Abstract
Background/Aims
Statistical geneticists commonly use certain two-locus penetrance models because these models are familiar and mathematically tractable. We investigate whether and under what circumstances these two-locus penetrance models correspond to models of causation.
Methods
We describe a sufficient component cause model for a hypothetical disease with two genetic causes. We then use the potential outcomes framework to determine the expected two-locus penetrances from this causal model and contrast them with commonly used two-locus penetrance models (additive, heterogeneity, and multiplicative penetrance models, as formulated by Risch [Am J Hum Genet 1990;46:222–228]).
Results
Conventional additive and multiplicative models can correspond to any two-locus causal model only when certain very specific algebraic relationships hold. The heterogeneity model corresponds to a two-locus causal model only if the model stipulates that no disease cases are caused by the combined presence of the causal genotypes at both loci (i.e. only when there is no causal gene-gene interaction). Hence the heterogeneity model provides a valid test of the null hypothesis of no gene-gene interaction, whereas the additive and multiplicative models do not.
Conclusion
We suggest that causal principles should provide the basis for statistical modeling in genetics.
Key Words: Causal models, Epidemiology, Genetics, Two-locus models, Penetrance, Additive models, Heterogeneity models, Multiplicative models, Epistasis
‘I see no greater impediment to scientific progress than the prevailing practice of focusing all of our mathematical resources on probabilistic and statistical inferences while leaving causal considerations to the mercy of intuition and good judgment.’
Judea Pearl, Causality, p. xiv [1].
Introduction
Statistical geneticists commonly use certain two-locus penetrance models when investigating the genetic causes of disease because these models are familiar and mathematically tractable. Previous research has investigated the properties [2], uses [3], and interpretation [4,5,6,7,8] of some of these models; however, no previous study has determined whether and under what circumstances they correspond to causal models. We investigate this correspondence in this paper. As described in the first paper of this two-part series [9], the use of causal models promises new insights for applied, theoretical, or methodological statistical genetics research, as it has for other quantitative fields such as economics, social sciences, and epidemiology. Causal models can be used to determine the patterns of disease risk expected from the number and frequency of disease-causing genes, the number and frequency of non-genetic causes, and their causal interactions.
As a motivating example, consider the assumption of a multiplicative penetrance model (described in detail below) to describe the expected two-locus penetrance matrix for a disease when gene-gene interaction underlies the genetic architecture. This model is summarized by Risch [10] and is regularly applied to investigate hypotheses of gene-gene interactions (see e.g. [11,12,13,14]). However, the pattern of disease that would, in fact, result from two genes acting in concert to cause disease has not been established [5,6,7,8]. In analyses using causal models, epidemiologists have found that when two risk factors are parts of the same causal mechanism (i.e. they ‘interact’ in a causal sense), risks in individuals with both factors are predicted to be greater than those predicted by an additive model, but not necessarily consistent with those predicted by a multiplicative model [15,16]. Thus, multiplicative penetrance models might not be consistent with models of causal gene-gene interaction.
Determining the two-locus penetrances resulting from causal models of disease-causing mechanisms such as genetic heterogeneity and gene-gene interaction will inform two important applications of penetrance models: (1) the validity of different penetrance-based models (e.g. multiplicative two-locus penetrance) for inferring the biological mechanisms that lead to disease (e.g. gene-gene interaction), and (2) approaches for simulating the expected joint distribution of genes and disease under an assumed biological mechanism.
In this paper we use Rothman's sufficient component cause (SCC) model [16,17] to describe the causes of a complex genetic disease. Rothman defines a sufficient cause as a set of minimal conditions and events that inevitably produces an outcome. The SCC framework recognizes that any given cause of a disease may be, indeed usually is, neither necessary nor sufficient. If an outcome has more than one sufficient cause, then no single sufficient cause is necessary for the outcome. We postulate a SCC model that includes the causal effects of genotypes at two unlinked loci, and incorporates reduced penetrance, phenocopies, genetic heterogeneity, and gene-gene interaction, as described in our accompanying paper [9]. We then use the potential outcomes (PO) framework [18] to determine the expected two-locus penetrances from this causal model and contrast them with 3 commonly used two-locus penetrance models (additive, heterogeneity, and multiplicative penetrance models described by Risch [10]). Finally, we discuss the results and implications of this comparison.
Definitions and Assumptions
Defining the Causal (SCC) Model
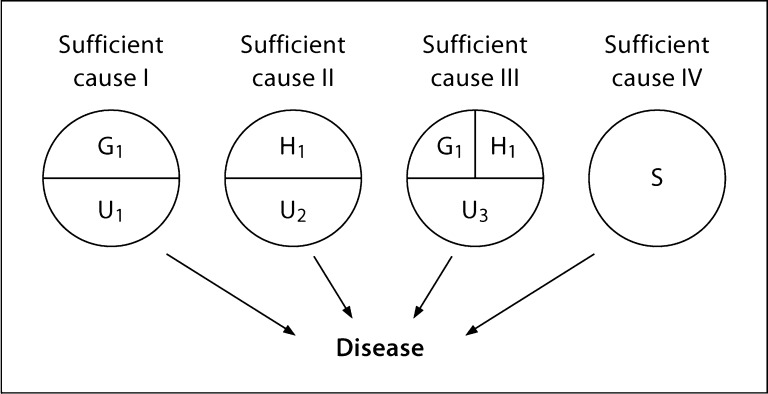
Our assumed SCC model includes two loci, G and H, where Gi and Hj denote the possible genotypes at these loci (fig. 1). We indicate the combinations of causal (a.k.a. susceptibility, predisposing, at-risk) genotypes at the respective loci with i = j = 1 and the combination of non-causal genotypes with i = j = 0. (Because this paper has considerably more mathematical development than [9], we have opted for this notation over the use of G– and H– for indicating i = j = 0 as in the first paper of this series.) Genetic heterogeneity is modeled by assuming that G1 and H1 participate in distinct sufficient causes (sufficient causes I and II in fig. 1), and gene-gene interaction by assuming they both participate in the same sufficient cause (sufficient cause III). As is generally assumed for complex diseases, neither locus alone or in combination is sufficient for disease, i.e. both the single- and two-locus genotypes demonstrate reduced penetrance, because each predisposing single- or two-locus genotype requires a causal partner (U1, U2, or U3) for disease to occur among the causal genotype carriers. We depict another characteristic of complex diseases, ‘phenocopies,’ by specifying sufficient cause IV, consisting of the single component cause S as a cause of disease that does not include genotypes G1 or H1 as component causes. (We have used S to denote this component cause, rather than X which was used in our companion paper [9], to avoid confusion with the notation used later in this paper.) Note that whether or not an individual is a phenocopy depends on the genotype of interest. For example, an individual who develops disease through sufficient cause II would be considered a phenocopy with respect to G1, whereas an individual who develops disease through sufficient cause IV would be considered a phenocopy with respect to both G1 and H1. U1, U2, U3 and S need not be specific entities or factors, but can represent sets of random or deterministic elements. Notably, the SCC model depicted in figure 1 assumes that neither G1 nor H1 has preventive effects on disease (i.e. assumes monotonicity).
Fig. 1.
A SCC model depicting a disease caused by genotypes at two loci, G and H, acting independently (sufficient causes I and II) and together (sufficient cause III). Sufficient cause IV represents the causes of disease that do not involve genotypes at the G, and H loci. G1 denotes the causal (i.e. predisposing) genotype at locus G, and H1 denotes the causal genotype at locus H. U1, U2, and U3 are the ‘causal partners’ required for the respective predisposing genotypes to cause disease (i.e. determinants of penetrance of G1 and H1).
Assumptions
First, we assume that the SCC model accurately describes a disease and its mechanism of causation. This assumption is required in order to use causal models as the basis for determining the expected joint distribution of genotype and disease. Second, we assume that the depictions of genetic heterogeneity and gene-gene interaction in the SCC model in figure 1 correspond to biological reality. This assumption is required to infer that comparison of predictions based on the SCC model depicted in figure 1 with those based on conventional statistical models provide valid information about measurement of underlying biological processes. To address our specific question about the expected two-locus penetrances for a disease caused by two genotypes (that may act together or separately to cause disease) and other genetic or non-genetic causes, we also assume that, taken together, these 4 sufficient causes explain the totality of disease in the population. That is, S represents the totality of sufficient causes of disease that do not include G or H as component causes. The only ways that G and H cause disease are in connection with sufficient causes I, II, and III.
We make additional assumptions that are not required to use SCC models but are often made in statistical genetics: linkage equilibrium between the two causal loci; independent distribution in the population of the component causes in the SCC model (G1, H1, U1, U2, U3, and S); dichotomous genotypes at the susceptibility loci; dichotomous outcomes; and that the component causes depicted in figure 1 only cause disease, i.e. do not prevent disease. We will address violations of the assumptions and extensions of the model in ‘Discussion’.
Penetrances according to the SCC Model
Having posited the SCC model depicted in figure 1, we determine the corresponding probability of disease conditioned on all possible one- or two-locus genotypes specified in the causal model, i.e. the penetrance matrix. Note that although we use a SCC model, we are estimating ‘statistical penetrance’ as defined in the first paper of this series [9] – i.e. the ‘proportion of individuals with disease among those who carry a specific genotype within a defined population.’ Given the SCC model in figure 1, the disease status of a person inheriting a particular genotype (e.g. G1) depends on two things: (1) whether the genotype completes a sufficient cause (e.g. whether U1 is present), and (2) whether any sufficient causes not including the genotype as a component cause have been completed (e.g. whether S is present). According to an SCC model, the probability of disease conditioned on a given genotype depends on the frequency of component causes other than that genotype. We refer to the component causes other than the genotype as ‘other causes.’ The combination of other causes to which an individual is exposed determines his/her disease status given each genotype combination [19].
Using the terminology of Robins and Greenland [18] and the PO framework, we define an individual's ‘response type’ as his/her pattern of disease outcomes, given each possible genotype combination. This illustrates a unique feature of causal models, in that the model is explicitly elaborated in terms of individual disease response rather than in terms of average risk across a population.
With respect to the two-locus penetrance matrix for genotypes at loci G and H, the genotypes of interest are the 4 mutually exclusive and exhaustive combinations of the binary genotypes: G0H0, G1H0, G0H1, and G1H1. In table 1, the Response type column provides a label for each pattern of disease across these 4 possible genotype combinations [18]. The Determinants of response type columnenumerates the possible combinations of ‘other causes’ (from fig. 1) that underlie an individual's response type, and the Probability of response type column gives the probability of each combination (i.e. each response type). The P[disease ∣ response type t, GiHj] column describes the disease outcome for each response type given each two-locus genotype. Note that the probability of disease for any individual conditioned on his/her observed genotype and ‘other causes’ is either 1 or 0.
Table 1.
The t response types, their determinants, P[response type: t], and P[disease | response type t, G;Hj]
| Response type (t) | Determinants of response type |
Probability of response type: rt=V [response type t] | P[disease|response type t, GiHj] |
||||||
|---|---|---|---|---|---|---|---|---|---|
| U1 | U2 | U3 | S | G1H1 | G0H1 | G1H0 | G0H0 | ||
| Immune (1) | absent | absent | absent | absent | r1 = (1 – u1)(1 – u2)(1 – u3)(1 – s) | 0 | 0 | 0 | 0 |
| G-causal (2) | present | absent | present or absent | absent | r2 = (u1)(1 – u2)(1 – s) | 1 | 0 | 1 | 0 |
| H-causal (3) | absent | present | present or absent | absent | r3 = (1 – u1)(u2)(1 – s) | 1 | 1 | 0 | 0 |
| GH-causal (4) | absent | absent | present | absent | r4 = (1 – u1)(1 – u2)(u3)(1 – s) | 1 | 0 | 0 | 0 |
| Parallel (5) | present | present | present or absent | absent | r5 = (u1)(u2)(1 – s) | 1 | 1 | 1 | 0 |
| Inevitable (6) | present or absent | present or absent | present or absent | present | r6 = s | 1 | 1 | 1 | 1 |
| Σ = 1 | 1 | 1 | 1 | 1 | |||||
The ‘Immune’ row consists of individuals who do not have ‘other causes’ U1, U2, U3, or S. Given the SCC model in figure 1, these individuals will not be diseased, regardless of their genotypes at loci G and H. The ‘G-causal’ row consists of individuals who have U1 but not U2 or S. These individuals will have disease when G1 is present and will not have disease when G1 is absent, regardless of their genotype at locus H, and independently of whether they have U3 or not. The ‘H-causal’ types are defined similarly. ‘GH-causal’ types have U3 but not U1, U2, or S and will have disease only when both G1 and H1 are present, but not when either or both are absent. ‘Parallel’ types have U1 and U2 (U3 may be present or absent) but not S and will have disease when G1 or H1 is present, but will not have disease when both are absent. Finally, ‘Inevitable’ individuals have S, and whether or not S occurs in conjunction with U1, U2, or U3, these individuals will have disease for all 4 possible combinations of genotypes at loci G and H. (The type we call ‘Inevitable’ is often called ‘Doomed’ [20]; we have replaced this term to avoid any negative connotation.)
Given our assumption that the component causes, including genotypes, are independently distributed in the population, these 6 response types will be distributed randomly among the different genotypes and vice versa. The probability of disease among individuals conditioned on their two-locus genotype is described by:
| (1) |
From equation 1 and table 1, we derive the probability of disease conditioned on each two-locus genotype. These equations make up the causal model-based two-locus penetrance matrix, where the lower-case u1, u2, u3, and s equal the probability that an individual has component causes U1, U2, U3, and S, respectively:
| (2) |
To illustrate the derivation of these probabilities, consider the quantity A. From table 1, P[disease ∣ response type t, G1H1] = 1 for G-causal, H-causal, GH-causal, Parallel, and Inevitable response types, which occur with probabilities r2, r3, r4, r5 and r6, respectively, whereas P[disease ∣ response type t, G1H1] = 0 for the Immune response type, which occurs with probability r1. That is, individuals with G1 and H1 will get disease if they represent any response type other than Immune. Thus
P[disease ∣ G1H1] = r2 + r3 + r4 + r5 + r6.
But also
so A = P[disease ∣ G1H1] = 1 – r1 = 1 – (1 – u1)(1 – u2) (1 – u3)(1 – s). Similar reasoning yields the other relationships in equation 2.
For each of the 4 probabilities in equation 2, the rightmost expression is in ‘one-minus’ form. This facilitates probabilistic interpretations in terms of the probability of not being affected. For example, B, the probability of being affected if an individual has genotype G0H1 can be expressed as one minus the probability of not being affected due to either U2 or S. This deconstruction of two-locus penetrances into response-type probabilities illuminates the causal components of penetrance. As stated above, these equations satisfy the usual definition of penetrance, i.e. the probability of disease conditioned on a genotype.
Mathematical Comparisons with Commonly Used Penetrance Assumptions for Two-Locus Models
Using the causal model-based definitions of genetic heterogeneity, gene-gene interaction, proportion susceptible and phenocopy illustrated in figure 1 and explained in the first paper of this series [9], we have derived formulae that describe the relative contributions of these phenomena to two-locus penetrances. From the SCC model and equation 2, one can see that the two-locus penetrances observed in a population will vary depending on: the frequency of causes other than the 2 genotypes of interest (s); whether 2 genotypes demonstrate causal genetic heterogeneity (e.g. u1 > 0, u2 > 0, u3 = 0 vs. u1 > 0, u2 = 0, u3 = 0); whether 2 genetic causes demonstrate causal gene-gene interaction (u3 > 0 vs. u3 = 0); and whether 2 genetic causes demonstrate both causal genetic heterogeneity and causal gene-gene interaction (e.g. u1 > 0, u2 > 0, u3 > 0 vs. u1 > 0, u2 = 0, u3 = 0). We now compare this causal model-based penetrance matrix with the following more familiar (non-causal-based) two-locus penetrance models.
Risch presented several penetrance models for a disease given a two-locus genotype, denoted by ωij and defined as ωij = P[disease ∣ GiHj] [10]. He presented these models for situations in which 2 loci either act independently to cause disease (i.e. two-locus causal genetic heterogeneity) or act together to cause disease (i.e. two-locus causal gene-gene interaction).
He described the additive model for a disease with underlying two-locus causal heterogeneity. The additive model assumes that ‘penetrance factors’ or ‘summands’ xi and yj exist such that
| (3) |
As defined by Risch [10], the values xi and yj are not intended to represent single-locus or marginal penetrances. Rather, they are abstract, unobservable quantities – mathematical constructs – designed to characterize the relationships among the various penetrances. For example, the additive model necessarily implies ω11 + ω00 = ω10 + ω01, or, in our context, P[disease ∣ G1H1] + P[disease ∣ G0H0] = P[disease ∣ G1H0] + P[disease ∣ G0H1], as we will show below.
The genetic heterogeneity model is also intended to represent causal heterogeneity, and differs from the additive model by correcting for the fact that xi + yj can theoretically exceed 1, whereas the probability of disease given a two-locus genotype cannot. The heterogeneity formula specifies
| (4) |
.
The multiplicative model, described by Risch [10] for diseases with underlying two-locus causal gene-gene interaction or epistasis, specifies that ‘penetrance factors’ xi and yj can be defined such that
| (5) |
Again, these penetrance factors are mathematical constructs.
Whether additive, heterogeneity, or multiplicative models accurately represent the patterns of disease risk that result from causal genetic heterogeneity or from causal gene-gene interaction has not been investigated before. In order to determine this, we now evaluate the correspondence between these models and the two-locus penetrances A–D when causal genetic heterogeneity (e.g. u1 > 0, u2 > 0) and causal gene-gene interaction (u3 > 0) are features of the SCC model.
Additive Penetrance Models
Table 2a shows a penetrance matrix that connects the causal two-locus penetrances in equation 2 with Risch's penetrance factors in equation 3. For example, for the additive model to correctly describe the two-locus penetrances, A = ω11 = x1+ y1, and similarly for B, C, and D. From table 2a, (A + D) and (B + C) must both equal x1 + x0 + y1 + y0. Thus, under this additive model, (A + D) must equal (B + C). In terms of component cause frequencies, this equality implies:
| (6) |
Table 2.
Mathematical relationships among two-locus penetrances (A, B, C, D) and penetrance factors xi and yj in 3 common models
| a | Additive model: ωij = yi (eq. 3) | |
|---|---|---|
| G1 | G0 | |
| H1 | x1 + y1 = A | x0 + y1 = B |
| H0 | x1 + y0 = C | x0 + y0 = D |
| b | Heterogeneity model: ωij = 1 – (1 – xi)(1 – yj) (eq. 4) | |
| G1 | G0 | |
| H1 | 1 – (1 – x1) × (1 – y1) = A | 1 – (1 – x0) × (1 – y1) = B |
| H0 | 1 – (1 – x1) × (1 – y0) = C | 1 – (1 – x0) × (1 – y0) = D |
| c | Multiplicative model: ωij = xiyj (eq. 5) | |
| G1 | G0 | |
| H1 | x1y1 = A | x0y1 = B |
| H0 | x1y0 = C | x0y0 = D |
Derived from the general two-locus SCC model depicted in figure 1.
The only non-trivial solution to this equation is
| (7) |
where u1 + u2 ≤ 1. All other solutions would require setting both sides of equation 6 to zero, thus implying either s = 1 (i.e. everyone in the population is ‘Inevitable’ type and has the disease), and/or at least one of u1 and u2 equals zero, which would mean we no longer had a two-locus model. (Details are in ‘Appendix’.)
In Risch [10] and elsewhere, the additive penetrance model is used to describe the disease in the population when genetic heterogeneity is a feature of the genetic architecture, i.e. when a disease has 2 independently acting genetic causes. If sufficient causes I and II together reflect our biological concept of heterogeneity, and sufficient cause III reflects our biological concept0 of gene-gene interaction, then using the additive model to describe a disease with genetic heterogeneity requires the assumption that gene-gene interaction is also a feature of the genetic architecture.
Table 3 summarizes the sets of assumptions required to make the respective models consistent with each other. The table reveals that in order for causal heterogeneity (i.e. u1 > 0 and u2 > 0) to be described using an additive two-locus penetrance model, gene-gene interaction must also occur (i.e. u3 > 0), unless all disease can be attributed either to: (1) a single-locus, meaning the disease would have happened in the absence of the causal genotype at the second locus because the entire population is susceptible to the causal genotype at the first locus (i.e. u1 or u2 = 1), or (2) a set of causes (other than the 2 genotypes of interest, i.e. sufficient cause IV) that produces disease in 100% of the population (i.e. s = 1). Thus, using the additive model to imply lack of interaction (or independence) in the causal sense of these words is misleading.
Table 3.
Sets of assumptions about the SCC model and the frequency of component causes Uj, U2, U3, and S in figure 1 that render the model consistent with additive, heterogeneity, and multiplicative two-locus penetrance models
| Statistical two-locus penetrance model | Constraint on probability of component cause in population |
Causal single locus | Causal genetic heterogeneity | Causal gene-gene interaction | Pheno-copies | Comments | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| u1 | u2 | u3 | s | |||||||
| Two-locus additive penetrance model xi + yj = ωij |
- |
- |
- |
1 |
allowed |
allowed* |
allowed* |
required |
entire population diseased due to S |
|
| 1 | 0 | - | <1 | required | not allowed | allowed* | allowed | fully penetrant single-locus model with or without phenocopies | ||
| 0 |
1 |
- |
<1 |
|||||||
| 0 | - | 0 | <1 | allowed | not allowed | not allowed | allowed | single-locus model with or without phenocopies | ||
|
- |
0 |
0 |
<1 |
|||||||
| 0 < u1 + u2 < 1 | <1 | not allowed | required | required | allowed | causal genetic heterogeneity; requires causal gene-gene interaction with or without phenocopies; u values constrained | ||||
| Two-locus heterogeneity penetrance model xi + yj – xiyj = ωij |
- |
- |
- |
1 |
allowed* |
allowed* |
allowed* |
required |
entire population diseased due to S |
|
| – | 1 | – | <1 | allowed | allowed* | allowed* | allowed | fully penetrant single-locus model with or without phenocopies (no genetic heterogeneity or gene-gene interaction) | ||
| 1 |
– |
– |
<1 |
|||||||
| – | – | 0 | <1 | allowed | allowed | not allowed | allowed | causal genetic heterogeneity; requires absence of causal gene-gene interaction with or without phenocopies | ||
| Two-locus multiplicative penetrance model xiyj =ωij | – |
– |
– |
1 |
allowed* |
allowed* |
allowed* |
required |
entire population diseased due to S |
|
| 0 | – | 0 | <1 | allowed | not allowed | not allowed | allowed | single-locus model with or without phenocopies | ||
| – |
0 |
0 |
<1 |
|||||||
| 0 | – | – | 0 | allowed | not allowed | allowed | not allowed | causal gene-gene interaction; requires absence of both causal heterogeneity and phenocopies | ||
| – |
0 |
– |
0 |
|||||||
| 0 < u1 + u2 < 1 | not allowed | required | required | required | causal gene-gene interaction; requires causal genetic heterogeneity with gene-gene interaction; u and s constrained | |||||
Although allowed, the mechanism does not have a ‘causal effect’, i.e. average causal effect = 0 as defined in [9]; – indicates unconstrained.
In summary, under our assumptions, the only way this additive (non-causally based) two-locus model can correspond to a causal two-locus model (aside from degenerate cases, such as when everyone in the population is affected, or when the genotype at one locus is fully penetrant) is when the proportion susceptible to the G1H1 genotype, u3, has a specific algebraic relationship to the proportions susceptible to the G1H0 and G0H1 genotypes, u1 and u2 as defined by equation 7.
Heterogeneity Penetrance Models
Table 2b shows a penetrance matrix that connects the causal two-locus penetrances in equation 2 with Risch's penetrance factors in equation 4. From table 2b we can see that Risch's penetrance factors must satisfy a certain algebraic relationship in order to fit equations 2. The table shows that (1 – A) × (1 – D) and (1 – B) × (1 – C) must both equal (1 – x1)(1 – y1)(1 – x0)(1 – y0) and therefore must equal each other. In terms of component cause frequencies, this implies:
| (8) |
(details in ‘Appendix’).
As with the additive model, there is only one non-trivial solution – in this case, where u3 = 0. That is, the heterogeneity model in equation 4 implies that no cases of this disease are due to causal gene-gene interaction (ruling out the trivial cases where at least one genotype is 100% penetrant or where everyone in the population is affected due to causes other than the 2 genotypes of interest). When u3 = 0, the probabilities u1, u2, and s are unconstrained between 0 and 1; hence the heterogeneity model holds for any combination of values of u1, u2, and s. If, together, sufficient causes I and II reflect our biological concept of genetic heterogeneity and sufficient cause III reflects our biological concept of gene-gene interaction, then assuming the two-locus heterogeneity model to describe a disease with biological genetic heterogeneity with or without phenocopies also assumes that gene-gene interaction is not part of the causal model.
In summary, the only way this heterogeneity penetrance model can correspond to a causal two-locus model (aside from degenerate cases such as everyone being affected, or one fully-penetrant genotype) occurs when u3, the proportion susceptible to G1H1, is zero, that is, when no cases of disease are caused by the combined presence of the causal genotypes at both loci G and H, i.e. no causal gene-gene interaction. Moreover, when some individuals in the population are GH-susceptible, i.e. u3 > 0, the two-locus penetrance among those with both causal genotypes (i.e. Risch's ω11) will exceed the two-locus penetrance predicted by the genetic heterogeneity model, x1 + y1 – x1y1. Given our assumptions, the excess penetrance will equal the proportion of GH-causal types (r4)in the population.
Multiplicative Penetrance Models
Table 2c shows a penetrance matrix that connects the causal two-locus penetrances in equation 2 with Risch's penetrance factors in equation 5. For example, A = ω1 = x1y1. For this model to describe the two-locus penetrances correctly, AD and BC must both equal x1y1x0y0 and must therefore equal each other. In terms of component cause frequencies, this becomes
| (9) |
The only non-trivial solution to this equation occurs when
| (10) |
with u3 > 0 and u1 + u2 ≤ 1 (details in ‘Appendix’). Note that equation 10 differs from equation 7 only by the factor s on the left-hand side of the equality.
Our definition of causal gene-gene interaction requires u3 > 0. Thus, equation 10 demonstrates that the multiplicative model requires the assumption that u1, u2, and s are all greater than zero, meaning that some individuals in the population will be susceptible to genotypes at each locus acting alone, i.e. causal genetic heterogeneity. Additionally, the proportion of individuals susceptible to one or the other of the interacting genotypes alone (i.e. due to sufficient cause I or II) must be less than unity.
In summary, under our assumptions, the only way this multiplicative model can correspond to a causal two-locus model of epistasis is for the causal parameters to satisfy the particular algebraic relationship in equation 10.
Application of the Causal Framework to Other Genetic Models
Above, we focused on Risch's models [10] because of their relative generality and because we want to illustrate the principles of using the causal framework to think about genetic models of complex diseases. These principles can also be applied to other models. For example, Vieland and Huang [4] define heterogeneity as the case in which the penetrance in individuals with both factors equals what one would expect with independent effects, i.e., in their notation, the case in which fAB = 1 – (1 – fA)(1 – fB). This is just a special case of Risch's heterogeneity model, arising when (in our notation) D = 0, or when (in Risch's notation) x0 = y0 = 0. Some of the subsequent controversy over Vieland and Huang (especially in [21]) focused on their choice of that definition, rather than on any of the mathematical results.
Sepulveda et al. [22] present another approach for two-locus models altogether, one in which alleles act independently to influence penetrance. Within this approach, they consider several different models (‘independent action’, ‘inhibition’, and ‘cumulative’ models), with both ‘dominant’ and ‘recessive’ inheritance, although they use these terms in non-standard ways. To consider just one example, in their ‘dominant’ independent action model, we have expressed their penetrances in ‘one-minus’ form (table 4). Here, πA and πB represent, respectively, the penetrance component of the A (first locus) or B (second locus) allele, and πext represents the ‘penetrance’ of any external factors. Expressing the probabilities this way reveals that this model can readily be expressed in causal terms, with A, B, and ‘external factors’ each included in a separate causal pie (cf. our equations 2). Full exploration of all the models in Sepulveda et al. [22] is beyond the scope of the current paper, but we have shown here how to approach that task.
Table 4.
Penetrance as a function of genotype in the ‘dominant’ independent action model in Sepulveda et al. [22]
| BB | Bb | bb | |
|---|---|---|---|
| AA | 1 – (1 – πA)2(1 – πB)2(1 – πext) | 1 – (1 – πA)2(1 – πB)(1 – πext) | 1 – (1 – πA)2(1 – πext) |
| Aa | 1 – (1 – πA)(1 – πB)2(1 – πext) | 1 – (1 – πA)(1 – πB)(1 – πext) | 1 – (1 – πA)(1 – πext) |
| aa | 1 – (1 – πB)2(1 – πext) | 1 – (1 – πB)(1 – πext) | 1 – (1 – πext) = πext |
Discussion
Additive two-locus penetrance models are frequently assumed to correspond to a causal model of genetic heterogeneity (lack of epistasis), whereas multiplicative models are assumed to correspond to a causal model of gene-gene interaction. Our results demonstrate that these assumptions are incorrect: both of these models actually correspond to a disease characterized by both causal gene-gene interaction and causal genetic heterogeneity. This suggests that neither model can be used for hypothesis testing, e.g. to reject the hypothesis of no epistasis in a complex disease.
Risch's additive two-locus penetrance model [10] corresponds to a causal model of genetic heterogeneity only under the assumptions that (1) there are individuals in the population who have disease due to the combination of genotypes at loci G and H (i.e. causal gene-gene interaction), and (2) the proportion of such individuals equals a specific algebraic function of the proportion of individuals susceptible to either locus alone, as in equation 7.
Similarly, a multiplicative two-locus penetrance model describes the two-locus penetrance for a disease with gene-gene interaction only under the assumptions that (1) there are individuals in the population who are susceptible to the causal genotype at locus G regardless of the genotype at locus H and vice versa, and (2) the proportion of such individuals equals a specific algebraic function of the proportion of individuals susceptible to the causal genotypes at loci G and H acting together and the proportion of phenocopies, as determined by equation 10.
In contrast, under the assumptions of this paper (the specified SCC model, 2 binary genotypes, a binary outcome, independent distribution of genetic and non-genetic causes, and our definitions of causal gene-gene interaction and causal genetic heterogeneity), Risch's heterogeneity penetrance model [10] perfectly describes the joint distribution of two causal genotypes in the absence of epistasis, i.e. when no individual in the population requires susceptibility genotypes at both loci for disease occurrence (u3 = 0). This suggests that the heterogeneity penetrance model is most appropriate for testing the null hypothesis of no epistasis. Further, our finding that when u3 > 0, the excess penetrance above that expected under the heterogeneity model is equal to the proportion of GH-causal types in the population (r4) suggests that this model might be useful for developing a method to assess the degree to which epistasis contributes to disease susceptibility.
Likewise, Risch's heterogeneity penetrance model [10] can be used to generate simulated two-locus genotypes and disease risk for diseases with causal genetic heterogeneity or causal gene-gene interaction. Causal gene-gene interaction can be simulated by specifying a value of P[disease ∣ G1H1] in excess of that predicted by the genetic heterogeneity model. Specifying P[disease ∣ G1H1] not in excess of that predicted by genetic heterogeneity indicates no causal gene-gene interaction. On the other hand, data simulated according to additive and multiplicative models may describe causal genetic heterogeneity and causal gene-gene interaction, but only under the highly constrained conditions described above and in table 3.
The use of response types, as implemented here, provides the foundation for future investigations with different or less restrictive assumptions. For example, we assumed that all causal genotypes and other component causes were distributed independently. Relaxation of this assumption (e.g. for genetic variants in linkage disequilibrium, or genetic variants associated with specific environmental exposures) could be accommodated by changing the probabilities of the response types in table 1. Similarly, our causal model assumed that when G1 and H1 are in separate sufficient causes they have different causal partners. Future work could investigate a model specifying identical causal partners for G1 and H1.
These findings challenge the notion that ‘there is not a precise correspondence between biological models of epistasis and those that are more statistically motivated’ [5], suggesting instead that familiar models used in statistical genetics do have biological interpretations, under certain assumptions and explicit definitions. Our results suggest that causal principles, not mathematical constructs, should provide the basis of evaluating assumptions in statistical genetics research when the goal of such models is to identify the causes of diseases. Any statistical measure used to identify causes makes assumptions about the underlying causal model. Here, we have demonstrated the assumptions for additive, multiplicative, and heterogeneity penetrance models given our assumed causal model. But more generally, any statistic used in the field (e.g. recombination fraction, genotype-relative risk, recurrence risk among relatives) makes assumptions about the underlying causal model.
Causal models provide the basis of an emerging class of powerful methods in other quantitative fields and have been demonstrated to apply to concepts in human genetics [9]. We used an SCC model, which provides a single lens through which to view the effects of not only genetic heterogeneity and gene-gene interaction but also gene frequency, mode of inheritance, phenocopy frequency, and penetrance on disease risk in families and in populations.
Risch [10] also used the multiplicative and additive penetrance models, as well as an identity-by-descent distribution, as the basis for determining the magnitude of increased risk of disease among relatives (λR) when disease is caused by two-locus epistasis or genetic heterogeneity, respectively. His λR and related estimates are frequently used as the basis for power calculations. We are currently using approaches similar to those applied here to model disease risk in families, in order to derive λR estimates and other family-based measures from fundamental principles of causation.
Appendix
Here we derive equations 6–10, i.e. the formulas for the relationships between the causal models and the commonly used penetrance assumptions for two-locus models.
Additive Penetrance Models – Equations 6 and 7
As noted in the text, we must have A + D = B + C in order for an additive model as in equation 3 to hold. From equation 2, this implies r2 + r3 + r4 + r5 + r6 + r6 = r3 + r5 + r6 + r2 + r5 + r6; which reduces to
| (A.1) |
From table 1, substitute component cause frequencies for response-type probabilities into (A.1):
| (A.2) |
.
which is equation 6. The only non-trivial solution arises when none of s, u1, or u2 equals unity, and neither u1 nor u2 equals zero. In this case, divide by (1 – s)(1 – u1)(1 – u2) to yield equation 7. Also note that since u3 in equation 7 represents a probability, it cannot exceed unity, and therefore u1 and u2 in equation 7 must satisfy the constraint u1u2 ≤ (1 – u1)(1 – u2), i.e.
| (A.3) |
To obtain the trivial solutions discussed in the text, set both sides of (A.2) to zero, and now this equation will be true (1) if s = 1; (2)if u3 = 0 and one of (u1, u2) equals zero, or (3) if one of (u1, u2) equals zero and the other equals unity. These correspond to the trivial cases discussed in the text.
Heterogeneity Penetrance Models – Equation 8
As noted in the text, we must have (1 – A)(1 – D) = (1 – B) (1 – C) in order for a heterogeneity model as in equation 4 to hold. From equation 2, this implies
| (A.4) |
which is equation 8. The only non-trivial solution arises when none of s, u1, or u2 equals unity: divide through by (1 – u1)(1 – u2)(1 – s)2 to yield 1 – u3 = 1, i.e. u3 = 0, as discussed in the text. To obtain the trivial solutions discussed in the text, set both sides of (A.4) to zero, and now this equation will be true if any one of s, u1, or u2 equals unity.
Multiplicative Penetrance Models – Equations 9 and 10
As noted in the text, we must have AB = BC in order for a multiplicative model as in equation 5 to hold. From equation 2, this implies
| (A.5) |
which is equation 9. For ease of notation, let νi = 1 – ui for all i, and let t = 1 – s. Equation A.5 becomes
Multiplying out the terms, and collecting all terms containing ν1ν2 on the left-hand-side, yields
| (A.6) |
One solution to equation A.6 is the trivial one in which t = 0, i.e. s = 1; that is, everyone in the population is ‘Inevitable’ and has the disease. Having accounted for that solution, divide both sides by t and convert back to u-s notation:
| (A.7) |
The only non-trivial solution arises when neither side of equation A.7 equals zero or unity. In this case, divide both sides by (1 – u1)(1 – u2), to yield
which simplifies to equation 10. Since u3 and s in equation 10are probabilities, their product cannot exceed unity; therefore, u1 and u2 must satisfy the constraint in equation A.3. Trivial solutions arise (1) when both sides of equation A.7 equal zero, which occurs when u1 + u2 = 1 and at least one of u1, u2, or u3s equals unity, or (2) when both sides of equation A.7 equal unity, which occurs when u1, u2, and u3s all equal zero.
Acknowledgements
A.M.M. was supported in part by NIMH grant T32-MH065213. This work was also supported by NIH grants R01-NS043472, R01-NS036319, R01-NS053998, and RC2-NS070344 (to R.O.); and NIMH grant R01-MH048858 (to S.E.H.). We are grateful to Sharon Schwartz, PhD, for instructive insights and critical comments.
References
- 1.Pearl J. Causality: Models, Reasoning, and Inference. Cambridge: New York, Cambridge University Press; 2000. [Google Scholar]
- 2.Li W, Reich J. A complete enumeration and classification of two-locus disease models. Hum Hered. 2000;50:334–349. doi: 10.1159/000022939. [DOI] [PubMed] [Google Scholar]
- 3.Strauch K, Fimmers R, Baur MP, Wienker TF. How to model a complex trait. 2. Analysis with two disease loci. Hum Hered. 2003;56:200–211. doi: 10.1159/000076394. [DOI] [PubMed] [Google Scholar]
- 4.Vieland VJ, Huang J. Two-locus heterogeneity cannot be distinguished from two-locus epistasis on the basis of affected-sib-pair data. Am J Hum Genet. 2003;73:223–232. doi: 10.1086/376563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cordell HJ. Epistasis: what it means, what it doesn't mean, and statistical methods to detect it in humans. Hum Mol Genet. 2002;11:2463–2468. doi: 10.1093/hmg/11.20.2463. [DOI] [PubMed] [Google Scholar]
- 6.Elston RC, Song D, Iyengar SK. Mathematical assumptions versus biological reality: myths in affected sib pair linkage analysis. Am J Hum Genet. 2005;76:152–156. doi: 10.1086/426872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartlett CW, Vieland VJ, Bartlett J, Bell JT, Bhattacharjee S, Clerget-Darpoux F, Bush WS, Edwards TL, Gao G, Halder I, Huang Y, Kotti S, Larkin EK, Li H, Motsinger AA, Mukhopadhyay N, Namkung J, Park T, Ritchie MD, Stein CM, Zhou JY. Discussing gene-gene interaction: warning–translating equations to English may result in jabberwocky. Genet Epidemiol. 2007;31(Suppl 1):S61–S67. doi: 10.1002/gepi.20281. [DOI] [PubMed] [Google Scholar]
- 8.Cordell HJ. Detecting gene-gene interactions that underlie human diseases. Nat Rev Genet. 2009;10:392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madsen AM, Hodge SE, Ottman R. Causal models for investigating complex disease: I. A primer. Hum Hered. 2011;72:54–62. doi: 10.1159/000330779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Risch N. Linkage strategies for genetically complex traits. I. Multilocus models. Am J Hum Genet. 1990;46:222–228. [PMC free article] [PubMed] [Google Scholar]
- 11.Wang S, Zhao H. Sample size needed to detect gene-gene interactions using linkage analysis. Ann Hum Genet. 2007;71:828–842. doi: 10.1111/j.1469-1809.2007.00367.x. [DOI] [PubMed] [Google Scholar]
- 12.Pinto D, Kasteleijn-Nolst Trenite DG, Cordell HJ, Mattheisen M, Strauch K, Lindhout D, Koeleman BP. Explorative two-locus linkage analysis suggests a multiplicative interaction between the 7q32 and 16p13 myoclonic seizures-related photosensitivity loci. Genet Epidemiol. 2007;31:42–50. doi: 10.1002/gepi.20190. [DOI] [PubMed] [Google Scholar]
- 13.Kallberg H, Padyukov L, Plenge RM, Ronnelid J, Gregersen PK, van der Helm-van Mil AH, Toes RE, Huizinga TW, Klareskog L, Alfredsson L, Epidemiological Investigation of Rheumatoid Arthritis study group Gene-gene and gene-environment interactions involving HLA-DRB1, PTPN22, and smoking in two subsets of rheumatoid arthritis. Am J Hum Genet. 2007;80:867–875. doi: 10.1086/516736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hodge SE. Some epistatic two-locus models of disease. I. Relative risks and identity-by-descent distributions in affected sib pairs. Am J Hum Genet. 1981;33:381–395. [PMC free article] [PubMed] [Google Scholar]
- 15.Darroch J. Biologic synergism and parallelism. Am J Epidemiol. 1997;145:661–668. doi: 10.1093/oxfordjournals.aje.a009164. [DOI] [PubMed] [Google Scholar]
- 16.Rothman KJ, Greenland S. Modern Epidemiology. ed 2. Philadelphia: Lippincott-Raven; 1998. [Google Scholar]
- 17.Rothman KJ. Causes. Am J Epidemiol. 1976;104:587–592. doi: 10.1093/oxfordjournals.aje.a112335. [DOI] [PubMed] [Google Scholar]
- 18.Robins JM, Greenland S. Identifiability and exchangeability for direct and indirect effects. Epidemiology. 1992;3:143–155. doi: 10.1097/00001648-199203000-00013. [DOI] [PubMed] [Google Scholar]
- 19.Greenland S, Poole C. Invariants and noninvariants in the concept of interdependent effects. Scand J Work Environ Health. 1988;14:125–129. doi: 10.5271/sjweh.1945. [DOI] [PubMed] [Google Scholar]
- 20.Greenland S, Robins JM. Identifiability, exchangeability, and epidemiological confounding. Int J Epidemiol. 1986;15:413–419. doi: 10.1093/ije/15.3.413. [DOI] [PubMed] [Google Scholar]
- 21.Cordell HJ. Affected-sib-pair data can be used to distinguish two-locus heterogeneity from two-locus epistasis. Am J Hum Genet. 2003;73:1468–1471. doi: 10.1086/380312. author reply 1471–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sepulveda N, Paulino CD, Carneiro J, Penha-Goncalves C. Allelic penetrance approach as a tool to model two-locus interaction in complex binary traits. Heredity. 2007;99:173–184. doi: 10.1038/sj.hdy.6800979. [DOI] [PubMed] [Google Scholar]