SUMMARY
The signaling adapter p62 is a critical mediator of important cellular functions owing to its ability to establish interactions with various signaling intermediaries. Here we identify raptor as an interacting partner of p62. Thus, p62 is an integral part of the mTORC1 complex, and is necessary to mediate amino acid signaling for the activation of S6K1 and 4EBP1. p62 interacts in an amino acid-dependent manner with mTOR and raptor. In addition, p62 binds the Rags proteins and favors formation of the active Rag heterodimer that is further stabilized by raptor. Interestingly, p62 colocalizes with Rags at the lysosomal compartment and is required for the interaction of mTOR with Rag GTPases in vivo, and for translocation of the mTORC1 complex to the lysosome, a crucial step for mTOR activation.
INTRODUCTION
The adapter protein p62 (also known as sequestosome 1) is a signaling hub initially identified as a partner of the atypical PKCs (aPKCs; PKCζ and PKCλ/ι) (Sanchez et al., 1998), interacting through the PB1-domain. This multidomain platform interacts selectively with different signaling proteins to regulate multiple cellular functions including cell survival, inflammation, apoptosis and autophagy (Moscat and Diaz-Meco, 2009a; Moscat et al., 2006). The cellular location of the signaling event also contributes to its specificity and plasticity in the modulation of cell functions.
Immunostaining of p62 reveals a clear, punctate pattern consistent with p62 being localized into cytosolic speckles, or aggregates, formed of PB1-driven p62 oligomers and p62-aPKC complexes, as well as polyubiquitin-conjugated proteins (Jin et al., 2009; Moscat et al., 2006; Pankiv et al., 2007; Sanz et al., 2000). We have previously shown that p62 colocalizes with Rab-7, suggesting that it might play a role in receptor trafficking to the lysosomal compartment (Sanchez et al., 1998). It has also been determined that these speckles are signal-organizing centers where p62 could catalyze the formation of higher-order complexes that favor the mechanism of action of different signaling molecules such as TRAF6 (Sanz et al., 2000), or caspase-8 (Jin et al., 2009), to modulate the survival/apoptosis decision point. However, the factors that determine which complex is formed at a given time, and within a specific cell context, remain to be identified.
In an attempt to identify novel components integrating the p62 signaling hub, we have initiated a proteomics approach. Here we demonstrate that raptor interacts with p62, which uncovers an unanticipated role for p62 in the mTOR pathway. Raptor is part of mTORC1 (Kim et al., 2002), one of the two multiprotein complexes, which also include mTORC2, in which the mTOR signaling network is organized (Guertin and Sabatini, 2007). mTORC1 is a major driver of cell growth, and is commonly deregulated in cancer (Sabatini, 2006). Upstream signals that trigger this complex include growth factors, insulin, hypoxia, intracellular energy levels, and amino acid availability (Sarbassov et al., 2005). Recent results have started to shed light on the mechanism whereby amino acids activate mTORC1. That is, the Rag GTPases have been shown to interact with mTORC1 and to be amino acid-specific regulators of this cascade through the translocation of mTORC1 to a lysosomal compartment (Kim et al., 2008; Sancak et al., 2010; Sancak et al., 2008), indicating that a signaling pathway essential for cell growth and survival could be regulated through the selective compartmentalization of its components inside the cell. Of note, we show here that p62 is another essential piece of the mTORC1 complex through its interaction with raptor and the Rags proteins. Importantly, we also show that p62 is required for mTORC1 compartmentalization and activation through regulation of its recruitment to the lysosome. This has important implications for the tumorigenic role of p62.
RESULTS
Identification of raptor as a p62-interacting protein
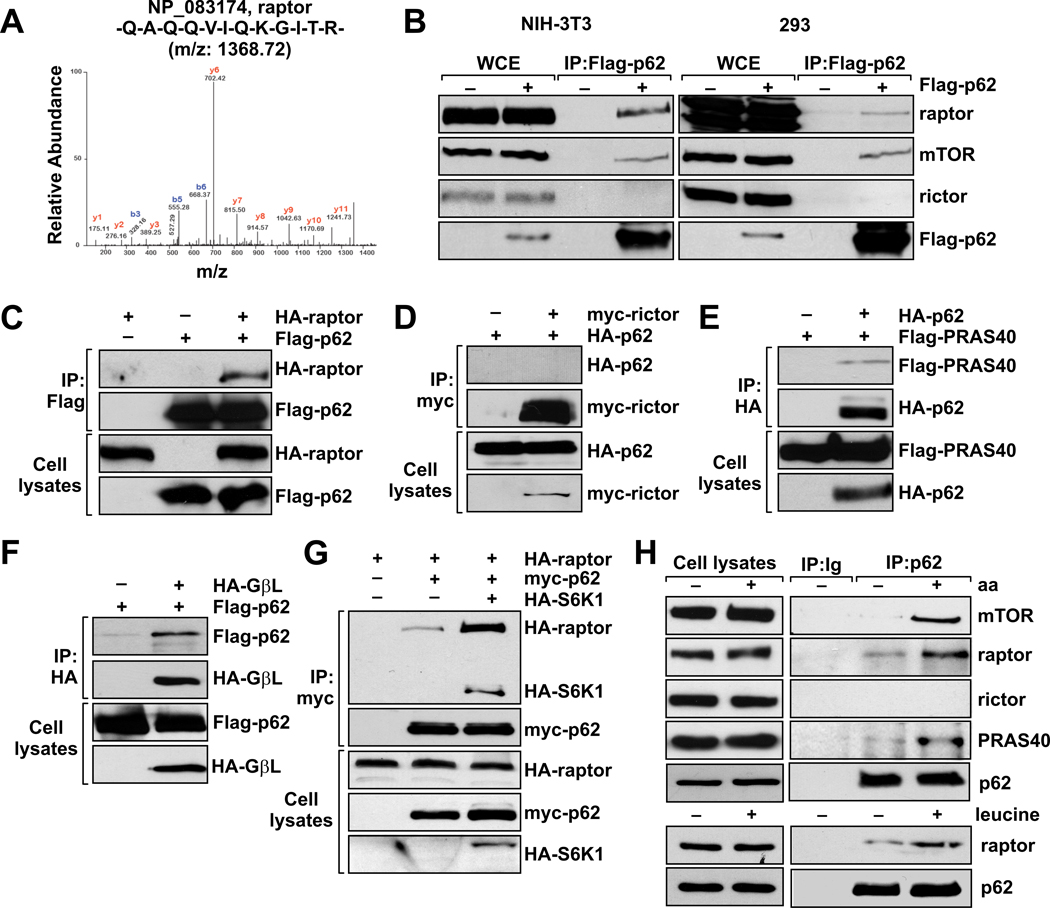
To identify novel partners of p62, we generated NIH-3T3 cells stably expressing Flag-tagged p62. Flag-bound immunoprecipitates from these cells were subjected to LC/MS/MS analysis, which led to the identification of raptor as a protein associated with p62 (Fig. 1A). To further validate the p62-raptor interaction, we asked whether endogenous raptor is associated with p62 immunoprecipitated from NIH-3T3 Flag-p62 extracts. Fig. 1B (left panel) shows a reproducible interaction between the two proteins. This interaction was also detected in the human cell line HEK-293 (Fig. 1B, right panel). Because raptor interacts with mTOR (Kim et al., 2002), we next tested whether p62 could be part of the mTOR complex or if the p62-raptor complex constitutes a different scaffold platform. Fig. 1B shows that mTOR was also recovered in the p62-bound immunoprecipitates, suggesting an unexpected link between p62 and the mTOR pathways.
Figure 1. p62 interacts with Raptor and components of the mTORC1 complex, but not of mTORC2.
(A) LC/MS/MS spectra of the purified Flag-p62 peptides corresponding to raptor from NIH-3T3 cells stably expressing Flag-p62. (B) Lysates and Flag immunoprecipitates from NIH-3T3 and 293 cells stably expressing Flag-p62 were analyzed to detect the indicated proteins. In (C) through (G) 293 cells were transfected with the indicated cDNAs in expression vectors, cell lysates were prepared, and lysates and HA-, Flag-, or myc-tagged immunoprecipitates were analyzed by immunoblotting to quantify the specified ectopic proteins. (H) Endogenous interaction of p62 with components of the mTOR signaling pathway in response to amino acids or leucine. Cell lysates and p62 immunoprecipitates from amino acid- or leucine-treated HEK293T cells were analyzed for the levels of specified proteins. These results are representative of three experiments.
mTOR exists in two distinct multiprotein complexes referred to as mTOR complex 1 (mTORC1) and 2 (mTORC2) (Guertin and Sabatini, 2007). Both complexes contain mTOR, but mLTS8/GbL, raptor and PRAS40 are specific components of mTORC1. The proteins rictor, mSin1 and protor are specific to mTORC2 (Guertin and Sabatini, 2007). To determine whether p62 is a specific component of mTORC1, and not of mTORC2, we looked for rictor in p62 immunoprecipitates. As shown in Fig. 1B, we did not detect any interaction between p62 and rictor, indicating that p62 selectively associates with mTORC1 and not mTORC2. To further test the specificity of p62’s association with mTORC1, we co-expressed epitope-tagged p62 with epitope-tagged raptor, PRAS40, GβL, and rictor, and analyzed their interactions. Figs. 1C–F show that p62 co-immunoprecipitated with all the mTORC1 components, but not those of mTORC2. Of note, when p62 was co-transfected with raptor and S6K1, a ternary complex containing these proteins was also detected, and the p62-raptor interaction was further stabilized in the presence of S6K1, most probably to favor an active complex (Fig. 1G). These results identify p62 as a component of mTORC1 and suggest that it might be critical to the mechanism of action of this pathway.
We next sought to identify the region(s) of p62 responsible for the interaction with raptor. For this, we expressed full-length HA-tagged raptor in combination with several myc-tagged fragments of p62 in HEK293 cells. The C-terminal region of p62 (amino acids 230–440) containing the UBA domain did not interact with raptor, whereas the N-terminal region (amino acids 1–266) interacted as strongly as the full-length protein (Fig. S1A). Deletion of the UBA domain from the full-length protein did not affect the interaction (Fig. S1B), in agreement with the lack of binding activity of the C-terminal portion of p62 (Fig. S1A). As the N-terminal region of p62 contains the PB1 domain, the ZZ-type zinc finger domain, and part of the TRAF6-binding domain (TB), we expressed different mutants of these interacting modules with raptor, and determined their contribution to the association between the two proteins. Deletion of the TB domain did not impair the interaction with raptor (Fig. S1B). In addition, a point mutation in the PB1 domain (K7A), which disrupts the interaction between p62 and other PB1-containing proteins, such as the aPKCs or NBR1, as well as the homodimerization of p62 (Moscat et al., 2006), did not affect this interaction (Fig. S1C). Also, a mutant expressing only the PB1 domain was not able to bind raptor (Fig. S1C), consistent with the idea that the PB1 domain is not involved in the p62-raptor interaction. Likewise, deletion of the ZZ domain had no effect on binding (Fig. S1D), suggesting that none of the known domains are responsible for this interaction. To further map the region that is required for binding, we shortened the N-terminal region further. Fragments encompassing amino acids 62–266 and 115–230 were sufficient to interact with raptor (Fig. S1E). The shortest fragment, 115–230, contains the ZZ domain (122–167), and this domain is not required for interaction with raptor (Fig. S1E). Thus, the region between the ZZ and the TB domains (167–230) most probably harbors the sequence responsible for the association between p62 and raptor (Fig. S1F).
p62 is required for the amino acid response
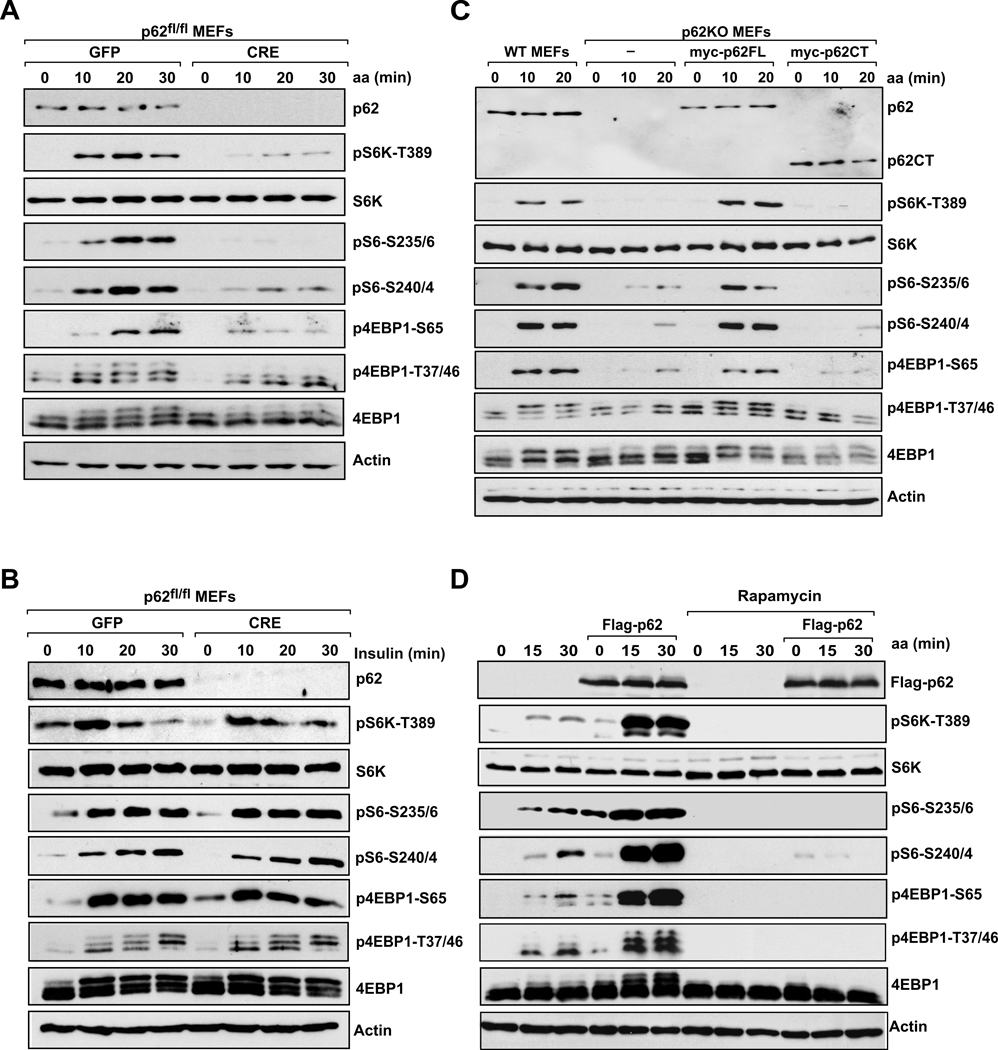
mTORC1, a central regulator of cell growth and protein synthesis, acts by channeling upstream signals, generated by growth factors and amino acids, towards downstream targets (Hay and Sonenberg, 2004). Therefore, we next determined whether the interaction between p62 and mTORC1 could be detected in endogenous proteins and whether or not a well-established activator of the pathway, such as amino acids, could modulate this endogenous association. Fig. 1H shows that endogenous p62 interacts with endogenous mTOR, raptor, and PRAS40, but not with rictor, and that this interaction is promoted by the addition of amino acids. The branched-chain amino acid, leucine, which stimulates mTORC1, is also sufficient to induce the formation of this endogenous complex (Fig. 1H). To investigate the physiological role of the p62-mTORC1 interaction in vivo, we next asked whether p62 is required for phosphorylation of the downstream targets S6K and 4EBP1 (Hay and Sonenberg, 2004) in response to insulin and amino acid stimulation. To do that, p62-floxed mouse embryo fibroblasts (p62fl/fl MEFs) were infected with Cre or GFP control adenoviruses. Fig. 2A shows that this strategy effectively led to the complete loss of p62 in these cells. Interestingly, whereas p62 deletion had no effect on insulin-activated S6K1, measured by the phosphorylation of S6K1 or its substrate S6 (Fig. 2B), p62 was selectively required for sensing nutrient signaling to S6 kinase activation (Fig. 2A). No effect on AKT activation in response to insulin was observed in p62-deleted cells (Fig. S2A), in agreement with the specific interaction of p62 with mTORC1 but not with mTORC2 (Fig. 1B). Interestingly, phosphorylation of 4EBP1, the other well-characterized translational regulator controlled by mTORC1 (Hsieh et al., 2010), was also reduced in p62-deficient cells in response to amino acids (Fig. 2A). To rule out any potential toxic effect of Cre infection, we repeated these experiments in MEFs from constitutive p62 KO mice. As shown in Fig. 2C, amino acid-induced phosphorylation of S6K1 and 4EBP1 was also dramatically impaired in p62 KO MEFs as compared with WT controls. Interestingly, reconstitution of these p62 KO cells with myc-tagged full-length p62 restored mTORC1 activation (Fig. 2C), whereas a p62CT mutant that does not bind raptor (Fig. S1F), failed to do so (Fig. 2C). In addition, p62 was also required for leucine-induced phosphorylation of S61 and 4EBP1 (Fig. S2B). These results suggest that p62 is a key regulator of mTORC1 signaling in vivo, and places p62 specifically in the nutrient-sensing pathway through its interaction with raptor. To check the relevance of these findings to human cells, we selectively knocked down p62 by lentiviral infection with shRNA in HEK293 cells and measured S6K1 and 4EBP1. Consistent with the MEF results, reduction in the levels of endogenous p62 by two different shRNAs decreased nutrient signaling to both S6K1 and 4EBP1 in human cells as compared with cells infected with a non-targeted control shRNA (shNT) (Fig. S2C). These data suggest that activation of the mTORC1 pathway in response to amino acids depends on endogenous p62 function, in both mouse and human cells.
Figure 2. p62 is essential for mTORC1 activation in response to amino acids.
Effects of p62 depletion on activation of the mTORC1 pathway in response to (A) amino acids or (B) insulin. p62fl/fl MEFs were infected with GFP or CRE adenoviruses, and then stimulated with amino acids or insulin. Lysates were analyzed for levels of the specified proteins. (C) WT and p62KO MEFs, reconstituted with p62FL or p62CT, were treated as described in (A). Cell lysates were prepared and analyzed by immunoblotting for the levels of the indicated proteins. (D) 293 cells stably expressing Flag-p62 were deprived of amino acids and serum for 50 min, pretreated with rapamycin for 30 min, and then stimulated with amino acids for the indicated durations. Cell lysates were analyzed for the levels of the specified proteins. These results are representative of three experiments.
We next determined whether the simple overexpression of p62 was sufficient to promote mTORC1 activation. Stable expression of p62 had only a minor effect on S6K1 and 4EBP1 phosphorylation in the absence of amino acids; however, p62 cooperated with amino acid stimulation to sustain a higher activation of the mTORC1 pathway (Fig. 2D). Of note, rapamycin treatment completely blocked p62-enhanced S6K1 and 4EBP1 phosphorylation, consistent with p62 functioning upstream of mTOR (Fig. 2D). These results are consistent with p62 being an integral component of nutrient signaling, and indicate that overexpression of p62 may favor a more efficient activation of the pathway.
The p62-interacting proteins, the aPKCs and NBR1, are dispensable for nutrient sensing
Because p62 itself is a scaffold protein that binds important signaling intermediaries through different domains (Moscat and Diaz-Meco, 2009a), we next asked whether any of these partners of p62 were functionally required to activate mTORC1. We focused on the role of the PB1-domain interacting proteins, the aPKCs (PKCζ and PKCλ) and NBR1 (Kirkin et al., 2009a; Moscat et al., 2006). As this domain is not required for the interaction with raptor, through its PB1, p62 might bring these other intermediaries to the mTORC1 complex. Therefore, to investigate the potential involvement of PKCζ, PKCλ and NBR1, we used knockout cells for each of these p62-interacting proteins and determined whether or not they were required for activation of the mTORC1 pathway in response to amino acid stimulation. Interestingly, unlike p62, none of these p62 partners were required for activation of this pathway (Fig. S2D–F). This indicates that p62’s function in nutrient sensing is independent of its interaction with the aPKCs or NBR1, and points to the possibility that different specialized p62-containing complexes might have different functions.
p62 influences cell size and autophagy in response to nutrients
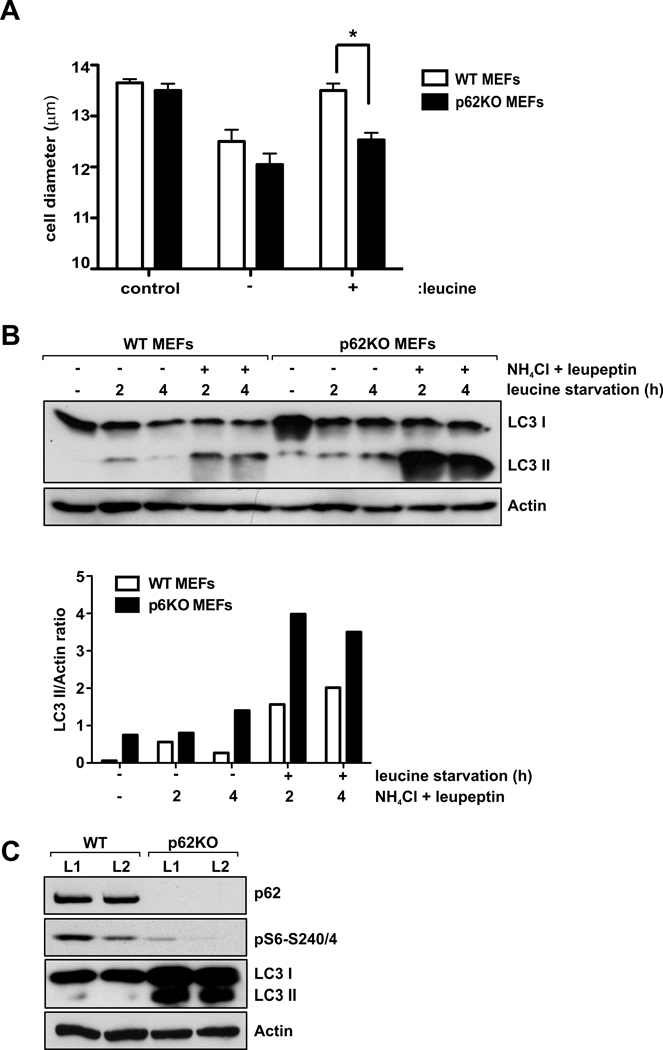
A well-known function of the mTORC1 signaling cascade is the positive regulation of cell size and subsequent impact on cell growth (Fingar et al., 2002; Hay and Sonenberg, 2004). Consistent with p62 being a positive regulator of mTORC1, we found that p62 depletion prevented leucine-induced increase in cell size (Fig. 3A). Inhibition of mTOR signaling by withdrawing nutrients or by adding rapamycin activates autophagy (Sabatini, 2006). We hypothesized that p62 deletion would also activate autophagy in response to starvation. As shown in Fig. 3B, leucine deprivation in MEFs induced autophagy as measured by accumulation of the conjugated form of LC3 (LC3-II), a commonly used method for detecting autophagic activity as LC3-II associates with the pre-autophagosomal and autophagosomal membranes (Mizushima et al., 2010). Interestingly, p62 KO cells had higher basal levels of LC3-II, compatible with an increase in autophagic function. This was confirmed by enhanced autophagic flow in the presence of the lysosomal inhibitors ammonium chloride and leupeptin in p62 KO cells during leucine withdrawal (Fig. 3B). This is also consistent with the increased accumulation of LC3-II in the livers of p62 KO mice in response to fasting, concomitant with a reduction in S6K1 activation (Fig. 3C). These results suggest that p62 could regulate autophagy in response to nutrient starvation, most probably through the inhibition of mTORC1. Interestingly, we observed that RNAi inhibition of a p62 ortholog in the nematode C. elegans led to an increase in LC3-positive puncta (Fig. S3), indicating that p62 reduction, like TOR inactivation (Hansen et al., 2008), can increase autophagy, and that such a link may be conserved. Collectively, these results reinforce the idea that p62 has a primary and functionally relevant role in the mTORC1 pathway.
Figure 3. p62 regulates cell size and autophagy.
(A) Cell size was determined in WT and p62KO MEFs deprived of leucine for 24 h and re-stimulated with leucine for 24 h. Error bars specify the standard deviation between three independently performed experiments. *p<0.05. (B) WT and p62KO MEFs were deprived of leucine and serum for 2 and 4 hours in the absence or presence NH4Cl and leupeptin. Cell lysates were analyzed for the levels of the specified proteins. Graphs represent the densitometric LC3-II/actin ratios. (C) Protein extracts from livers of 24h fasted WT and p62KO mice were analyzed for the levels of the specified proteins. These results are representative of three experiments.
p62 interacts with the Rag proteins
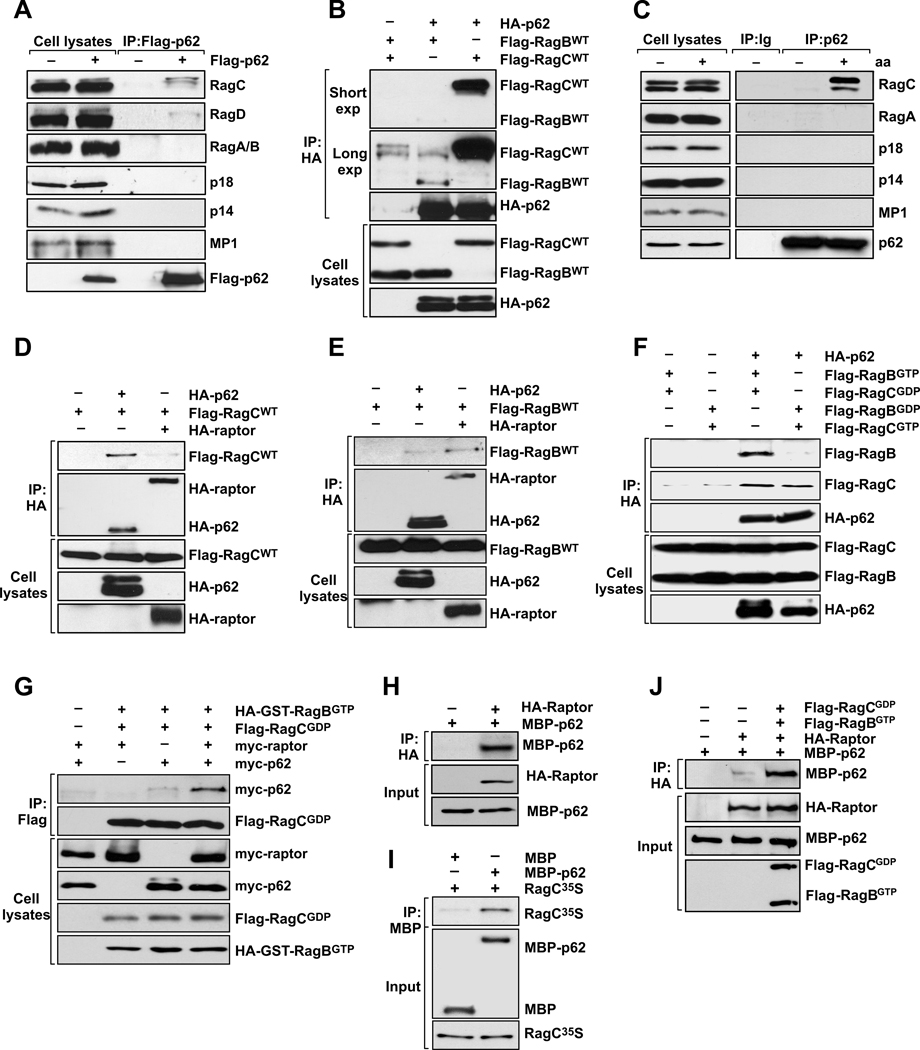
The evidence suggesting a critical role for p62 in amino acid signaling to mTORC1 raises the question of where p62 might function in this pathway. As p62 is required for the amino acid response, and because the Rag GTPases have recently been identified as key mediators in this process (Kim et al., 2008; Sancak et al., 2010; Sancak et al., 2008), we asked whether p62 interacted with the Rag proteins. We used stably expressing Flag-p62 cells to assess interactions between p62 and the Rag proteins. In mammals, there are four Rag proteins (RagA, RagB, RagC, and RagD) that form heterodimers consisting of RagA or RagB with RagC or RagD. RagA and RagB, like RagC and RagD, are highly similar and functionally redundant (Hirose et al., 1998; Schurmann et al., 1995). Therefore, we used antibodies for RagC, Rag D, and RagA/B for immunoblotting to analyze the association of p62 with endogenous Rag GTPases. Interestingly, we detected a specific interaction of p62 with RagC and RagD (Fig. 4A), but no interaction with endogenous RagA/B (Fig. 4A). To further test this interaction, we expressed p62 with RagB or RagC in HEK293 cells. p62 interacted with both RagC and RagB (Fig. 4B), although, in keeping with the results of Fig. 4A, p62 interacted more strongly with RagC than with RagB (Fig. 4B). Of note, the endogenous interaction of p62 with RagC was also promoted in response to amino acids (Fig. 4C). Since the Rag proteins have been shown to interact with the Ragulator complex to act as a lysosomal docking for the mTORC1 complex (Sancak et al., 2010), we next asked whether p62 was able to interact with the Ragulator components, MP1, p14, and p18. We did not find any of the Ragulator proteins associated with Flag-p62 immunoprecipitates (Fig. 4A) or in the endogenous p62 complex (Fig. 4C) indicating that p62, in conjunction with the Rag proteins, could be an alternative docking element for mTORC1.
Figure 4. Amino acid-induced regulation of the p62-Rag complex.
(A) Cell lysates and Flag-p62 immunoprecipitates from NIH-3T3 cells stably expressing Flag-p62 were analyzed by immunoblotting to determine levels of the specified proteins. (B) p62 preferentially binds to RagC. 293 cells were transfected with the indicated cDNAs in expression vectors, cell lysates were prepared, and lysates and HA- or Flag-tagged immunoprecipitates were analyzed by immunoblotting to determine amounts of the specified ectopic proteins. (C) Endogenous interaction of p62 with Rags and components of Ragulator in response to amino acids. Cell lysates and p62 immunoprecipitates from amino acid-treated 293 cells were analyzed for the levels of specified proteins. In (D) to (G), 293 cells were transfected with the indicated cDNAs in expression vectors, cell lysates were prepared, and lysates and HA- or Flag-tagged immunoprecipitates were analyzed by immunoblotting to determine amounts of the specified ectopic proteins. (H) Recombinant MBP-p62 was incubated with HA-raptor purified from HEK293T cells, and the ability of raptor to bind directly to p62 was determined in pulldown experiments employing HA beads. (I) In vitro translated RagC was incubated with MBP or MBP-p62 and its ability to bind directly to p62 was determined in pulldown experiments employing amylose beads. (J) Recombinant MBP-p62 was incubated with HA-raptor purified from HEK293T cells, in the absence or presence of RagBGTP or RagCGDP, and the ability of raptor to bind directly to p62 was determined in pulldown experiments employing HA beads. These results are representative of three experiments.
As the Rag proteins also bind raptor (Kim et al., 2008; Sancak et al., 2008), to better understand the topology of these interactions, we next tested whether RagC and RagB had any binding preference for p62 or raptor. Figs. 4D and E show that RagC binds more robustly to p62 than to raptor, whereas RagB shows a preference for raptor. Furthermore, p62, like raptor, interacted preferentially with the active heterodimer (RagBGTP/RagCGDP), and not with the inactive one (RagBGDP/RagCGTP) (Fig. 4F). This interaction with the active heterodimer was further enhanced by the presence of raptor (Fig. 4G). Furthermore, p62 binds directly with both raptor (Fig. 4H) and RagC (Fig. 4I) in vitro, and the presence of the active dimer increases in vitro raptor-p62 binding (Fig. 4J).
p62 regulates formation of the active Rag heterodimer
As the Rags function as heterodimers (Sekiguchi et al., 2001), and it is known that p62 is able to homodimerize (Moscat et al., 2006), it is possible that p62 might favor the formation of the RagB/C heterodimer, which could be further stabilized by raptor. In fact, results in Fig. 5A show that p62 increases formation of the active heterodimer between RagBGTP and RagCGDP. In addition, Rag proteins could also be important for p62-raptor interaction. To test this, we used specific lentiviral shRNAs for RagC and RagD to knock down Rag GTPases. As previously shown (Sancak et al., 2008), loss of RagC and RagD also caused loss of RagA and RagB, as the Rag proteins were unstable when not in the heterodimer form (Fig. 5B). Amino acid stimulation induced association of p62 with raptor in cells treated with non-targeted lentiviral shRNA (NT) and this interaction was abolished in cells lacking Rag proteins (Fig. 5B). Collectively, these results link p62 to the Rag GTPases as members of the nutrient-sensing pathway and suggest that p62 might function in this pathway by helping to stabilize the active heterodimer in association with raptor.
Figure 5. p62 promotes formation of the active Rag heterodimer.
(A) p62 stabilizes the Rag heterodimer. HEK293T cells were transfected with the indicated cDNAs in expression vectors, cell lysates were prepared, and lysates and Flag-tagged immunoprecipitates were analyzed by immunoblotting to detect the specified ectopic proteins. The graph shows a quantification of the RagB/C dimer formation. Results are represented as means ± standard deviations of three different experiments. *p<0.05. (B) p62 immunoprecipitates from 293 cells, infected with lentiviral shRNA for RagC or RagD, or non-targeting (NT) shRNA, were analyzed by immunoblotting to determine levels of the specified proteins. (C) Cell lysates and RagC immunoprecipitates from WT and p62 KO immortal MEFs reconstituted with p62FL or p62CT were analyzed by immunoblotting to determine levels of specified proteins. (D) Effects of p62 on GTP loading of RagB. The percentage of GTP bound to RagB is indicated for each sample. A portion of cell lysate was taken before immunoprecipitation to probe for expression. (E) p62fl/fl MEFs, infected with GFP or CRE adenovirus, were transfected with Flag-RagBGTP and RagCGDP. Cells were starved for 4 h and restimulated with amino acids for 10 min. Cell lysates were analyzed by immunoblotting to determine levels of the specified proteins. These results are representative of three experiments.
Based on these results linking p62 to the Rags, and on the fact that interaction of these GTPases with mTOR is a critical step in the activation of the mTORC1 pathway, we next asked whether p62 was necessary for the association between mTOR and the Rags. Interestingly, Fig. 5C shows that p62 is required for the mTOR-RagC interaction, which was abolished in p62 KO MEFs. In addition, the formation of the RagC/RagA heterodimer was also impaired in p62 KO cells (Fig. 5C). Importantly, both interactions were restored by reintroduction of full-length p62 but not the p62CT mutant (Fig. 5C). The Rag-Ragulator complex was not affected by the loss of p62 (data not shown). These results suggest that p62 might be controlling mTOR’s association with the Rags by regulating formation of the active Rag heterodimer, as this is the form that preferentially binds mTOR. Because the GTP-loading state of the Rags is a critical factor in activation of the heterodimer (Kim et al., 2008; Sancak et al., 2008), and therefore for mTOR binding, we determined whether p62 could modulate the GTP loading of the Rag proteins as part of the mechanism of promoting mTORC1 activation. As shown in Fig. 5D, p62 overexpression was in fact able to modulate the amount of GTP bound to RagB, thus mimicking the effect of amino acid stimulation. This was dependent on Rag binding as the p62CT mutant was not able to promote this effect (Fig. 5D). Collectively, these results placed p62 as an important mediator of mTORC1 activation in nutrient sensing that acts by linking mTORC1 to the Rag proteins by modulating Rag heterodimer formation and its GTP-dependent activation. This is consistent with a model in which p62 acts upstream of the Rags. Indeed, Fig. 5E demonstrates that the expression of a constitutively active Rag dimer in p62 KO cells is able to rescue impaired mTORC1 activation in response to amino acids.
p62 colocalizes with mTORC1 and is required for mTOR recruitment to lysosomes
Activation of the mTORC1 pathway by amino acids has been shown to provoke the translocation of mTORC1 from an unidentified vesicular compartment to a membrane-bound compartment containing Rab7, a marker of both late endosomes and lysosomes (Sancak et al., 2008). More recent data show that the precise location of mTORC1 is at the lysosomal surface, characterized by LAMP2-positive staining (Sancak et al., 2010). We have previously shown that p62 localizes with Rab7, displaying a similar punctate staining pattern (Sanchez et al., 1998). Therefore, it is plausible that p62 colocalizes with mTORC1 in the same membrane compartment. To test this possibility, we asked whether p62 was colocalized with the two main mTORC1 components, mTOR and raptor. HEK293 cells were cotransfected with HA-p62 along with either myc-TOR or myc-raptor. Staining with the corresponding anti-tag antibodies showed colocalization of p62 with both mTOR (Fig. S4A) and raptor (Fig. S4B). In addition, p62 was also colocalized with RagC (Fig. S4C). Consistent with this, p62 deletion mutants that did not interact with raptor (Fig. S1A) or RagC (Fig. S4D) did not colocalize with RagC (Fig. S4C). Since p62 interacts with the Rag proteins, which are required for the recruitment of mTORC1 to the lysosome (Sancak et al., 2010; Sancak et al., 2008), we asked whether p62 impinges on the amino acid signaling cascade by regulating mTORC1 localization. First, we used confocal immunofluorescence to determine whether p62 was colocalized with LAMP2. Double staining of endogenous p62 and LAMP2 revealed colocalization of the two proteins (Fig. 6A and S5A), which was independent of nutrient availability, as the same colocalization was observed with or without amino acid stimulation (Fig. 6A). These results are reminiscent of the lysosomal localization of the Rag GTPases, which is independent of amino acid availability (Sancak et al., 2010), and prompted us to test whether p62 colocalizes with the Rag GTPases in this lysosomal compartment. Interestingly, endogenous p62 colocalized with endogenous RagC in an amino acid-independent manner (Fig. 6B and S5A). These results demonstrate that p62 and the Rag GTPases interact and form a signaling complex in the same cellular compartment.
Figure 6. p62 regulates mTOR translocation to the lysosomal compartment.
Starved or restimulated 293 cells were co-immunostained for p62 and (A) LAMP2, (B) RagC, (C) mTOR, or (D) raptor. (E) p62fl/fl MEFs infected with GFP or CRE adenovirus and overexpressing activated RagB were starved for 4 h and then restimulated with amino acids for 10 min. Afterwards, they were co-immunostained for mTOR and LAMP2. (F) Images of WT and p62 KO immortal MEFs co-immunostained for RagC and LAMP2. In all images, inserts show selected fields that were magnified two or five times to show areas of staining overlap (Merge). Scale bars=10 µm. Images are representative of three experiments.
We next examined whether endogenous p62 was also colocalized with the main components of mTORC1. mTOR and raptor displayed the characteristic punctate staining and amino acid addition promoted their translocation to the well-characterized lysosomal compartment (Fig. 6C–D and S5A). Of note, colocalization of p62 with both mTOR and raptor was highly increased upon amino acid stimulation (Fig. 6C–D and S5A). These results are in keeping with p62 being an integral part of the mTORC1 multiprotein complex in vivo. Because the translocation of mTOR to LAMP2-positive membranes is a critical step for the activation of the mTORC1 pathway in the amino acid response, and because p62 is required for the full activation of this cascade, it seemed possible that p62 might be necessary for mTOR recruitment to the lysosome. Remarkably, upon amino acid stimulation of p62 KO cells, mTOR retained a diffuse punctate staining pattern and its colocalization with LAMP2 was severely diminished, as compared with that in WT cells (Fig. 6E and S5B). Of note, expression of a constitutively active mutant of RagB was able to rescue mTOR localization to the lysosomes in p62 KO cells (Fig. 6E). Furthermore, RagC localization to the lysosomes was also altered in these KO cells (Fig. 6F). This is in keeping with the finding that p62 acts upstream of the Rags and is required for Rag activation and interaction with mTOR (Fig. 5). In addition, reconstitution of p62 KO cells with full-length myc-p62 but not with the p62CT mutant rescued mTOR recruitment to the lysosomes (Fig. S5B). Collectively, these results indicate that p62 is a required functional docking site for mTORC1 activation to signal nutrient sensing.
p62 drives tumorigenesis through mTORC1 activation
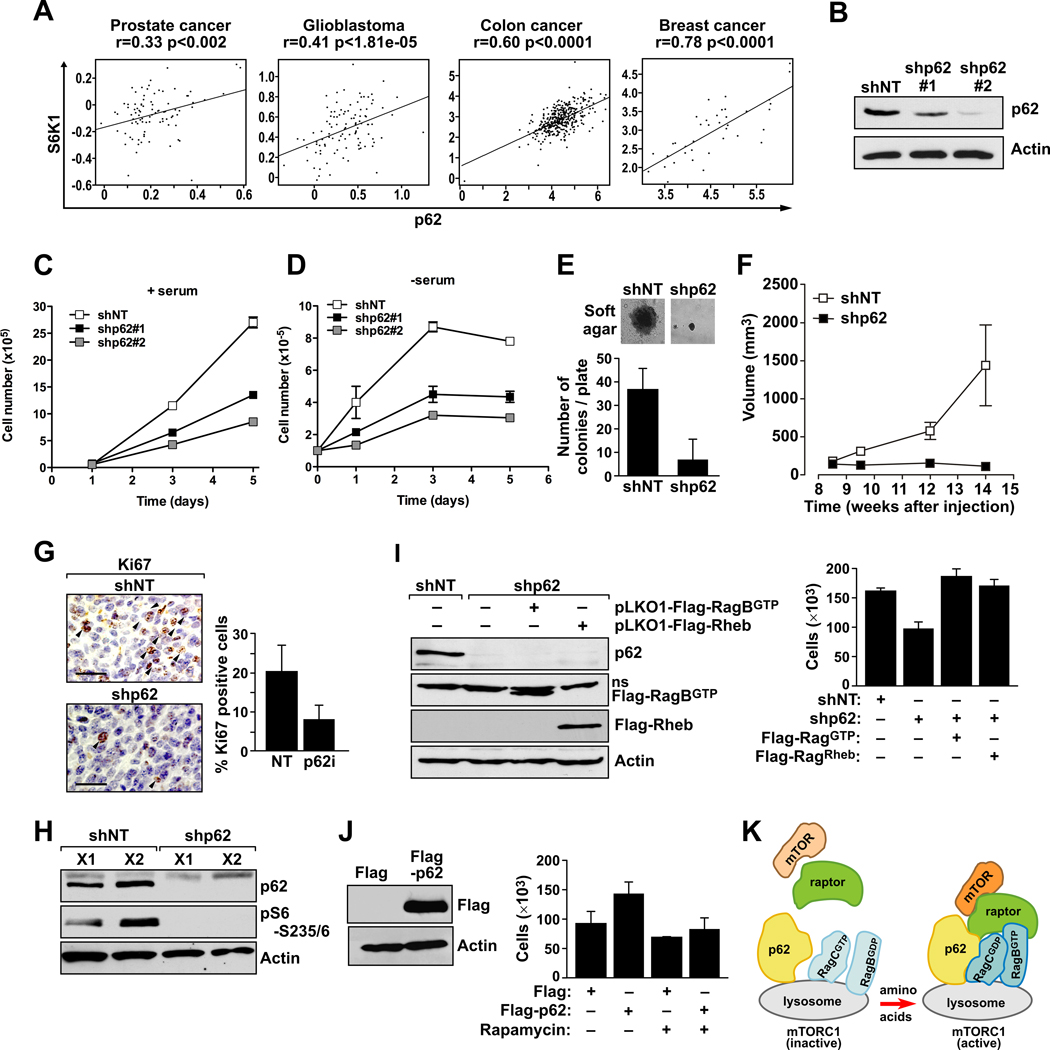
mTORC1 is a central integrator of upstream signals to regulate protein synthesis and cell growth, and it is increasingly apparent that it is commonly deregulated in cancer (Guertin and Sabatini, 2007). On the other hand, p62 is an important signaling hub frequently overexpressed in human tumors (Duran et al., 2008; Moscat et al., 2009). Here, we have identified p62 as a critical component of the mTORC1 pathway, which prompted us to investigate whether there is any correlation between the expression of p62 and mTORC1 in human tumors. To do this, we examined the levels of p62 and S6K1 (as a downstream kinase of mTORC1) mRNA in databases with transcriptional profiles of human tumors (Rhodes et al., 2004; Sun et al., 2006; Yu et al., 2004). Interestingly, p62 levels significantly correlated with S6K1 in multiple human cancers, including glioblastoma and prostate, breast, and colon cancer (Fig. 7A), suggesting that the p62-mTORC1 cassette could be a relevant target in cancer treatment.
Figure 7. p62 drives tumorigenesis through activation of the mTORC1 pathway.
(A) Positive correlation between p62 and S6K1 mRNA levels in human cancers. (B) Cap2 cells were infected with lentiviral shRNAs including non-targeting shRNA (shNT), and two different lentiviral shRNAs for p62 (shp62#1 and shp62#2). After selection with puromycin, levels of p62 were analyzed by immunoblot. (C) Growth curve of shNT and shp62 cells under normal growing conditions or (D) in the absence of serum. Results are shown as means ± standard deviations. (E) Soft-agar growth of shNT and shp62 Cap2 cells, and quantification of the number of colonies at 21 days. The experiment shown in panel E is representative of another two with similar results. The graph in panel E shows means ± standard deviations. (F) Suspensions of shNT and shp62 Cap2 cells, as described in (B), were intradermally injected into each flank of nude mice, and tumors were allowed to develop for 15 days. Tumor size was measured twice per week. Results are means ± standard deviations (n = 5). (G) Immunohistochemical analysis of Ki67 expression in shNT and shp62 Cap2 cell xenografts (left). Graph shows the quantitation of Ki67-positive cells in these tumors (right). Results are the means ± standard deviations of counts from 10 different fields per mouse, with a total of 5 mice per condition. (H) Cell lysates from these tumors (X1 and X2) were analyzed by immunoblotting to determine amounts of the specified proteins. (I) Stably expressing Flag-RagBGTP or Flag-Rheb prostate cancer cells were infected with shNT or shp62 lentiviral vectors. Cell lysates were analyzed by immunoblotting to determine levels of specified proteins (left). Cell viability was determined at 3 days after serum withdrawal (right). Results are shown as means ± standard deviations. (J) Stably expressing Flag-p62 cells were analyzed by immunoblotting to determine levels of the specified proteins (left). Cell viability was determined at 3 days after serum withdrawal (right) in the presence or absence of rapamycin. Results are shown as means ± standard deviations. (K) Model for the role of p62 in amino acid signaling towards mTORC1 activation.
To determine if p62 is in fact a key component in tumorigenesis through the mTORC1 pathway, we used prostate cancer cells grown in vitro and as xenografted tumors in mice. We first tested whether knocking down p62 in the CaP2 prostate cancer cell line (a PTEN-null murine prostate epithelial cell line) impaired their ability to form tumors when injected into nude mice. shRNAs targeting p62 (shp62) or a control shRNA (shNT) were delivered by lentivirus and stably expressed in CaP2 cells by selection with puromycin. As shown in Fig. 7B, p62 shRNAs depleted p62 levels with different efficiencies. Interestingly, knockdown of p62 in these cells reduced proliferation in the presence or absence of serum (Fig. 7C and D) to a degree consistent with the level of p62 depletion. Of note, the capacity of CaP2 to form colonies in soft agar, a marker of cell transformation, was also impaired in p62 knock-down cells (Fig. 7E). These results support the notion that p62 is important for the full proliferative and tumorigenic properties of cancer cells. To determine whether p62 depletion affected the ability of CaP2 cells to form solid tumors in vivo, we injected CaP2 cells transduced with shNT or shp62 subcutaneously into nude mice and monitored tumor formation for 12 weeks (Fig. 7F). Importantly, CaP2 cells expressing shNT formed tumors that were visibly larger than the tumors formed by CaP2 cells expressing shp62 (Fig. 7F). This requirement for p62 was also observed with regard to tumor formation by xenografted shp62-treated TRAMP-C1 cells (Fig. S6A–B). Immunohistochemical analysis (IHC) of these samples revealed a marked reduction in cell proliferation, as measured by Ki67, in the small tumors of the shp62 cells as compared with the control group (Fig. 7G). Interestingly, this reduction in proliferation highly correlated with the complete inhibition of S6K1 activation as measured both by western blot (Fig. 7H) and IHC (Fig. S6C), suggesting that mTORC1 pathway could be an important mediator of p62 actions in cancer cells. To further test the functional contribution of mTORC1 activation, we tested whether the expression of a constitutively active mutant of RagB or the overexpression of Rheb, a well-known activator of mTORC1 (Laplante and Sabatini, 2009), was sufficient to rescue the cell proliferation defect observed in shp62 cells. Fig. 7I shows that, in fact, both activated RagB and Rheb were able to rescue cell growth, indicating that inhibition of the mTORC1 pathway is responsible for the reduction in cell proliferation induced by p62 loss in cancer cells. Furthermore, overexpression of p62 in these cells enhanced proliferation, and this proliferative advantage in p62 overexpressers was blocked by treatment with rapamycin (Fig. 7J). These results suggest that p62 is a critical component of the mTORC1 complex in cancer cells, as it is in normal cells, which has important implications for the deregulation of the mTORC1 cascade in cancer and its potential targeting during cancer therapeutics.
DISCUSSION
Specificity and crosstalk are two important aspects in understanding how cell signaling in the context of complex networks generates precise cellular responses to a myriad of different stimuli. Key to this process are protein scaffolds, which are multidomain proteins that assemble specific signaling complexes in different cellular locations to assure a spatially and temporally controlled signal (Zeke et al., 2009). p62 is an adapter involved in the regulation of intracellular signaling and is implicated in critical physiological and pathological settings such as the activation of NF- B and metabolic control (Moscat and Diaz-Meco, 2009a,b). Through its multidomain structure, p62 acts as a hub in cell signaling (Moscat and Diaz-Meco, 2009a). For example, in the NF-κB signaling pathway, p62 controls osteoclastogenesis by interacting with the E3 ubiquitin ligase TRAF6, which is also critical for T-cell differentiation and tumor progression (Duran et al., 2004; Martin et al., 2006; Duran et al., 2008). On the other hand, p62 controls adipogenesis and obesity via interaction with ERK1, and apoptosis through binding to caspase-8 (Rodriguez et al., 2006; Lee et al., 2010; Jin et al., 2009). In addition, p62 interacts with ubiquitin through the UBA domain, and is involved in autophagy via interaction with LC3 (Pankiv et al., 2007; Kirkin et al., 2009b).
The identification of raptor as a p62-associated protein uncovered an unexpected link between p62 and the mTOR pathway. mTOR is itself a scaffold protein that exists in two main complexes, mTORC1 and mTORC2, which channel different signals within the cell (Sabatini, 2006). Here we show that p62 is specifically part of mTORC1, and not mTORC2, through its interaction with raptor. Interestingly, p62 is selectively required for mTORC1 activation in response to amino acids but not to other stimuli such as insulin. Thus, p62 is required for S6K and 4EBP1 phosphorylation upon amino acid stimulation, which is consistent with its ability to promote the association of downstream kinases, such as S6K1, with mTORC1. Thus, as a scaffold, p62 helps to propagate the signal by stabilizing the raptor complex with downstream substrates. This function of p62 in nutrient sensing seems to be independent of other p62-interacting proteins, as the deletion of any of the aPKCs (PKCζ or PKCλ/ι) or of NBR1, both PB1-mediated p62 interactors (Kirkin et al., 2009a; Moscat et al., 2006), had no effect on mTORC1 activation in response to amino acids. These results suggest that p62 might be integrating different complexes to fulfill specific functions, although we cannot rule out that the aPKCs or NBR1, or both, could be part of the same mTORC1 complex, although with redundant roles.
Another important mechanism to control specificity during cell signaling is to modulate the intracellular location of the signal. That is, signaling adapters can provide a docking platform in a specific cellular compartment to promote efficient propagation of the signal in that particular topographic context. In this regard, recently published results showed that the nutrient-sensing response is coupled to lysosomal targeting of the mTORC1 complex (Sancak et al., 2008; Sancak et al., 2010). The Rag GTPases were recently identified as amino acid-specific regulators of this pathway (Sancak et al., 2008; Kim et al., 2008). In the presence of amino acids, Rag heterodimers containing GTP-bound RagA/B interact with the raptor subunit of mTORC1, and mediate its translocation to the lysosomal surface, in proximity to its activator Rheb (Sancak et al., 2008; Sancak et al., 2010). Here, we show that p62 binds the Rags proteins and raptor, and favors the formation of the active Rag heterodimer, which is further stabilized by raptor (Fig. 7K). Because p62 is able to form oligomers, this could be a mechanism to further tune the activation of the pathway by regulating Rag dimerization. This is also reminiscent of p62’s activation of different pathways by modifying the aggregation of interactors such as TRAF6 or caspase-8 (Sanz et al., 2000; Duran et al., 2008; Jin et al., 2009). In both cases, the promotion of aggregation is mediated by the induction of polyubiquitination (Moscat and Diaz-Meco, 2009a,b). It is not known whether there are additional modifications that regulate Rag dimerization by the p62/Raptor complex, and this question warrants further work to fully unravel this newly identified regulatory step.
Our results show that p62 is part of a docking platform of the mTORC1 complex in the lysosomal compartment. p62 colocalizes with LAMP2 (a lysosomal marker) and the Rags. Remarkably, p62 is required for the interaction of mTOR with the Rag GTPases in vivo, and for the translocation of the mTORC1 complex to the lysosomal surface, a crucial step in mTOR activation. These findings are in keeping with our initial observation that p62 was located in a Rab7-positive late-endosomal compartment (Sanchez et al., 1998), and demonstrates that it is a critical regulator of the trafficking of signaling complexes from their inactive locations to sites where they are maximally activated. These results are also consistent with evidence showing that endosomal trafficking regulates mTORC1 function, and that the full activation of this pathway requires the integrity of the endocytic network. The identification of Ragulator adds complexity to the regulation of mTOR trafficking (Sancak et al., 2010). We have not detected endogenous interactions between p62 and any of the subunits of Ragulator, indicating that p62 is an alternative docking site for the modulation of mTORC1 activation. Thus, different signaling speckles may contain different molecules to allow a more localized and spatially controlled signal.
In conclusion, the identification of raptor as a p62-associated protein sheds light on the link between p62 and the mTOR pathway, specifically in sensing nutrient availability. Therefore, p62 can act to nucleate different signaling molecules to ensure the efficiency and selectivity of the signal transduction process. Given that the components of mTORC1 are commonly deregulated in cancer, the recent identification of p62 as a critical step in this pathway may help in the design of better targeted therapies for the treatment of tumors in which mTOR is altered.
EXPERIMENTAL PROCEDURES
Materials, Mice, Cell lines, and Plasmids
See the Supplemental Experimental Procedures.
Identification of raptor as a p62-interacting protein
Raptor was detected in anti-Flag-p62 immunoprecipitates prepared from HEK-293 cells stably expressing Flag-p62. Proteins eluted from the affinity matrix with the Flag peptide were resolved by SDS-PAGE and silver stained. Gel bands were excised and digested with trypsin overnight. The resulting peptides were analyzed by mass spectrometry in the UC Proteomics Laboratory (see the Supplemental Experimental Procedures).
Amino acid, insulin, and leucine stimulation of cells
Amino acid, insulin, and leucine starvation of HEK-293, HEK-293T, HeLa, and MEF cells were performed and cells were processed for biochemical and immunofluorescence assays as described in Supplemental Experimental Procedures.
Cell lysis and immunoprecipitations
Cell lysate preparation and immunoprecipitations were done as described in the Supplemental Experimental Procedures.
Supplementary Material
ACKNOWLEDGEMENTS
This work was funded by NIH Grants R01CA134530 (M.T.D.-M.), R01CA132847 (J.M.), a Department of Defense Grant DoD-PC080441 (M.T.D.-M.), and an American Federation for Aging Research Research Award (M.H.). We are grateful for the skilled research assistance of Emily Kellner, Lyndsey Bolanos, and Joyce Chu. We thank Maryellen Daston for editing this manuscript and Glenn Doerman for the art work.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures and six figures.
REFERENCES
- 1.Duran A, Linares JF, Galvez AS, Wikenheiser K, Flores JM, Diaz-Meco MT, Moscat J. The signaling adaptor p62 is an important NF-kappaB mediator in tumorigenesis. Cancer Cell. 2008;13:343–354. doi: 10.1016/j.ccr.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Duran A, Serrano M, Leitges M, Flores JM, Picard S, Brown JP, Moscat J, Diaz-Meco MT. The atypical PKC-interacting protein p62 is an important mediator of RANK-activated osteoclastogenesis. Dev Cell. 2004;6:303–309. doi: 10.1016/s1534-5807(03)00403-9. [DOI] [PubMed] [Google Scholar]
- 3.Fingar DC, Salama S, Tsou C, Harlow E, Blenis J. Mammalian cell size is controlled by mTOR and its downstream targets S6K1 and 4EBP1/eIF4E. Genes Dev. 2002;16:1472–1487. doi: 10.1101/gad.995802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guertin DA, Sabatini DM. Defining the role of mTOR in cancer. Cancer Cell. 2007;12:9–22. doi: 10.1016/j.ccr.2007.05.008. [DOI] [PubMed] [Google Scholar]
- 5.Hansen M, Chandra A, Mitic LL, Onken B, Driscoll M, Kenyon C. A role for autophagy in the extension of lifespan by dietary restriction in C. elegans. PLoS genetics. 2008;4:e24. doi: 10.1371/journal.pgen.0040024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev. 2004;18:1926–1945. doi: 10.1101/gad.1212704. [DOI] [PubMed] [Google Scholar]
- 7.Hirose E, Nakashima N, Sekiguchi T, Nishimoto T. RagA is a functional homologue of S. cerevisiae Gtr1p involved in the Ran/Gsp1-GTPase pathway. J Cell Sci. 1998;111(Pt 1):11–21. doi: 10.1242/jcs.111.1.11. [DOI] [PubMed] [Google Scholar]
- 8.Hsieh AC, Costa M, Zollo O, Davis C, Feldman ME, Testa JR, Meyuhas O, Shokat KM, Ruggero D. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer Cell. 2010;17:249–261. doi: 10.1016/j.ccr.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR, Ashkenazi A. Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell. 2009;137:721–735. doi: 10.1016/j.cell.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 10.Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- 11.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–945. doi: 10.1038/ncb1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009a;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 13.Kirkin V, McEwan DG, Novak I, Dikic I. A role for ubiquitin in selective autophagy. Mol Cell. 2009b;34:259–269. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 14.Laplante M, Sabatini DM. mTOR signaling at a glance. J Cell Sci. 2009;122:3589–3594. doi: 10.1242/jcs.051011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee SJ, Pfluger PT, Kim JY, Nogueiras R, Duran A, Pages G, Pouyssegur J, Tschop MH, Diaz-Meco MT, Moscat J. A functional role for the p62-ERK1 axis in the control of energy homeostasis and adipogenesis. EMBO Rep. 2010;11:226–232. doi: 10.1038/embor.2010.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin P, Diaz-Meco MT, Moscat J. The signaling adapter p62 is an important mediator of T helper 2 cell function and allergic airway inflammation. EMBO J. 2006;25:3524–3533. doi: 10.1038/sj.emboj.7601250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moscat J, Diaz-Meco MT. p62 at the crossroads of autophagy, apoptosis, and cancer. Cell. 2009a;137:1001–1004. doi: 10.1016/j.cell.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moscat J, Diaz-Meco MT. To aggregate or not to aggregate? A new role for p62. EMBO Rep. 2009b;10:804. doi: 10.1038/embor.2009.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moscat J, Diaz-Meco MT, Albert A, Campuzano S. Cell signaling and function organized by PB1 domain interactions. Mol Cell. 2006;23:631–640. doi: 10.1016/j.molcel.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 21.Moscat J, Diaz-Meco MT, Wooten MW. Of the atypical PKCs, Par-4 and p62: recent understandings of the biology and pathology of a PB1-dominated complex. Cell Death Differ. 2009;16:1426–1437. doi: 10.1038/cdd.2009.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 23.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rodriguez A, Duran A, Selloum M, Champy MF, Diez-Guerra FJ, Flores JM, Serrano M, Auwerx J, Diaz-Meco MT, Moscat J. Mature-onset obesity and insulin resistance in mice deficient in the signaling adapter p62. Cell Metab. 2006;3:211–222. doi: 10.1016/j.cmet.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 25.Sabatini DM. mTOR and cancer: insights into a complex relationship. Nat Rev Cancer. 2006;6:729–734. doi: 10.1038/nrc1974. [DOI] [PubMed] [Google Scholar]
- 26.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez P, De Carcer G, Sandoval IV, Moscat J, Diaz-Meco MT. Localization of atypical protein kinase C isoforms into lysosome-targeted endosomes through interaction with p62. Mol Cell Biol. 1998;18:3069–3080. doi: 10.1128/mcb.18.5.3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanz L, Diaz-Meco MT, Nakano H, Moscat J. The atypical PKC-interacting protein p62 channels NF-kappaB activation by the IL-1-TRAF6 pathway. EMBO J. 2000;19:1576–1586. doi: 10.1093/emboj/19.7.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 31.Schurmann A, Brauers A, Massmann S, Becker W, Joost HG. Cloning of a novel family of mammalian GTP-binding proteins (RagA, RagBs, RagB1) with remote similarity to the Ras-related GTPases. J Biol Chem. 1995;270:28982–28988. doi: 10.1074/jbc.270.48.28982. [DOI] [PubMed] [Google Scholar]
- 32.Sekiguchi T, Hirose E, Nakashima N, Ii M, Nishimoto T. Novel G proteins, Rag C and Rag D, interact with GTP-binding proteins, Rag A and Rag B. J Biol Chem. 2001;276:7246–7257. doi: 10.1074/jbc.M004389200. [DOI] [PubMed] [Google Scholar]
- 33.Sun L, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, Passaniti A, Menon J, Walling J, Bailey R, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 34.Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, McDonald C, Thomas R, Dhir R, Finkelstein S, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 35.Zeke A, Lukacs M, Lim WA, Remenyi A. Scaffolds: interaction platforms for cellular signalling circuits. Trends Cell Biol. 2009;19:364–374. doi: 10.1016/j.tcb.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.