quantitative PCR analysis, primers were designed to span transcript specific exon junctions and primer sequences can be found in Table S4. Real-time PCR was performed and data was normalized to Act5C. For microarray analysis, cDNA was digested with RNase A for 30 min at 37°C and cleaned by running through a PCR purification column (Qiagen). Sample labeling, hybridization, and data extraction were performed by the FSU-Nimblegen Microarray facility (http://www.bio.fsu.edu/nimblegen_microarray.php) using Nimblegen 12×135K arrays. Two biological replicates were performed for each sample. To determine genes that change in expression after 20-HE induction, we used ArrayStar software (DNASTAR, Inc.) and included a statistical cutoff of 10% FDR generated with a moderated t-test with Benjamini Hochberg multiple testing correction and a fold-change of 1.5.
Supplementary Material
Highlights.
CP190 recruitment to DNA is dramatically reduced during the heat-shock response
DNA localization of CP190, Su(Hw), CTCF and BEAF is regulated during 20HE treatment
CP190 mediates chromatin looping after 20HE treatment
CP190 is necessary for proper regulation of 20HE non-responsive genes
Acknowledgments
We would like to thank Dr. Elissa Lei for sharing antibodies and RNAi protocols. We also thank The Genomic Services Lab at the HudsonAlpha Institute for Biotechnology for their help in performing Illumina sequencing of ChIP-Seq samples. This work was supported by U.S. Public Health Service Award GM35463 from the National Institutes of Health.
Footnotes
ACCESSION NUMBERS
ChIP-seq and gene expression data are deposited in NCBI’s Gene Expression Omnibus (GEO) under accession number GSE30740 and GSE30686 respectively. Data can also be found under SuperSeries accession number GSE30741.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashburner M. Sequential gene activation by ecdysone in polytene chromosomes of Drosophila melanogaster. II. The effects of inhibitors of protein synthesis. Dev Biol. 1974;39:141–157. doi: 10.1016/s0012-1606(74)80016-3. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Bonner JJ. The induction of gene activity in drosophilia by heat shock. Cell. 1979;17:241–254. doi: 10.1016/0092-8674(79)90150-8. [DOI] [PubMed] [Google Scholar]
- Burtis KC, Thummel CS, Jones CW, Karim FD, Hogness DS. The Drosophila 74EF early puff contains E74, a complex ecdysone-inducible gene that encodes two ets-related proteins. Cell. 1990;61:85–99. doi: 10.1016/0092-8674(90)90217-3. [DOI] [PubMed] [Google Scholar]
- Bushey AM, Dorman ER, Corces VG. Chromatin insulators: regulatory mechanisms and epigenetic inheritance. Mol Cell. 2008;32:1–9. doi: 10.1016/j.molcel.2008.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushey AM, Ramos E, Corces VG. Three subclasses of a Drosophila insulator show distinct and cell type-specific genomic distributions. Genes Dev. 2009;23:1338–1350. doi: 10.1101/gad.1798209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M, Corces VG. The ubiquitin ligase dTopors directs the nuclear organization of a chromatin insulator. Mol Cell. 2005;20:105–116. doi: 10.1016/j.molcel.2005.08.031. [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Gaszner M, Felsenfeld G. Insulators: exploiting transcriptional and epigenetic mechanisms. Nat Rev Genet. 2006;7:703–713. doi: 10.1038/nrg1925. [DOI] [PubMed] [Google Scholar]
- Gauhar Z, Sun LV, Hua S, Mason CE, Fuchs F, Li TR, Boutros M, White KP. Genomic mapping of binding regions for the Ecdysone receptor protein complex. Genome Res. 2009;19:1006–1013. doi: 10.1101/gr.081349.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerasimova TI, Byrd K, Corces VG. A chromatin insulator determines the nuclear localization of DNA. Mol Cell. 2000;6:1025–1035. doi: 10.1016/s1097-2765(00)00101-5. [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Gdula DA, Gerasimov DV, Simonova O, Corces VG. A Drosophila protein that imparts directionality on a chromatin insulator is an enhancer of position-effect variegation. Cell. 1995;82:587–597. doi: 10.1016/0092-8674(95)90031-4. [DOI] [PubMed] [Google Scholar]
- Gerasimova TI, Lei EP, Bushey AM, Corces VG. Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Mol Cell. 2007;28:761–772. doi: 10.1016/j.molcel.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagege H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forne T. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat Protoc. 2007;2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- Ivaldi MS, Karam CS, Corces VG. Phosphorylation of histone H3 at Ser10 facilitates RNA polymerase II release from promoter-proximal pausing in Drosophila. Genes Dev. 2007;21:2818–2831. doi: 10.1101/gad.1604007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuan PF, Chun H, Keles S. CMARRT: a tool for the analysis of ChIP-chip data from tiling arrays by incorporating the correlation structure. Pac Symp Biocomput. 2008:515–526. [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei EP, Corces VG. RNA interference machinery influences the nuclear organization of a chromatin insulator. Nat Genet. 2006;38:936–941. doi: 10.1038/ng1850. [DOI] [PubMed] [Google Scholar]
- Maeda RK, Karch F. Making connections: boundaries and insulators in Drosophila. Curr Opin Genet Dev. 2007;17:394–399. doi: 10.1016/j.gde.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Majumder P, Gomez JA, Boss JM. The human major histocompatibility complex class II HLA-DRB1 and HLA-DQA1 genes are separated by a CTCF-binding enhancer-blocking element. J Biol Chem. 2006;281:18435–18443. doi: 10.1074/jbc.M601298200. [DOI] [PubMed] [Google Scholar]
- Mohan M, Bartkuhn M, Herold M, Philippen A, Heinl N, Bardenhagen I, Leers J, White RA, Renkawitz-Pohl R, Saumweber H, et al. The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. EMBO J. 2007;26:4203–4214. doi: 10.1038/sj.emboj.7601851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negre N, Brown CD, Shah PK, Kheradpour P, Morrison CA, Henikoff JG, Feng X, Ahmad K, Russell S, White RA, et al. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 2010;6:e1000814. doi: 10.1371/journal.pgen.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver D, Sheehan B, South H, Akbari O, Pai CY. The chromosomal association/dissociation of the chromatin insulator protein Cp190 of Drosophila melanogaster is mediated by the BTB/POZ domain and two acidic regions. BMC Cell Biol. 2010;11:101. doi: 10.1186/1471-2121-11-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai CY, Lei EP, Ghosh D, Corces VG. The centrosomal protein CP190 is a component of the gypsy chromatin insulator. Mol Cell. 2004;16:737–748. doi: 10.1016/j.molcel.2004.11.004. [DOI] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riddiford LM, Cherbas P, Truman JW. Ecdysone receptors and their biological actions. Vitam Horm. 2000;60:1–73. doi: 10.1016/s0083-6729(00)60016-x. [DOI] [PubMed] [Google Scholar]
- Robbins MJ, Starr KR, Honey A, Soffin EM, Rourke C, Jones GA, Kelly FM, Strum J, Melarange RA, Harris AJ, et al. Evaluation of the mGlu8 receptor as a putative therapeutic target in schizophrenia. Brain Res. 2007;1152:215–227. doi: 10.1016/j.brainres.2007.03.028. [DOI] [PubMed] [Google Scholar]
- Segraves WA, Hogness DS. The E75 ecdysone-inducible gene responsible for the 75B early puff in Drosophila encodes two new members of the steroid receptor superfamily. Genes Dev. 1990;4:204–219. doi: 10.1101/gad.4.2.204. [DOI] [PubMed] [Google Scholar]
- Stevens B, Alvarez CM, Bohman R, O’Connor JD. An ecdysteroid-induced alteration in the cell cycle of cultured Drosophila cells. Cell. 1980;22:675–682. doi: 10.1016/0092-8674(80)90543-7. [DOI] [PubMed] [Google Scholar]
- Wendt KS, Peters JM. How cohesin and CTCF cooperate in regulating gene expression. Chromosome Res. 2009;17:201–214. doi: 10.1007/s10577-008-9017-7. [DOI] [PubMed] [Google Scholar]
- Xu Z, Wei G, Chepelev I, Zhao K, Felsenfeld G. Mapping of INS promoter interactions reveals its role in long-range regulation of SYT8 transcription. Nat Struct Mol Biol. 2011;18:372–378. doi: 10.1038/nsmb.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.