SUMMARY
NADH is a key metabolic cofactor whose sensitive and specific detection in the cytosol of live cells has been difficult. We constructed a fluorescent biosensor of cytosolic NADH-NAD+ redox state by combining a circularly permuted GFP T-Sapphire with a bacterial NADH-binding protein Rex. Although the initial construct reported [NADH] × [H+]/[NAD+], its pH sensitivity was eliminated by mutagenesis. The engineered biosensor, Peredox, reports cytosolic NADH:NAD+ ratios and can be calibrated with exogenous lactate and pyruvate. We demonstrated its utility in several cultured and primary cell types. We found glycolysis opposed the lactate dehydrogenase equilibrium to produce a reduced cytosolic NADH-NAD+ redox state. We also observed different redox states in primary mouse astrocytes and neurons, consistent with hypothesized metabolic differences. Furthermore, using high-content image analysis, we monitored NADH responses to PI3K pathway inhibition in hundreds of live cells. As an NADH reporter, Peredox should enable better understanding of bioenergetics.
INTRODUCTION
Nicotinamide adenine dinucleotide (reduced: NADH or oxidized: NAD+) is a key cofactor for electron transfer in metabolism. Reduction-oxidation (redox) reactions catalyzed by various NAD(H)-dependent dehydrogenases are vital for biochemical processes such as glycolysis and mitochondrial metabolism. In addition, NADH-NAD+ redox has been implicated in the regulation of embryonic development and aging (Dumollard et al., 2007; Chen et al., 2009), as well as in pathological conditions such as diabetes, cancer, and epilepsy (Eto et al., 1999; Zhang et al., 2006; Garriga-Canut et al., 2006).
To assess cellular NADH-NAD+ redox state, there have been two general approaches. Chemical methods infer the NADH:NAD+ ratio indirectly from the concentrations of redox couples such as lactate and pyruvate (Williamson et al., 1967). However, this requires the use of cell extracts and is thus incompatible with studying dynamics in intact, individual cells. A less invasive optical approach monitors cellular NAD(P)H autofluorescence. Under ultraviolet excitation, NADH and another related cofactor NADPH are fluorescent, whereas their oxidized counterparts NAD+ and NADP+ are not (Chance et al., 1962). As NADH and NADPH give identical autofluorescence signals, they are collectively denoted as NAD(P)H, although the two cofactors govern distinct metabolic reactions (Klingenberg and Bücher, 1960). Since NADH is present at higher levels than NADPH in many tissues, the NAD(P)H signal has often been interpreted as changes in NADH (Lowry et al., 1957; Klingenberg and Bücher, 1960; Chance et al., 1962). However, data from simultaneous imaging of both NAD(P)H and flavoprotein autofluorescence underscore the ambiguity of the NAD(P)H signal, suggesting that the NAD(P)H signal may primarily report protein-bound NADPH instead of NADH (Rocheleau et al., 2004). In addition to the signal ambiguity, in general, cytosolic and mitochondrial NAD(P)H autofluorescence have been assumed to reflect glycolysis and oxidative metabolism, respectively (Patterson et al., 2000; Shuttleworth et al., 2003; Kasischke et al., 2004; Gordon et al., 2008). In practice, mitochondrial signals dominate the measurements, whereas cytosolic signals are small and difficult to separate from the bright mitochondrial signals. For more specific and sensitive detection of cytosolic NADH-NAD+ redox state, a fluorescent NADH biosensor would be valuable.
One strategy to create genetically encoded fluorescent biosensors involves circularly permuted fluorescent proteins (cpFPs), derived from green fluorescent proteins (GFPs). Circular permutation joins the original N and C termini with a peptide linker and creates new termini near the chromophore (Baird et al., 1999). A specific detector or binding protein fused to the new N and C termini creates conformational coupling between binding and fluorescence. With this strategy, fluorescent biosensors have been engineered to report calcium, hydrogen peroxide, cyclic 3′,5′-guanosine monophosphate (cGMP), and ATP:ADP ratio (Nagai et al., 2001; Belousov et al., 2006; Nausch et al., 2008; Berg et al., 2009). For a detector domain, we chose the redox sensing repressor Rex, a bacterial NADH-binding protein that links metabolic state to gene expression (Brekasis and Paget, 2003). Rex is a homodimer; each subunit comprises an N-terminal domain and a C-terminal NADH-binding domain (Sickmier et al., 2005). Upon NADH binding, Rex adopts a closed instead of an open conformation (Sickmier et al., 2005; Wang et al., 2008; McLaughlin et al., 2010). Here, we found that integration of a cpFP T-Sapphire into Rex yielded a fluorescent sensor of NADH. The initial construct (and likely the native Rex protein) reported [NADH] × [H+]/[NAD+]. By eliminating its pH dependence via targeted mutagenesis, we constructed a genetically encoded fluorescent biosensor of cytosolic NADH:NAD+ ratios. This reporter, Peredox, revealed cytosolic NADH-NAD+ redox dynamics in mammalian cells upon metabolic challenges. With transient or stable expression, we demonstrated the utility of Peredox in a variety of primary and cultured cell types. We found different cytosolic NADH-NAD+ redox states in primary mouse cortical astrocytes versus neurons, as well as changes in cytosolic NADH-NAD+ redox in cultured epithelial cells upon perturbation of PI3K pathway signaling.
RESULTS
A cpFP inserted into T-Rex reports [NADH] × [H+]/[NAD+]
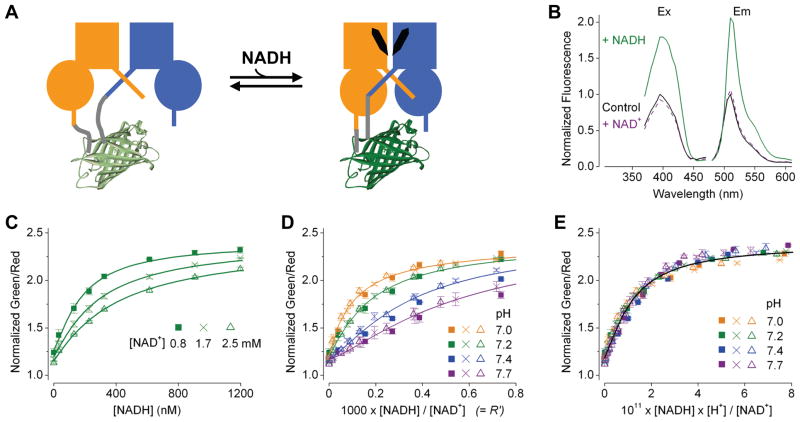
To engineer an NADH biosensor, we inserted a cpFP variant of T-Sapphire (Zapata-Hommer and Griesbeck, 2003) into a tandem dimer of Rex from Thermus aquaticus (T-Rex), between the two subunits. By PCR we created a library of peptide linkers for the cpFP insertion, expressed the protein library in bacteria, and assayed the purified proteins for fluorescence responses. We found that a cpFP inserted into T-Rex could couple conformational changes with fluorescence to report NADH (Figures 1A and S1 available online). A construct, named P0, exhibited an increase in green fluorescence upon NADH application (Figure 1B). Its spectra were similar to that of T-Sapphire, with an excitation peak around 400 nm and an emission peak around 510 nm. While NADH application enhanced the green fluorescence, it did not change the red fluorescence of a tandemly attached mCherry (Figure S2A; Shaner et al., 2004), which was included to normalize the signal for protein expression. The affinity of P0 for NADH was less than 5 nM (Figure S2B), which was surprising given that the estimated concentration of free NADH in the cytosol is in the hundreds of nanomolar (Zhang et al., 2002). In addition to NADH, NAD+ can bind to Rex. Unlike NADH, application of NAD+ yielded only minimal change in fluorescence (Figure 1B). However, increasing NAD+ concentration effectively lowered the sensor’s apparent affinity for NADH (Figure 1C), indicating that NAD+ competes with NADH for binding. According to this competitive scheme, when the concentrations of NADH and NAD+ exceed their affinity constants, the sensor’s steady state fluorescence response would report the NADH:NAD+ ratio (Figure 1D; Berg et al., 2009). Although conventionally the ratio R = [NAD+]/[NADH] is reported, we plotted the response against the alternative ratio R′ = [NADH]/[NAD+] × 1000. The R′ at which the response is half maximal is called the ‘KR′’ of the sensor, analogous to the dissociation constant (Kd) of a receptor. At pH 7.2, P0 had ~8000-fold higher affinity for NADH than for NAD+; its half maximal response corresponded to a KR′ of 0.12. Because P0 was constructed with a pH resistant GFP T-Sapphire, its fluorescence in the unoccupied state or in the NADH-bound state was pH resistant as expected. Nevertheless, the ability of NAD+ to compete with NADH for binding did vary substantially with pH (Figure 1D). With higher pH, competition by NAD+ became stronger, further lowering the sensor’s apparent affinity for NADH and increasing KR′. Remarkably, this pH dependence could be described as a strict dependence of the sensor on [NADH] × [H+]/[NAD+] (Figure 1E). Indeed, this expression is a component of the equilibrium constant of any NAD(H)-dependent dehydrogenase, and it is proportional to the ratio of the redox couple. For instance, when the lactate dehydrogenase (LDH) reaction is at equilibrium, the lactate:pyruvate ratio is proportional to [NADH] × [H+]/[NAD+].
Figure 1. Characterization of purified P0, a Rex-cpFP chimera.
(A) Schematic showing the sensor design, with a cpFP (PDB: 3evp) interposed between the two T-Rex subunits (blue and orange), and a change of fluorescence upon binding of NADH (black). (B) Excitation and emission spectra in the control condition (solid black), after addition of 100 μM NAD+ (dash purple), or 100 μM NAD+ and 0.2 μM NADH (solid green), normalized to the peak intensity in the control condition. For excitation spectra, emission was measured at 510 ± 5 nm; for emission spectra, excitation was at 400 ± 2.5 nm. (C) Green to red fluorescence ratios at the indicated [NAD+] at pH 7.2 plotted against [NADH]. (D) Fluorescence ratios at the indicated [NAD+] and pH plotted against R′. (E) Fluorescence ratios at the indicated [NAD+] and pH plotted against [NADH] × [H+]/[NAD+]. Fluorescence ratios (mean ± SEM, n = 3) were normalized to the control condition in the absence of pyridine nucleotides at pH 7.2 at 25°C.
Optimization eliminates the pH sensitivity of the NADH biosensor
Many factors other than metabolism perturb cellular pH. Although P0 reports the interesting quantity [NADH] × [H+]/[NAD+], its pH sensitivity renders the signal non-specific to cytosolic NADH-NAD+ redox and thus not highly useful. Nonetheless, this quantity [NADH] × [H+]/[NAD+] suggests that, similar to NADH-binding dehydrogenases, there is a single proton or charge transfer for each NAD+ (or NADH) binding event. We reasoned that the highly conserved residue Tyr98, located near the nicotinamide moiety of the NADH molecule in the T-Rex structure (Sickmier et al., 2005), might participate in the proton transfer upon NAD+ binding (Jörnvall et al., 1995). We then made a protein library with various mutations on Tyr98. In addition, to speed up kinetics (see below), we performed error-prone PCR mutagenesis and screened for improved sensor variants. The best product of the screen contained the mutations Tyr98Trp and Phe189Ile in the first subunit and Tyr98Trp in the second subunit.
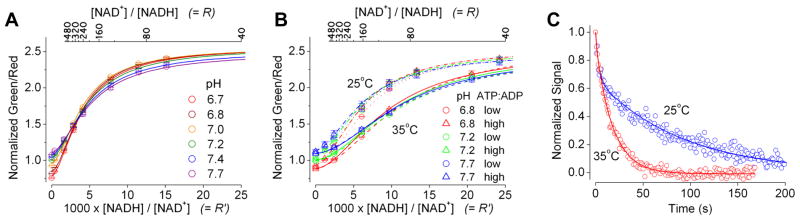
This circularly permuted GFP based sensor of NADH-NAD+ redox, named Peredox, demonstrated notable improvements in pH resistance and kinetics for cytosolic NADH-NAD+ redox sensing. While Peredox retained the spectral properties of the original P0 construct (Figures S3A and S3B), it was far more resistant to pH changes in the physiological range (Figures 2A and S3C). For an increase of one pH unit, which produced a 10-fold increase of KR′ in P0 (Figure 1D), the KR′ of Peredox changed only slightly by ~20%, with a crossover point (signal insensitive to pH) at R′ ≈ 3 (Figure 2A). Upon NADH saturation, Peredox displayed an increase of ~150% in fluorescence response (i.e. a 2.5-fold increase). Due to its lower affinities for NAD+ and NADH, Peredox reported the cytosolic NADH level, partly compensated for the NAD+ level, rather than strictly the NADH:NAD+ ratio. The NADH:NAD+ ratio required for a half maximal response was somewhat resistant to changes in the overall [NADH + NAD+]: a 3-fold change in the NAD+ pool size in the physiological range produced a ~2-fold change in the sensor midpoint for NADH:NAD+ ratio (Figure S3D). Peredox was specific against other metabolites structurally related to NADH, such as NADPH, NADP+, ADP ribose, nicotinamide, β-nicotinamide mononucleotide, AMP, and adenosine. Not only was there no change in its fluorescence response upon addition of these metabolites (Figure S3E), but also the NADH:NAD+ titrations in their presence were similar to that of control (Figures S3F and S3G). The main interference came from ADP and ATP, with an apparent affinity (Ki) in the millimolar range (Figure S3H), consistent with published data on B-Rex (Wang et al., 2008). In Peredox, while the specificity of binding NADH over ADP or ATP was roughly 30,000-fold, NADH is far less abundant than ADP or ATP in typical intracellular environments. Although physiological levels of ADP and ATP could compete with NADH and NAD+ for binding, ADP and ATP act similarly to each other, at various pH’s and temperatures; also, the interference from ADP and ATP is less prominent with increasing NAD+ (Figures 2B and S3I–L). Thus, while Peredox might be slightly sensitive to changes in the total adenine nucleotide pool size, its response is not affected by changes in the cellular energy charge, or the ATP:ADP ratio. While metabolic challenges often lead to energy deprivation and decrease in cytosolic ATP:ADP ratio, changes in cytosolic adenine nucleotide pool size are minimal (Schwenke et al., 1981; Malaisse and Sener, 1987) and would not be expected to interfere with Peredox response. In addition, Peredox exhibited much improved kinetics over P0. To determine the rate of NADH dissociation from the biosensor, we added the LDH enzyme with pyruvate to consume the free NADH and monitored the response over time. While NADH dissociation from P0 was slow, with a time constant of 25 minutes at 25°C, NADH dissociation from Peredox was much faster, with a time constant of 50 seconds at 25°C and 16 seconds at 35°C (Figure 2C).
Figure 2. Characterization of purified Peredox.
(A) Green to red fluorescence ratios at the indicated pH, plotted against R′ or R (above the plot), with 80 μM NAD+ at 25°C. (B) Fluorescence ratios at the indicated pH, temperature, and ATP:ADP ratios (low, 0.3; high, 3.6; with total adenine nucleotides of 4.6 mM), plotted against R′ or R (above the plot), with 80 μM NAD+. Fluorescence ratios (mean ± SEM, n = 3) were normalized to the control condition in the absence of pyridine nucleotides at pH 7.2. (C) Kinetics of fluorescence signal upon addition of pyruvate and LDH to Peredox pre-equilibrated with saturating NADH at 25°C or 35°C, normalized to initial and final values.
Peredox reports cytosolic NADH-NAD+ redox state in mammalian cells
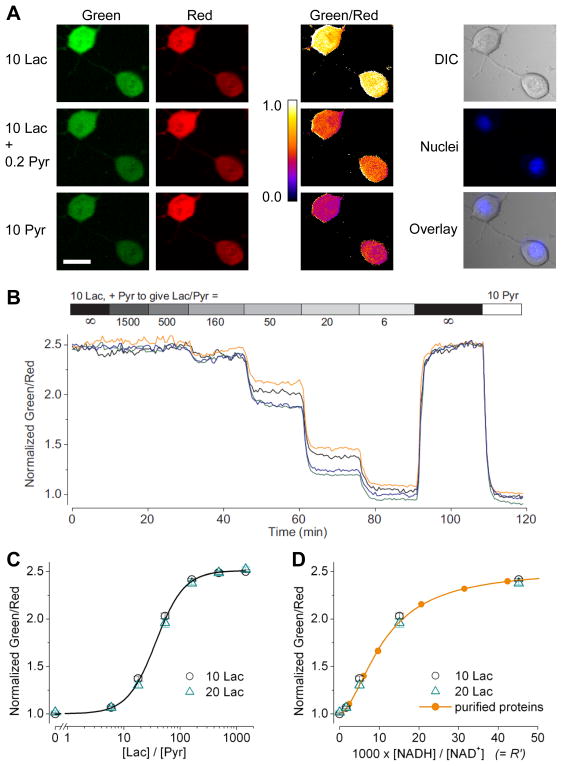
After characterizing Peredox-mCherry as a purified protein, we validated its utility in mammalian cells. We expressed Peredox-mCherry in cultured mouse neuroblastoma Neuro-2a cells and monitored its fluorescence response. Under confocal microscopy, the sensor fluorescence in both green and red images was fairly uniform through the cell (Figure 3A). The pixel-by-pixel ratio of the green image divided by the red image appeared consistent, with no apparent difference observed between cytosolic and nuclear signals. To test whether Peredox could report cytosolic NADH-NAD+ redox state, we varied the concentrations of lactate and pyruvate in the extracellular solution, thereby altering the intracellular concentrations of these metabolites (Bücher et al., 1972). Interconversion between lactate and pyruvate catalyzed by endogenous LDH should lead to concomitant exchange between cytosolic NADH and NAD+ species. Note that cells were not permeabilized, and glucose was absent in the extracellular solution. With widefield time-lapse microscopy, we monitored changes in fluorescence response of Peredox. In addition, we established that cellular background autofluorescence was far weaker and did not interfere with Peredox fluorescence signal (Figure S4). In the presence of lactate (10 mM), the green to red fluorescence ratio of Peredox was maximal (Figure 3B). After adding incremental amounts of pyruvate, we observed a stepwise decrease in the green to red fluorescence ratio, with millimolar pyruvate reducing the ratio down to 40% of the maximal value. This signal change was consistent with oxidation of NADH into NAD+ and a concurrent reduction of pyruvate into lactate. After each solution switch, the signal adjusted rapidly and arrived at a new steady state in a few minutes. The steady state fluorescence response of Peredox depended systematically on the lactate:pyruvate ratio in the extracellular solution (Figure 3C); the lactate:pyruvate ratio required for a half maximal response was ~40. To test the robustness of this ratio-sensing behavior, we repeated the experiments with different total concentrations of lactate and pyruvate. In each case, the lactate:pyruvate ratio, rather than their absolute concentrations, determined the response. We could also plot the Peredox response to the extracellular lactate:pyruvate ratio as a function of predicted NADH:NAD+ ratio (Figure 3D), by using the known equilibrium constant of the LDH reaction (Williamson et al., 1967) and assuming a constant physiological pH of 7.4. For comparison we also show the data from purified Peredox proteins, with total adenine nucleotide of 4.6 mM and free NAD+ of 80 μM, consistent with published estimates (Veech et al., 1979; Zhang et al., 2002). In both the dynamic range and the relative affinities for NADH and NAD+, there is good agreement between the sensor responses in cells and from purified proteins. Therefore, Peredox reports cytosolic NADH:NAD+ ratios in mammalian cells. Furthermore, since in the absence of glucose we could use exogenous lactate and pyruvate to set cytosolic NADH:NAD+ redox, it appears that lactate and pyruvate equilibrate readily between extracellular and intracellular environments, and that the LDH reaction is approximately at equilibrium.
Figure 3. Imaging cytosolic NADH-NAD+ redox state in various extracellular lactate:pyruvate ratios.
(A) Left: Confocal green and red fluorescence images of two cultured mouse neuroblastoma Neuro-2a cells expressing Peredox supplied with 10 mM lactate, 10 mM lactate and 0.2 mM pyruvate, or 10 mM pyruvate. Scale bar 20 μm. Middle: Pseudocolored pixel by pixel green to red ratio images. The slight edge effect seen is likely due to the optical z-shift of 1.5 μm between the green and red confocal images. Right: Widefield differential interference contrast (DIC), nuclear staining, and the overlay image. (B) Time course of fluorescence ratios of four Neuro-2a cells perfused with the indicated lactate:pyruvate ratios. (C) Steady state fluorescence responses of Neuro-2a cells plotted against extracellular lactate:pyruvate ratios, with lactate of 10 mM or 20 mM (mean ± SEM, n = 12–15 cells from two independent experiments). (D) Steady state fluorescence responses in (D) plotted against the predicted R′, by assuming the LDH reaction was at equilibrium and a pH of 7.4. Data of purified Peredox proteins were with 80 μM NAD+ and 4.6 mM total adenine nucleotides at 35°C. Line fitted with a logistic function using a Hill coefficient of 1.8.
Glycolysis opposes the LDH equilibrium to produce a reduced cytosolic NADH-NAD+ redox state in cultured cells
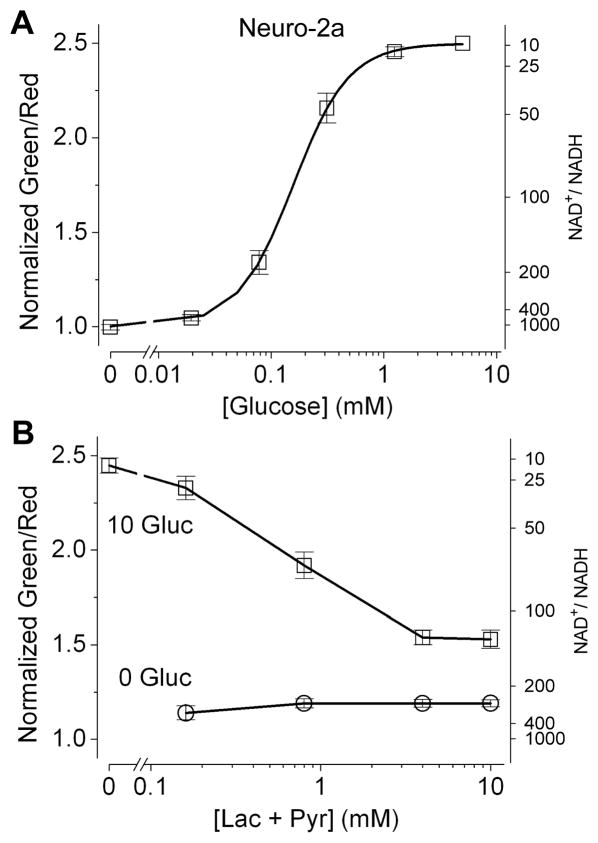
In addition to LDH, glucose metabolism via glycolysis at the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) reaction would be expected to affect cytosolic NADH-NAD+ redox state. In cultured Neuro-2a cells supplied with glucose alone, we found the sensor signal to be maximal (Figure S5). To test whether such strongly reduced cytosolic NADH-NAD+ redox state depended on glucose metabolism, we performed the following experiments. First, we varied the concentration of glucose in the extracellular solution (Figure S5A). Not only was the signal minimal in the absence of glucose, but also its steady state fluorescence response depended on the glucose supply; the glucose concentration required for a half maximal response was ~0.2 mM (Figure 4A). Second, when we applied iodoacetate, which irreversibly inhibits GAPDH, the signal promptly decreased (Figure S5B), indicating a decline in cytosolic NADH consistent with glycolytic inhibition. Therefore, the reduced cytosolic NADH-NAD+ redox state in cultured Neuro-2a cells supplied with glucose depended on both the presence of glucose and the GADPH reaction. Nevertheless, given that lactate and pyruvate could equilibrate across the cell membrane readily, intracellular lactate and pyruvate might be washed away in cells that were perfused with glucose alone. If availability of intracellular pyruvate were to become limiting for the LDH reaction, this could account for the reduced NADH-NAD+ redox state in these cultured cells. To address this concern, we systematically varied the total concentrations of extracellular lactate and pyruvate (while keeping a constant lactate:pyruvate ratio of 10), with or without supplying glucose (Figure 4B). On one hand, when glucose was absent, Peredox indicated a cytosolic NAD+:NADH ratio of ~300 across a wide range of total lactate and pyruvate concentrations. On the other hand, when glucose was present, cytosolic NADH-NAD+ redox state became significantly more reduced. While this observation appeared more pronounced at lower concentrations of lactate and pyruvate, it held across a wide concentration range. With physiological amounts of lactate and pyruvate (totaling 0.8 to 4 mM; Williamson et al., 1967), glucose metabolism reduced cytosolic NAD+:NADH ratios by 2- to 4-fold to 70–130, when availability of intracellular pyruvate should not be limiting. We obtained similar results in another cultured tumor cell line, rat glioma C6 (Figure S6). In both cases, the redox status of cytosolic NADH-NAD+ was no longer determined by the LDH reaction alone, but rather by a balance between the LDH reaction and glycolysis.
Figure 4. Cultured mouse neuroblastoma Neuro-2a cells supplied with glucose show a more reduced cytosolic NADH-NAD+ redox state.
(A) Steady state fluorescence responses of Neuro-2a cells plotted against concentrations of extracellular glucose (mean ± SEM, n = 19 cells from two independent experiments). (B) Steady state fluorescence responses of Neuro-2a cells plotted against total concentrations of extracellular lactate and pyruvate, with a constant lactate:pyruvate ratio of 10, and glucose of 10 mM or 0 mM (mean ± SEM, n = 7 cells from three independent experiments). For the alternate y axis, the predicted NAD+:NADH ratio was calculated from purified protein measurements. p < 0.001 (paired t-test) for all conditions in 10 mM vs. 0 mM glucose.
Primary cultured cortical astrocytes and neurons differ in their cytosolic NADH-NAD+ redox states
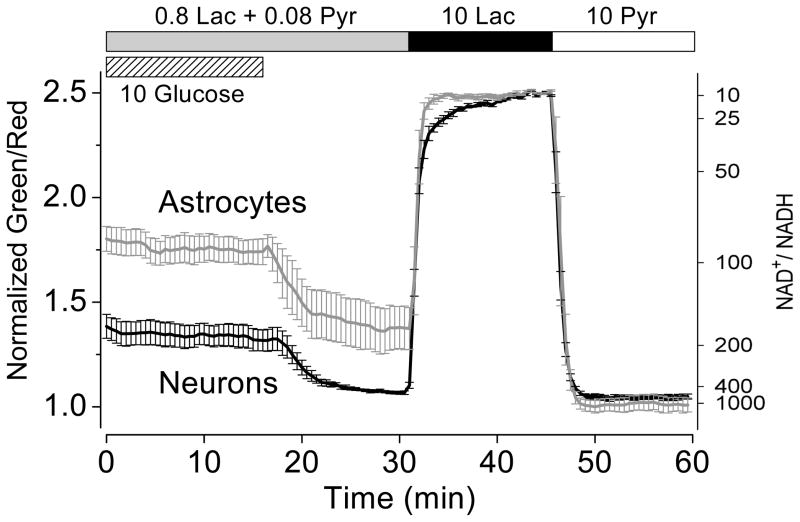
Glucose metabolism has been proposed to differ in astrocytes and neurons (Pellerin and Magistretti, 1994; Herrero-Mendez et al., 2009). To investigate whether their cytosolic NADH-NAD+ redox states differ, we expressed Peredox-mCherry in primary cultured mouse cortical astrocytes and neurons. However, we observed bright red puncta that appeared to be lysosomal aggregates due to accumulation of mCherry (Katayama et al., 2008); these red puncta rendered the normalized signal unreliable. To circumvent this problem, we used a nuclear targeted version (Nagai et al., 2001) of Peredox to measure nuclear NADH signals. Since there is presumably no diffusion barrier for free NADH and NAD+ between nuclear and cytosolic compartments (and we saw no difference between nuclear and cytosolic signals in cultured cells), our nuclear signals should indicate cytosolic NADH-NAD+ redox state (Zhang et al., 2002). When measured under the same conditions, primary astrocytes showed significantly more reduced cytosolic NADH-NAD+ redox states than primary neurons (Figure 5). Furthermore, upon glucose withdrawal, both cell types promptly showed a decline in cytosolic NADH. For both primary astrocytes and neurons, similar to cultured Neuro-2a cells (Figure 3B), perfusion with lactate (10 mM) alone yielded maximal fluorescence response, whereas the signal became minimal upon pyruvate (10 mM) perfusion.
Figure 5. Primary cultured mouse cortical astrocytes and neurons differ in their cytosolic NADH-NAD+ redox states.
Time course of fluorescence ratios of primary mouse cortical astrocytes or neurons expressing Peredox-NLS and perfused with solutions as indicated (mean ± SEM, n = 4–5 cells from four independent experiments). For the alternate y axis, the predicted NAD+:NADH ratio was calculated from purified protein measurements. p < 0.01 (paired t-test) for astrocytes vs. neurons prior to the 10 mM lactate condition.
In stably-expressed MCF-10A cells, Peredox reports cytosolic NADH decrease upon PI3K pathway inhibition
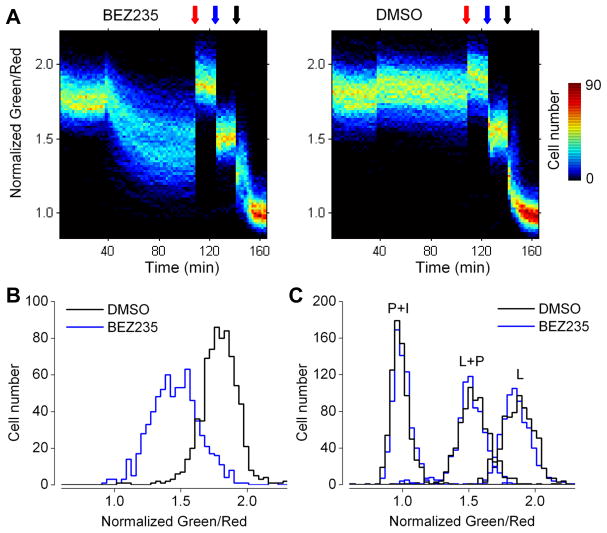
Given that glucose metabolism is regulated by growth factor signaling including the PI3K/Akt/mTOR pathway (Vander Heiden et al., 2009, Sengupta et al., 2010), we explored the effects of PI3K pathway signaling on glucose metabolism as indicated by the cytosolic NADH-NAD+ redox state. To this end, we generated a cultured mammary epithelial MCF-10A cell line stably expressing nuclear targeted Peredox-mCherry; the nuclear targeting allowed us to distinguish individual cells easily and enabled the use of high-content image analysis to monitor hundreds of cells concurrently in a multi-well imaging experiment (Jones et al., 2008). At the baseline condition, MCF-10A cells showed reduced cytosolic NADH-NAD+ redox states (Figure 6A). Upon the application of a dual inhibitor of PI3K and mTOR, NVP-BEZ235 (Maira et al., 2008), we observed a decrease in cytosolic NADH level in nearly all cells over the course of an hour (Figure 6B). This NADH decline was consistent with the expected glycolytic inhibition as a consequence of inhibiting PI3K pathway signaling. Cytosolic NADH-NAD+ redox states were unaffected in cells treated with the DMSO vehicle control. Afterwards, as a control, we applied reagents that were known to perturb cytosolic NADH-NAD+ redox states: application of lactate (20 mM) alone, lactate (20 mM) and pyruvate (1 mM), and pyruvate (20 mM) with the glycolytic inhibitor iodoacetate (0.4 mM) could poise these cells to high, medium, and low cytosolic NADH levels, respectively. The fluorescence responses to these control treatments were remarkably similar between the drug-treated and the control groups (Figure 6C).
Figure 6. In stably-expressed mammary epithelial MCF-10A cells, Peredox reports cytosolic NADH decrease upon PI3K pathway inhibition.
(A) Heat maps of normalized fluorescence ratios of MCF-10A cells stably expressing Peredox-NLS plotted against time. After 38 minutes of baseline, cells were treated with 1 μM NVP-BEZ235 (left) or DMSO (right). As a control, 20 mM lactate, 20 mM lactate and 1 mM pyruvate, and 20 mM pyruvate and 0.4 mM iodoacetate were applied at red, blue, and black arrows, respectively. For each group, ~700 cells from six fields in two experiments were collected. While the dynamic range was less than the usual 2.5-fold, this was likely due to inadequate control of extracellular lactate and pyruvate concentrations, as these cells were imaged in the 24-well plate formats and solutions were changed without rinsing, as opposed to imaging in a chamber under continuous perfusion of fresh solutions. Fluorescence ratios binned in increments of 0.012. (B) Histograms of normalized fluorescence ratios after one-hour treatment with DMSO (black) or NVP-BEZ235 (blue), binned in increments of 0.027. (C) Histograms of normalized fluorescence ratios from the indicated groups after calibration with 20 mM lactate (L), 20 mM lactate and 1 mM pyruvate (L+P), and 20 mM pyruvate and 0.4 mM iodoacetate (P+I).
DISCUSSION
We exploited the conformational change of the Rex protein upon NADH binding to engineer a cpFP-based biosensor of cytosolic NADH-NAD+ redox state. Although the initial construct reported [NADH] × [H+]/[NAD+], optimization via targeted mutagenesis eliminated its pH sensitivity. This genetically encoded fluorescent biosensor, Peredox, reported cytosolic NADH:NAD+ ratios in mammalian cells. Its fluorescence signal could be calibrated with exogenous lactate and pyruvate. With transient or stable expression, we demonstrated its utility in various cultured and primary cell types. In cultured mammalian cells, we found that glucose metabolism via glycolysis opposed the lactate dehydrogenase equilibrium to produce a reduced NADH-NAD+ redox state. We also observed different cytosolic NADH-NAD+ redox states in primary mouse astrocytes and neurons under identical conditions, consistent with their hypothesized metabolic differences. Finally, using Peredox stably expressed in cultured MCF-10A cells, we monitored changes in cytosolic NADH-NAD+ redox states from hundreds of individual cells upon inhibition of PI3K pathway signaling.
Peredox presents several advantages over conventional chemical methods and NAD(P)H autofluorescence in monitoring cytosolic NADH-NAD+ redox state. Using chemical methods, NADH and NAD+ can be quantified directly in cell extracts, but these measurements detect the total amounts of NAD(H) and ignore their subcellular location. As most cellular NADH resides in the mitochondria, cytosolic NADH determination with such methods can be complicated by substantial contamination from the mitochondrial NADH pool. Also, such direct chemical methods cannot distinguish between protein-bound and free species of NAD(H). These measurements of total NADH:NAD+ ratios often vastly overestimate the actual cytosolic free NADH:NAD+ ratios, owing to sequestration from cellular proteins that is greater for NADH than for NAD+. Considering that free NADH:NAD+ ratio is the pertinent parameter for biological NADH-NAD+ redox reactions, other chemical methods have been devised to extract and quantify redox couples such as lactate and pyruvate. Assuming that the LDH reaction is at equilibrium, the lactate:pyruvate ratios can be used to infer the free NADH:NAD+ ratios in the cytosol, assuming a constant known pH (Williamson et al., 1967). However, such indirect methods can be affected by changes in cellular pH as well as by contamination from the mitochondrial fraction. More importantly, the assumption that the LDH reaction is at equilibrium has not been examined. As we have shown, cultured tumor cells such as Neuro-2a and C6 supplied with glucose showed a reduced cytosolic NADH-NAD+ redox state, despite the abundance of exogenous lactate and pyruvate present at a ratio chosen to keep it oxidized. From a measurement standpoint, this indicates that the LDH reaction was likely not at equilibrium, and thus the measurement of cytosolic free NADH:NAD+ ratios using Peredox is likely to be more reliable than indirect methods using lactate:pyruvate ratios. Besides, unlike chemical methods, Peredox can be used to monitor cytosolic NADH-NAD+ redox states in intact, individual cells.
In comparison to the endogenous NAD(P)H autofluorescence, Peredox is a bit slower, but much more specific and sensitive in monitoring cytosolic NADH-NAD+ redox state. Given its kinetics, Peredox can report NADH-NAD+ redox dynamics with a time resolution of a few seconds. It may be difficult to use it to monitor faster events, such as the dip and the overshoot of endogenous NAD(P)H autofluorescence observed in brain slices (Shuttleworth et al., 2003; Kasischke et al., 2004). Nonetheless, NAD(P)H autofluorescence reflects the combined signal of NADH and NADPH and is therefore not specific for NADH alone (Avi-Dor et al., 1962; Rocheleau et al., 2004). Also, since the autofluorescence is contributed mostly by protein-bound NAD(P)H species, changes in intracellular environment such as pH could affect the NAD(P)H-protein interaction and the autofluorescence signal (Ogikubo et al., 2011). In contrast, Peredox detects free NADH and reports the biologically relevant NADH:NAD+ ratio, and its response is highly specific for NADH over NADPH. Furthermore, Peredox is roughly 100-fold brighter than NAD(P)H autofluorescence (Table S1). With Peredox we could reliably monitor cytosolic NADH, which is difficult to measure with autofluorescence imaging, particularly in the face of bright mitochondrial autofluorescence (nuclear autofluorescence has been used as a proxy for cytosolic autofluorescence; Patterson et al., 2000). Lastly, since NAD(P)H autofluorescence requires ultraviolet (one-photon) excitation, which could induce cell toxicity, two-photon microscopy is usually performed instead. Conversely, with a red-shifted excitation wavelength and brighter fluorescence, Peredox can be used in conventional one-photon microscopy, thereby simplifying the technical requirements for imaging experiments.
In designing Peredox, we have substantially minimized the interference of pH. In general, upon metabolic manipulations, changes in intracellular pH are common. This may interfere with biosensor measurements by altering the GFP fluorescence or the ligand-binding protein scaffold. Many cpFP-based fluorescent biosensors are pH sensitive, with pKa’s near the physiological range (Nagai et al., 2001; Belousov et al., 2006; Nausch et al., 2008; Berg et al., 2009). For some biosensors, a pH fluctuation of 0.3 units could be mistaken for the entire excursion of the sensor response. Fluorescence modulation in cpFP-based sensors has been attributed to a shift in the pKa of the chromophore upon ligand binding to the detector domain (Baird et al., 1999). Despite the apparent requirement for pH sensitivity inherent in this proposed mechanism, we decided to use a pH-resistant GFP. In doing so we hoped to modulate the cpFP’s quantum yield rather than its pKa upon NADH sensing. For this reason we chose the circularly permuted T-Sapphire (Zapata-Hommer and Griesbeck, 2003), a GFP variant notable for a low pKa of ~5. Its chromophore remains almost neutral in the ground-state and ionizes only upon excitation, leading to green emission with a substantial Stokes shift of ~110 nm. We optimized the sensor response in the pH insensitive regime of the GFP (corresponding to the physiological pH range in cells). The resulting biosensor is far more pH resistant and, unlike many other cpFP-based sensors, does not need pH measurement and correction, unless large pH changes are expected and high precision is needed. Moreover, the use of a pH resistant cpFP in the sensor design allowed us to uncover the inherent pH sensitivity of the NADH-binding protein scaffold. We initially conceived T-Rex as a sensor of the NADH:NAD+ ratio, so we were surprised to find that it depended on [NADH] × [H+]/[NAD+]. Given the tight hydrophobic nicotinamide-binding pocket, the implication of a single proton transfer upon NAD(H) binding in Rex was unexpected. We were nevertheless able to eliminate its intrinsic pH dependence via directed mutagenesis of the conserved residue Tyr98.
In using a genetically encoded biosensor, a potential concern is whether it will perturb the biological systems that it is used to measure; however, Peredox seems unlikely to perturb cellular free NADH by either catalysis or buffering. The dependence of Rex on [NADH] × [H+]/[NAD+] is reminiscent of the equilibrium constant of NADH-binding dehydrogenases, raising the question of whether Rex is not just an NADH-binding protein but also a redox enzyme. If the Rex-based biosensor were to have catalytic activity, it could alter the level of cellular NADH, and binding or release of NAD(H) might depend on the presence of substrate (Kumar et al., 2002). Yet, structural and biochemical evidence seems to suggest that Rex does not possess any catalytic activity. The T-Rex structure reveals a compact hydrophobic pocket enclosing the nicotinamide moiety of the NADH molecule, without a cavity for substrate accommodation or polar functional groups characteristic of redox enzymes (Sickmier et al., 2005). Also, most NADH-binding dehydrogenases exhibit some catalysis even with non-optimal substrates. We however observed no oxidation of NADH in Rex upon pyruvate application (50 mM for >20 hours), unlike the case for the eukaryotic transcriptional corepressor C-terminal binding protein (Kumar et al., 2002). Even if Rex were a redox enzyme, the mutation at the highly conserved Tyr98 residue would likely spoil any potential catalytic activity in Peredox. In addition, another concern was that Peredox might perturb the cellular free NADH pool by direct binding and buffering. However, this effect is likely to be negligible, as cytosolic NADH is probably buffered by the endogenous protein-bound pool, constituting ~95% of the total cytosolic NADH (Zhang et al., 2002). With a total cytosolic NADH concentration of 3 μM, biosensor expression (at 1–10 μM; Akerboom et al., 2009) is expected to change cellular NADH buffering capacity by roughly 4-fold or less.
To normalize Peredox measurements, a second FP mCherry was attached in tandem. In primary cultured cells, we found expression of RFP such as mCherry led to lysosomal aggregates as previously reported (Katayama et al., 2008). As these puncta preclude reliable comparison of the green-to-red fluorescence ratios, we instead used a nuclear targeted version of Peredox-mCherry. Possible alternative approaches to avoid puncta include the use of another FP such as mCitrine (Griesbeck et al., 2001) for normalization, or fluorescence lifetime imaging to obtain self-normalized measurements without using a second FP (Tantama et al., 2011).
Being genetically encoded, Peredox may be targeted to various cell types and compartments to monitor cellular and subcellular variations in NADH metabolism. However, Peredox with its current affinity would not be expected to work well in mitochondria, as mitochondrial NADH:NAD+ ratio has been estimated to be 100- to 1000-fold higher than cytosolic NADH:NAD+ ratio (Williamson et al., 1967). Consistent with this estimate, when we targeted Peredox to mitochondrial matrix, we did not observe any signal change upon metabolic challenges. Nevertheless, it may be possible to re-engineer its NADH binding sites for mitochondrial redox sensing.
Glycolysis opposes the lactate dehydrogenase equilibrium to produce a reduced cytosolic NADH-NAD+ redox state in cultured Neuro2a and C6 cancer cells. From a biological standpoint, the measured value of the cytosolic NADH-NAD+ redox is surprisingly reduced, corresponding to an NAD+:NADH ratio of <150, compared with a typical range of 200–700 from tissue estimates under a range of conditions (Williamson et al., 1967). Qualitatively, this is not unexpected: cancer cells are known to undergo aerobic glycolysis (Warburg, 1956), whereby most of the glucose is metabolized into lactate, and the production of lactate from pyruvate is presumably driven by elevated NADH:NAD+ ratios. However, at these extremely reduced redox values, it seems surprising that the GADPH reaction can proceed forward to establish net glycolysis and a net production of NADH (Cerdán et al., 2006). The reduced redox level is dependent on the presence of glucose, arguing against net gluconeogenic flux. Perhaps the net forward flux through glycolysis is driven by rapid consumption of intermediate metabolites via pathways downstream of the GAPDH reaction, in accordance with the high biosynthetic requirements of cancer cells (Vander Heiden et al., 2009). In addition, considering that the GADPH reaction has been proposed to be coupled to the subsequent step, the 3-phosphoglycerate kinase (PGK) reaction (Veech et al., 1979), future imaging experiments of both cytosolic NADH:NAD+ ratio and ATP:ADP ratio (Berg et al., 2009) could yield insights on how these parameters and the potential coupling between these two reactions regulate the state of glycolysis.
We found a difference in cytosolic NADH-NAD+ redox between primary cultures of cortical astrocytes and neurons, measured under identical conditions of extracellular glucose, lactate, and pyruvate. This redox difference, as observed in separately cultured cells, is consistent with the proposed redox gradient between astrocytes and neurons (Cerdán et al., 2006). More generally, this redox difference is consistent with the hypothesis that glucose metabolism in the brain is compartmentalized, with different cell types performing distinct metabolic functions. While cytosolic NADH-NAD+ redox states in cultured cells are likely to be different from in vivo, Peredox, being genetically encoded, should in the future allow us to monitor NADH metabolism in astrocytes and neurons more physiologically, such as in brain slice preparations and in vivo.
By expressing Peredox stably in a cultured cell line, we could monitor changes in cytosolic NADH-NAD+ redox states in hundreds of individual cells upon perturbation of PI3K pathway signaling. We demonstrated that Peredox can be used in a high-content imaging format to obtain kinetically detailed metabolic data from individual cells. In the future, Peredox should facilitate studies on how NADH-NAD+ redox metabolism is regulated by growth factor signaling pathways in live cells, as well as how individual cell responses correlate with the status of other signaling systems. As a reporter of cellular NADH-NAD+ redox state, Peredox should open the door to a better understanding of such potentially elaborate organization of metabolism and bioenergetics, as well as the consequences of such metabolic regulation for cellular signaling and physiology.
EXPERIMENTAL PROCEDURES
Gene Construction and Protein Characterization
Gene construction and characterization of the sensor are described in Supplemental Information. Sequences and plasmids for expression of Peredox have been deposited with Addgene (pending).
Cell Culture, Transfection, and Transduction
Cells were maintained in 95% air and 5% CO2 at 37°C. Mouse neuroblastoma Neuro-2a cells (CCL-131, American Type Culture Collection) were cultured in Minimal Essential Medium (MEM; all media and serum from Invitrogen) with 10% fetal bovine serum (FBS), 24 mM NaHCO3, 2 mM HEPES, and 1 mM sodium pyruvate. Rat glioma C6 cells (CCL-107, ATCC) were cultured in Dulbecco’s modified Eagle’s medium–nutrient mixture Ham’s F-12 (DMEM/F-12) with 2.5% FBS and 14 mM NaHCO3, pH 7.1 (NaOH). Neuro-2a and C6 cells were plated onto protamine coated coverslips 4–8 hr prior to transfection using Effectene (Qiagen). The next day, Neuro-2a and C6 cells were rinsed with phosphate buffered saline and maintained in MEM with 1% FBS, 24 mM NaHCO3, 2 mM HEPES, 1 mM sodium pyruvate, and 100 units/ml penicillin with 100 μg/ml streptomycin (pen-strep; Lonza). For Neuro-2a differentiation, 10–20 μM retinoic acid was added. Primary dissociated mouse cortical astrocytes (P0–P2) and neurons (E16–E18) were prepared as described (Bartlett and Banker, 1984) and seeded onto poly-D-lysine coated plates in MEM supplemented with 10% horse serum, 33 mM glucose, 2 mM glutamine (Lonza), 1 mM sodium pyruvate, and pen-strep for 3 hr. Then, primary astrocytes were maintained in DMEM/F-12 with 10% FBS, 24 mM NaHCO3, and pen-strep, and they were passed twice before transfection using Effectene on DIV (days in vitro) 21. Primary neurons were maintained in Neurobasal medium with B27 serum free supplements, 2 mM glutamine, and pen-strep before transfection using calcium phosphate on DIV 4. For stable expression of Peredox-NLS, mammary epithelial MCF-10A cells (CRL-10317, ATCC) were transduced using the retroviral pMSCV vector followed by puromycin selection. MCF-10A cells were cultured in DMEM/F-12 with 5% horse serum, 20 ng/ml epidermal growth factor (EGF), 0.5 μg/ml hydrocortisone, 100 ng/ml cholera toxin, 10 μg/ml insulin, and pen-strep, as described (Debnath et al., 2003).
Confocal and Widefield Time-lapse Microscopy
Cells were imaged 2–5 days after transient transfection. The extracellular solution contained (in mM): 121.5 NaCl, 25 NaHCO3, 2.5 KCl, 2 CaCl2, 1.25 NaH2PO4, and 1 MgCl2, bubbled with 95% air and 5% CO2, delivered at 2–3 ml/min at 33–35°C. Glucose, lactate, and pyruvate were supplemented as indicated. Primary astrocytes and neurons were in the initial condition for 40–60 min prior to image acquisition. Confocal images were acquired on an inverted Nikon Eclipse TE300 microscope mounted with an Andor Revolution Differential Spinning Disk (DSD) unit for optical sectioning controlled by iQ software. Excitation light from Prior Lumen Pro passed through Semrock 405/10 nm or 578/16 nm filters. The DSD unit contained a Chroma 59022bs dichroic with Omega 490 nm and 590 nm short pass filters. Emission light passed through a Chroma 59022m filter with Semrock 525/50 nm or 629/56 nm filters. Using the high sectioning mode, confocal images were acquired with ~4.5 μm thickness with 2 s exposure and 1×1 binning, using a Nikon 20×/0.75 Plan Apo objective. Nuclei were stained with InVitrogen SYBR-safe. Widefield images were acquired with a PCO Sensicam QE CCD camera mounted on an Olympus BX51 upright microscope, with a Zeiss 20×/0.5 Achroplan or an Olympus 60×/0.9 LUMPlanFl/IR objective. A TILL Photonics Polychrome IV monochromator with a 12.5 nm slit width was used with a Chroma 69002x exciter, a 69002bs dichroic, and a dual band Semrock FF01-524/628-25 emitter. Green and red images were acquired at 405 nm and 575 nm excitation every 20–30 s with 3–40 ms exposure and 4×4 binning. Using TILLvisION v4.0.1 image software or ImageJ, we subtracted background, set threshold to avoid ratioing artifacts, and generated a pixel-by-pixel green to red ratio image for each time point. Fluorescence response of each cell in 10 mM lactate was defined as 2.5, and data was normalized accordingly. For stable MCF-10A experiments, ~500 cells were plated onto the center of each well of a 24-well plate 2–3 days prior. Prior to image acquisition, cells were in phenol red-free RPMI supplemented with 5% BSA, EGF, hydrocortisone, cholera toxin, and insulin, at 37°C and 5% CO2 for 40–60 min. Cells were then treated with BEZ235 (1 μM) or the vehicle control DMSO and calibrated with lactate, pyruvate, and iodoacetate as indicated. Widefield images were acquired using a Nikon inverted Eclipse Ti microscope, equipped with a Nikon 20×/0.75 Plan Apo objective and an environmental chamber. Green fluorescence was obtained with a Chroma ET405/20x exciter, a T425LPXR dichroic, and an ET525/50m emitter; red fluorescence, a HQ575/50x exciter, a Q610LP dichroic, and a HQ640/50m emitter. Images were taken every 2 min with 100 ms exposure and 2×2 binning. Using a custom built MATLAB program (Jones et al., 2008), we subtracted background, set threshold and cell segmentation, and generated a green to red ratio for each cell for each time point. Data were analyzed using MATLAB (MathWorks) and Origin 6.0 (MicroCal).
Supplementary Material
Highlights.
Peredox reports the cytosolic NADH-NAD+ redox state in individual live cells.
Glycolysis opposes the LDH equilibrium in controlling cytosolic NADH-NAD+ redox.
Peredox is pH-resistant and works robustly in a high-content imaging format.
Acknowledgments
We thank T. Abramson for expert technical assistance; D. Clapham and B. Sabatini for providing GFP constructs; J. Flanagan for providing Neuro-2a; J. Berg, J. Brugge, L. Cantley, D. Clapham, J. Cohen, N. Danial, T. Schwarz, M. Vander Heiden, and members of the Yellen lab for their comments. This work was supported by the US National Institutes of Health (R01 NS055031 to G.Y.), an NIH postdoctoral fellowship (F32 NS066613 to M.T.), the U.S. Army Breast Cancer Research Program multidisciplinary postdoctoral fellowship (W81XWH-08-1-0609 to J.G.A.), as well as an Albert J. Ryan fellowship and a Stuart H. Q. and Victoria Quan predoctoral fellowship in neurobiology (to Y.P.H.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akerboom J, Rivera JDV, Guilbe MMR, Malavé ECA, Hernandez HH, Tian L, Hires SA, Marvin JS, Looger LL, Schreiter ER. Crystal structures of the GCaMP calcium sensor reveal the mechanism of fluorescence signal change and aid rational design. J Biol Chem. 2009;284:6455–6464. doi: 10.1074/jbc.M807657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avi-Dor Y, Olson JM, Doherty MD, Kaplan NO. Fluorescence of pyridine nucleotides in mitochondria. J Biol Chem. 1962;237:2377–2383. [Google Scholar]
- Baird GS, Zacharias DA, Tsien RY. Circular permutation and receptor insertion within green fluorescent proteins. Proc Natl Acad Sci U S A. 1999;96:11241–11246. doi: 10.1073/pnas.96.20.11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett WP, Banker GA. An electron microscopic study of the development of axons and dendrites by hippocampal neurons in culture. I. Cells which develop without intercellular contacts. J Neurosci. 1984;4:1944–1953. doi: 10.1523/JNEUROSCI.04-08-01944.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bücher T, Brauser B, Conze A, Klein F, Langguth O, Sies H. State of oxidation-reduction and state of binding in the cytosolic NADH-system as disclosed by equilibration with extracellular lactate-pyruvate in hemoglobin-free perfused rat liver. Eur J Biochem. 1972;27:301–317. doi: 10.1111/j.1432-1033.1972.tb01840.x. [DOI] [PubMed] [Google Scholar]
- Belousov VV, Fradkov AF, Lukyanov KA, Staroverov DB, Shakhbazov KS, Terskikh AV, Lukyanov S. Genetically encoded fluorescent indicator for intracellular hydrogen peroxide. Nat Methods. 2006;3:281–286. doi: 10.1038/nmeth866. [DOI] [PubMed] [Google Scholar]
- Berg J, Hung YP, Yellen G. A genetically encoded fluorescent reporter of ATP:ADP ratio. Nat Methods. 2009;6:161–166. doi: 10.1038/nmeth.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brekasis D, Paget MSB. A novel sensor of NADH/NAD+ redox poise in Streptomyces coelicolor A3(2) EMBO J. 2003;22:4856–4865. doi: 10.1093/emboj/cdg453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerdán S, Rodrigues TB, Sierra A, Benito M, Fonseca LL, Fonseca CP, García-Martín ML. The redox switch/redox coupling hypothesis. Neurochem Int. 2006;48:523–530. doi: 10.1016/j.neuint.2005.12.036. [DOI] [PubMed] [Google Scholar]
- Chance B, Cohen P, Jobsis F, Schoener B. Intracellular oxidation-reduction states in vivo. Science. 1962;137:499–508. doi: 10.1126/science.137.3529.499. [DOI] [PubMed] [Google Scholar]
- Chen S, Whetstine JR, Ghosh S, Hanover JA, Gali RR, Grosu P, Shi Y. The conserved NAD(H)-dependent corepressor CTBP-1 regulates Caenorhabditis elegans life span. Proc Natl Acad Sci U S A. 2009;106:1496–1501. doi: 10.1073/pnas.0802674106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Dumollard R, Ward Z, Carroll J, Duchen MR. Regulation of redox metabolism in the mouse oocyte and embryo. Development. 2007;134:455–465. doi: 10.1242/dev.02744. [DOI] [PubMed] [Google Scholar]
- Eto K, Tsubamoto Y, Terauchi Y, Sugiyama T, Kishimoto T, Takahashi N, Yamauchi N, Kubota N, Murayama S, Aizawa T, Akanuma Y, Aizawa S, Kasai H, Yazaki Y, Kadowaki T. Role of NADH shuttle system in glucose-induced activation of mitochondrial metabolism and insulin secretion. Science. 1999;283:981–985. doi: 10.1126/science.283.5404.981. [DOI] [PubMed] [Google Scholar]
- Garriga-Canut M, Schoenike B, Qazi R, Bergendahl K, Daley TJ, Pfender RM, Morrison JF, Ockuly J, Stafstrom C, Sutula T, Roopra A. 2-Deoxy-D-glucose reduces epilepsy progression by NRSF-CtBP-dependent metabolic regulation of chromatin structure. Nat Neurosci. 2006;9:1382–1387. doi: 10.1038/nn1791. [DOI] [PubMed] [Google Scholar]
- Gordon GRJ, Choi HB, Rungta RL, Ellis-Davies GCR, MacVicar BA. Brain metabolism dictates the polarity of astrocyte control over arterioles. Nature. 2008;456:745–749. doi: 10.1038/nature07525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griesbeck O, Baird GS, Campbell RE, Zacharias DA, Tsien RY. Reducing the environmental sensitivity of yellow fluorescent proteins. Mechanism and applications. J Biol Chem. 2001;276:29188–29194. doi: 10.1074/jbc.M102815200. [DOI] [PubMed] [Google Scholar]
- Herrero-Mendez A, Almeida A, Fernández E, Maestre C, Moncada S, Bolaños JP. The bioenergetic and antioxidant status of neurons is controlled by continuous degradation of a key glycolytic enzyme by APC/C-Cdh1. Nat Cell Biol. 2009;11:747–752. doi: 10.1038/ncb1881. [DOI] [PubMed] [Google Scholar]
- Jones TR, Kang IH, Wheeler DB, Lindquist RA, Papallo A, Sabatini DM, Golland P, Carpenter AE. CellProfiler Analyst: data exploration and analysis software for complex image-based screens. BMC Bioinformatics. 2008;15:482. doi: 10.1186/1471-2105-9-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jörnvall H, Persson B, Krook M, Atrian S, Gonzàlez-Duarte R, Jeffery J, Ghosh D. Short-chain dehydrogenases/reductases (SDR) Biochemistry. 1995;34:6003–6013. doi: 10.1021/bi00018a001. [DOI] [PubMed] [Google Scholar]
- Kasischke KA, Vishwasrao HD, Fisher PJ, Zipfel WR, Webb WW. Neural activity triggers neuronal oxidative metabolism followed by astrocytic glycolysis. Science. 2004;305:99–103. doi: 10.1126/science.1096485. [DOI] [PubMed] [Google Scholar]
- Katayama H, Yamamoto A, Mizushima N, Yoshimori T, Miyawaki A. GFP-like proteins stably accumulate in lysosomes. Cell Struct Funct. 2008;33:1–12. doi: 10.1247/csf.07011. [DOI] [PubMed] [Google Scholar]
- Klingenberg M, Bücher T. Biological oxidations. Annu Rev Biochem. 1960;29:669–708. doi: 10.1146/annurev.bi.29.070160.003321. [DOI] [PubMed] [Google Scholar]
- Kumar V, Carlson JE, Ohgi KA, Edwards TA, Rose DW, Escalante CR, Rosenfeld MG, Aggarwal AK. Transcription corepressor CtBP is an NAD+-regulated dehydrogenase. Mol Cell. 2002;10:857–869. doi: 10.1016/s1097-2765(02)00650-0. [DOI] [PubMed] [Google Scholar]
- Lowry OH, Roberts NR, Kapphahn JI. The fluorometric measurement of pyridine nucleotides. J Biol Chem. 1957;224:1047–1064. [PubMed] [Google Scholar]
- Maira SM, Stauffer F, Brueggen J, Furet P, Schnell C, Fritsch C, Brachmann S, Chène P, De Pover A, Schoemaker K, Fabbro D, Gabriel D, Simonen M, Murphy L, Finan P, Sellers W, García-Echeverría C. Identification and characterization of NVP-BEZ235, a new orally available dual phosphatidylinositol 3-kinase/mammalian target of rapamycin inhibitor with potent in vivo antitumor activity. Mol Cancer Ther. 2008;7:1851–1863. doi: 10.1158/1535-7163.MCT-08-0017. [DOI] [PubMed] [Google Scholar]
- Malaisse WJ, Sener A. Glucose-induced changes in cytosolic ATP content in pancreatic islets. Biochim Biophys Acta. 1987;927:190–195. doi: 10.1016/0167-4889(87)90134-0. [DOI] [PubMed] [Google Scholar]
- McLaughlin KJ, Strain-Damerell CM, Xie K, Brekasis D, Soares AS, Paget MSB, Kielkopf CL. Structural basis for NADH/NAD+ redox sensing by a Rex family repressor. Mol Cell. 2010;38:563–575. doi: 10.1016/j.molcel.2010.05.006. [DOI] [PubMed] [Google Scholar]
- Nagai T, Sawano A, Park ES, Miyawaki A. Circularly permuted green fluorescent proteins engineered to sense Ca2+ Proc Natl Acad Sci U S A. 2001;98:3197–3202. doi: 10.1073/pnas.051636098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nausch LWM, Ledoux J, Bonev AD, Nelson MT, Dostmann WR. Differential patterning of cGMP in vascular smooth muscle cells revealed by single GFP-linked biosensors. Proc Natl Acad Sci U S A. 2008;105:365–370. doi: 10.1073/pnas.0710387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogikubo S, Nakabayashi T, Adachi T, Islam MS, Yoshizawa T, Kinjo M, Ohta N. Intracellular pH sensing using autofluorescence lifetime microscopy. J Phys Chem B. 2011 doi: 10.1021/jp2058904. [DOI] [PubMed] [Google Scholar]
- Patterson GH, Knobel SM, Arkhammar P, Thastrup O, Piston DW. Separation of the glucose-stimulated cytoplasmic and mitochondrial NAD(P)H responses in pancreatic islet beta cells. Proc Natl Acad Sci U S A. 2000;97:5203–5207. doi: 10.1073/pnas.090098797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellerin L, Magistretti PJ. Glutamate uptake into astrocytes stimulates aerobic glycolysis: a mechanism coupling neuronal activity to glucose utilization. Proc Natl Acad Sci U S A. 1994;91:10625–10629. doi: 10.1073/pnas.91.22.10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocheleau JV, Head WS, Piston DW. Quantitative NAD(P)H/flavoprotein autofluorescence imaging reveals metabolic mechanisms of pancreatic islet pyruvate response. J Biol Chem. 2004;279:31780–31787. doi: 10.1074/jbc.M314005200. [DOI] [PubMed] [Google Scholar]
- Schwenke WD, Soboll S, Seitz HJ, Sies H. Mitochondrial and cytosolic ATP/ADP ratios in rat liver in vivo. Biochem J. 1981;200:405–408. doi: 10.1042/bj2000405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Peterson TR, Sabatini DM. Regulation of the mTOR complex 1 pathway by nutrients, growth factors, and stress. Mol Cell. 2010;40:310–322. doi: 10.1016/j.molcel.2010.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BNG, Palmer AE, Tsien RY. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Shuttleworth CW, Brennan AM, Connor JA. NAD(P)H fluorescence imaging of postsynaptic neuronal activation in murine hippocampal slices. J Neurosci. 2003;23:3196–3208. doi: 10.1523/JNEUROSCI.23-08-03196.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sickmier EA, Brekasis D, Paranawithana S, Bonanno JB, Paget MSB, Burley SK, Kielkopf CL. X-ray structure of a Rex-family repressor/NADH complex insights into the mechanism of redox sensing. Structure. 2005;13:43–54. doi: 10.1016/j.str.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Tantama M, Hung YP, Yellen G. Imaging intracellular pH in live cells with a genetically encoded red fluorescent protein sensor. J Am Chem Soc. 2011;133:10034–10037. doi: 10.1021/ja202902d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MGV, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veech RL, Lawson JW, Cornell NW, Krebs HA. Cytosolic phosphorylation potential. J Biol Chem. 1979;254:6538–6547. [PubMed] [Google Scholar]
- Wang E, Bauer MC, Rogstam A, Linse S, Logan DT, von Wachenfeldt C. Structure and functional properties of the Bacillus subtilis transcriptional repressor Rex. Mol Microbiol. 2008;69:466–478. doi: 10.1111/j.1365-2958.2008.06295.x. [DOI] [PubMed] [Google Scholar]
- Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- Williamson DH, Lund P, Krebs HA. The redox state of free nicotinamide-adenine dinucleotide in the cytoplasm and mitochondria of rat liver. Biochem J. 1967;103:514–527. doi: 10.1042/bj1030514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata-Hommer O, Griesbeck O. Efficiently folding and circularly permuted variants of the Sapphire mutant of GFP. BMC Biotechnol. 2003;3:5. doi: 10.1186/1472-6750-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Piston DW, Goodman RH. Regulation of corepressor function by nuclear NADH. Science. 2002;295:1895–1897. doi: 10.1126/science.1069300. [DOI] [PubMed] [Google Scholar]
- Zhang Q, Wang SY, Nottke AC, Rocheleau JV, Piston DW, Goodman RH. Redox sensor CtBP mediates hypoxia-induced tumor cell migration. Proc Natl Acad Sci U S A. 2006;103:9029–9033. doi: 10.1073/pnas.0603269103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.