Abstract
To better understand the genomic organization and evolution of Sox genes in vertebrates, we cytogenetically mapped Sox2 and Sox14 genes in cichlid fishes and performed comparative analyses of their orthologs in several vertebrate species. The genomic regions neighbouring Sox2 and Sox14 have been conserved during vertebrate diversification. Although cichlids seem to have undergone high rates of genomic rearrangements, Sox2 and Sox14 are linked in the same chromosome in the Etroplinae Etroplus maculatus that represents the sister group of all remaining cichlids. However, this genes are located on different chromosomes in several species of the sister group Pseudocrenilabrinae. Similarly the ancestral synteny of Sox2 and Sox14 has been maintained in several vertebrates, but this synteny has been broken independently in all major groups as a consequence of karyotype rearrangements that took place during the vertebrate evolution.
Keywords: Cichlidae, genome evolution, molecular cytogenetics, chromosome
Introduction
Sox genes, a gene family encoding transcription factors involved in a variety of development processes, are found throughout the animal kingdom (Guo et al. 2009). These genes are expressed in various phases of embryonic development and, among vertebrates, are involved with testis development, neural crest cell development, neurogenesis, oligodendrocyte development, and chondrogenesis (Kiefer 2007). Sox genes are characterized by the presence of a DNA-binding HMG (high mobility group) domain and are subdivided into ten subgroups (A–J) based upon their HMG box sequences (Bowles et al. 2000). The Sox B group (Sox1, Sox2, Sox3, Sox14 and Sox21) is of particular interest, since the members of this group play a major role in neural development and participate in the earliest events of central nervous system (CNS) differentiation in Drosophila, Xenopus, chicken and mouse (Collignon et al. 1996; Uchikawa et al. 1999; Hargrave et al. 2000; Kishi et al. 2000; McKimmie et al. 2005). Also they are most closely related to Sry and appear to be functionally conserved during evolution (McKimmie et al. 2005).
Sox1 and Sox2 (group B1 members/activators), and Sox14 and Sox21 (B2 members/repressors), are arranged in two pairs, each comprising one SoxB1 activator and one SoxB2 repressor. At least one pair of group B1 and group B2 Sox genes are recognizable in the genomes of lower animals, including sponge (Larroux et al. 2008), sea urchin (Howard-Ashby et al. 2006), ascidian (Satou and Satoh 2005) and amphioxus (Meulemans and Bronner-Fraser 2007), although their linkages have not been confirmed. The early state of SoxB evolution is perfectly represented by the Drosophila melanogaster genome, which shows that the orthologs of vertebrates Sox2 and Sox14 are clustered on the same chromosome (Wei et al. 2011). This organization was also found in other insect genomes, including Anopheles gambiae, Apis mellifera, Tribolium castaneum, Nasonia vitripennis and Bombyx mori, where Sox B genes are clustered on the same chromosome or scaffold assembly (Wei et al. 2011).
Among vertebrates, Sox B genes have arisen by rounds of whole genomic duplications, rearrangement and divergence from ancestral Sox B genes (Kirby et al. 2002; Guth and Wegner 2008). Linkage of Sox2 and Sox14 was previously observed in several mammals and in the chicken Gallus gallus (revised in Popovic and Stevanovic 2009). At the same time, Sox2 and Sox14 were not linked in other mammals and in the fish Danio rerio (revised in Popovic and Stevanovic 2009). In the cichlid fish O. niloticus, Sox2 and Sox14 genes mapped to different chromosomes corresponding to linkage group 17 (LG17) and LG23, respectively (Cnaani et al. 2007). Although both conditions of linkage and non linkage of Sox2 and Sox14 seem to occur in different vertebrates, the absence of information for non-mammalian species does not allows major conclusions. Okuda et al. (2006) suggest that the chromosomal organization of group B Sox genes in fishes is different from other vertebrates. It is not yet clear whether the duplication, coupled with functional divergence and physical dispersal occurred early in the radiation of vertebrates, or occurred more recently.
Cichlids have been used as model organisms to study a diversity of evolutionary mechanisms because they represent one of the most striking examples of rapid and convergent evolutionary radiation among vertebrates. Here, we cytogenetically mapped the distribution of Sox2 and Sox14 on the chromosomes of several cichlid species by fluorescence in situ hybridization (FISH) and performed a comparative mapping analyses of Sox2 and Sox14 orthologs in numerous vertebrate species based on the available genomic databases. Our results show that the linkage of Sox2 and Sox14 is maintained in many vertebrate taxa and that the separation of these genes onto different chromosomes seems to have occurred independently in all major vertebrate groups.
Materials and methods
Animals and sampling
Cichlid species used in this work were obtained from three sources. Cichlids from Lake Malawi were collected from the wild from 2005-2008 and maintained in the Tropical Aquaculture Facility (TAF) of the University of Maryland (UMD), College Park, MD, USA. South American species were collected from the wild in several Brazilian rivers. Additional species of uncertain origin were obtained from commercial sources in Botucatu, SP, Brazil, and were maintained in the Fish Room of the Laboratório Genômica Integrativa (FR-LGI) at São Paulo State University (UNESP), Botucatu (Table 1). All the specimens examined were fixed in formaldehyde and then stored in alcohol in the fish collections of TAF-UMD and FR-LGI.
Table 1.
Cichlids analyzed.
| Subfamily and wild distribution |
Groups or Tribes |
Species | 2n | Origin of specimens |
|---|---|---|---|---|
| Etroplinae (India and Madagascar) |
Etroplus maculatus | 46 | Petshop | |
|
| ||||
| Pseudocrenilabrinae (Africa) |
Tilapiine | Oreochromis niloticus | 44 | TAF-UMD |
| Oreochromis mossambicus | 44 | TAF-UMD | ||
| Oreochromis aureus | 44 | TAF-UMD | ||
| Tilapia mariae | 40 | TAF-UMD | ||
| Haplochromine | Haplochromis obliquidens | 44 | Petshop | |
| Metriaclima lombardoi | 44 | TAF-UMD | ||
| Astatotilapia burtoni | 40 | TAF-UMD | ||
| Labeotropheus trewavasae | 44 | TAF-UMD | ||
| Hemichromine | Hemichromis bimaculatus | 44 | Petshop | |
|
| ||||
| Cichlinae (America) |
Cichlini | Cichla kelberi | 48 | Araguaia River, Brazil |
| Astronotini | Astronotus ocellatus | 48 | Tiête River, Brazil | |
| Heroini | Symphysodon aequefasciatus | 48 | Petshop | |
| Geophagini | Geophagus brasiliensis | 48 | Tietê River, Brazil | |
Bacterial artificial chromosome (BAC) clones and probe labelling
Two BAC clones, from a genomic library of the Nile tilapia O. niloticus, containing the genes Sox2 from LG23 and Sox14 from LG17 (BAC IDs b04TI053B06 and b03TI079I04, respectively) (Cnaani et al. 2007) were used as probes for FISH. BAC extraction was conducted using the PhasePrep®™ BAC DNA Kit (Sigma-Aldrich, St Louis, MO, USA) according to supplier’s protocol. The BAC clones were labeled with biotin or digoxigenin (DIG) coupled nucleotides (Roche Applied Sciences, Indianapolis, IN, USA) using whole genome amplification (WGA2 &3) kits (Sigma-Aldrich), according to the supplier’s protocol. For double-color FISH, 16 μl of a hybridization mixture containing 50% deionized formamide, 2xSSC, 10% dextran sulfate, 10 μg of salmon sperm DNA, and 100 ng each of biotin and dig-labeled probes was prepared, denatured for 10 min at 65°C and immediately cooled on ice.
Chromosome preparation and FISH procedure
Chromosome preparations were obtained as previously described (Bertollo et al. 1978), and the slides with the chromosomes were air-dried, treated with pepsin (0.01% in 10 mM HCl) and dehydrated in an ethanol series one day before use. The slides were denatured in 70% formamide/2xSSC, pH 7 for 40s, and dehydrated in an ice-cold ethanol series. 16μl of probe mixture (containing 100 ng of each DNA probe) were hybridized under a 22 mm × 32 mm cover slip in a 37°C moist chamber for 48 h. Slides were washed two times for 5 min in 50% formamide/2xSSC, pH 7 at 43°C with agitation, then 10 min in 2xSSC, pH 7 at 42°C with continuous agitation. Hybridization signals were detected with avidin-fluoroscein isothiocyanate (FITC) and rhodamine-anti-DIG (Roche Applied Sciences, Indianapolis, IN, USA), according to the supplier’s protocol. After three washes of 2 min in phosphate buffer detergent (4xSSC/1% Tween-20), slides were mounted with antifade solution containing 4′,6-diamidino-2-phenyloindole (DAPI). Results were recorded with an Olympus BX61 microscope equipped with an Olympus digital camera DP71 and software Image-Pro MC 6.0.
Sequences similarity and comparative genomic database analyses
The sequences of an 883 nucleotide fragment of Sox2 and the entire coding sequence of Sox14 from Oreochromis niloticus were obtained from National Center for Biotechnology Information (NCBI) databases (www.ncbi.nlm.nih.gov/genbank/) with accession numbers EF431920-EF431927) (Cnaani et al. 2007). The similarity analyses between several vertebrates were done using nucleotide megablast at Basic Local Alignment Search Tool (BLAST) at NCBI (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
The chromosomal locations of Sox2 and Sox14 genes among several vertebrates were determined using information currently available in the public genomic databases of NCBI Map Viewer (http://www.ncbi.nlm.nih.gov/genomes), Sanger Institute Ensembl Database (http://www.ensembl.org) and BouillaBase-Comparative Genome Browsers (www.BouillaBase.org). The syntenic relationship analyzes of Sox2 and Sox14 genes were conducted using Genomicus genome browser (http://www.dyogen.ens.fr/genomicus/). Each gene was analyzed separated using human as reference species. The identification of syntenic genes among fish species using O. niloticus as a reference was determined using BouillaBase browser, since the gene prediction of O. niloticus is not yet available in the Genomicus browser.
Results
Comparative cytogenetic mapping
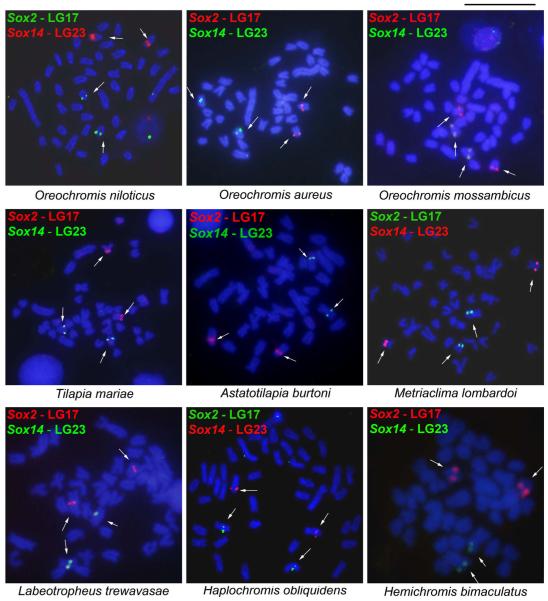
The Sox2 and Sox14 genes mapped to different chromosomes in O. niloticus (Figure 1), corresponding to LG17 and LG23 respectively, as expected (Cnaani et al. 2007). In the other eight Pseudocrenilabrinae species studied, Sox2 and Sox14 also mapped on two different chromosome pairs (Figure 1). The Sox2 (LG17) was located in the pericentromeric region of a medium subtelocentric/acrocentric (st/a) chromosome in all African cichlids investigated (Figure 1). Sox14 (LG23) showed variations in the chromosomal position being interstitially located on the long arm of a larger st/a chromosome in all tilapiine species (Figure 1) and in the haplochromine species (Figure 1). Exceptions were observed in Labeotropheus trewavasae (haplochromine), where Sox14 was located on a meta/submetacentric (m/sm) chromosome (Figure 1) as well as in the hemichromine, Hemichromis bimaculatus (Figure 1). B chromosomes were detected in Haplochromis obliquidens and Metriaclima lombardoi (haplochromines) as previously reported (Poletto et al. 2010a; 2010b), but no signal of Sox genes were detected in these B chromosomes (data not shown).
Fig. 1.
Cytogenetic mapping of Sox2 and Sox14 (arrows) in Pseudocrenilabrinae Cichlidae species showing their distribution in different chromosomes. The tilapiines include Oreochromis niloticus, Oreochromis mossambicus, Oreochromis aureus and Tilapia mariae; the haplocharomine includes Haplochromis obliquidens, Metriaclima lombardoi, Astatotilapia burtoni and Labeotropheus trewavasae; and the hemichromines is represented by Hemichromis bimaculatus. Scale bar 5μm.
In the Asian cichlid species, Etroplus maculatus (Etroplinae), the Sox2 and Sox14 were positioned on the two arms of a single small metacentric chromosome pair (Figure 2). In South American cichlids (Cichlinae) belonging to different tribes (Table 1), none of the BAC probes produced identifiable chromosomal signals, probably because the occurrence of rearrangements that could have differentiated the genomic blocks containing Sox2 and Sox14 in relation to the O. niloticus, the species source of the BAC clones used as the chromosome probes.
Fig. 2.
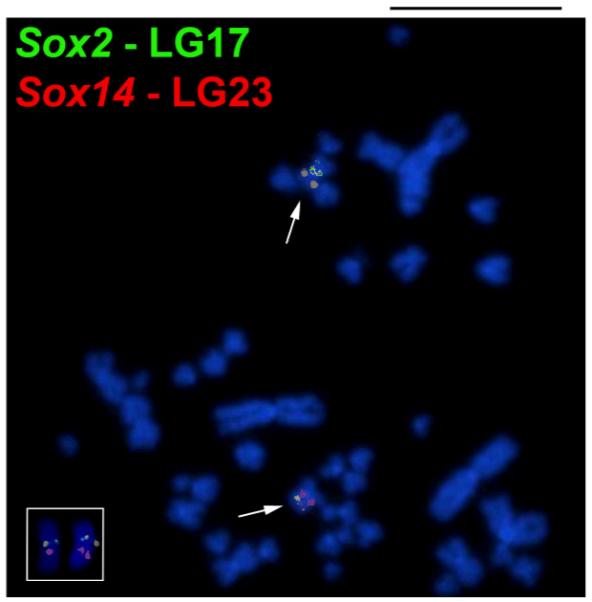
Cytogenetic mapping of Sox2 (green label) and Sox14 (red label) genes (arrows) in Etroplus maculatus. In the insert, a chromosome labeled pair of a second metaphase spread. Both genes are positioned on the same chromosome. Scale bar 5μm.
Sox genes of cichlids and comparative genomics
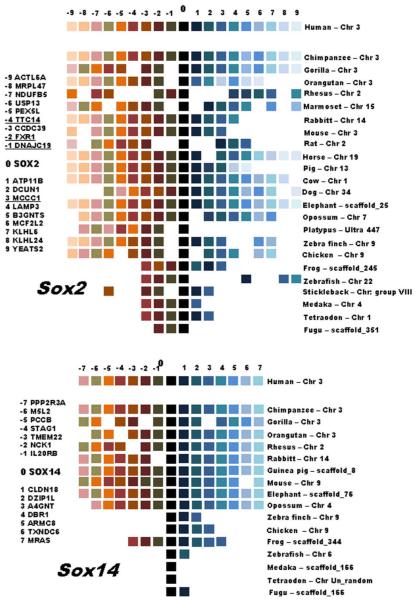
Analysis of similarity between Sox2 (EF431924.1) and Sox14 (EF431920.1) gene sequences of Oreochromis niloticus (Cnaani et al. 2007) indicates high levels of conservation among the homologs Sox genes of other vertebrate species, ranging from 79% to 100% for Sox2 (See Supplementary Material S1) and 78% to 100% for Sox14 (See Supplementary Material S2). Furthermore, based on web databases we have determined that Sox2-Sox14 are linked in several vertebrate species, including some mammals (Gorilla gorilla, Pongo abelli, Sus scrofa, Bos taurus and Ornithorhynchus anatinus) and the bird Gallus gallus (Table 2). On the other hand, Sox2 and Sox14 are located on different chromosomes in other vertebrates, including mammals (Callithrix jacchus, Canis familiaris, Equus cabalus, Rattus norvegicus, Mus musculus, Monodelphis domestica) and the fish Danio rerio (Table 2). The genomic position of Sox2 was also identified for the fish species Tetraodon nigroviridis, Gasterosteus aculeatus and Oryzias latipes, but no information was retrieved for Sox14 (Table 2), probably because more genome sequence and physical chromosome map data are available related to Sox2 than to Sox14. For several other species, it was impossible to determine the genomic organization of both Sox2 and Sox14 genes because the existing genome sequences are incomplete.
Table 2.
Chromosomal position of Sox2 and Sox14 genes in different vertebrate species and their nucleotide similarity level compared to O. niloticus. Chr, chromosome position; S On, Similarity to O. niloticus; AN, Accession number; NA, sequences for Sox genes are not available; NM, sequences of Sox genes are available but it was not possible to identify the genomic position. Species with linkage of Sox2 and Sox14 are highlighted in yellow, species in which Sox2 and Sox14 are unlinked are highlighted in blue, and the species with the linkage data not yet determined for Sox2 and Sox14 are highlighted in red.
| Major group/Species | Sox2 | Sox14 | ||||
|---|---|---|---|---|---|---|
| Chr | S On | AN | Chr | S On | AN | |
| Mammals | ||||||
| Homo sapiens * | 3 | 81% | NG_009080 | 3 | 79% | NM_004189 |
| Pan troglodytes * | 3 | 83% | XM_516895 | 3 | 79% | XM_526317 |
| Gorilla gorilla | 3 | ENSFM0050000027095 1 |
3 | ENSGGOG000000138 78 |
||
| Pongo abelii | 3 | 82% | XM_002814321 | 3 | 79% | XM_002814084 |
| Callithrix jacchus | 15 | 82% | XM_002807565 | 1 | 83% | XM_002742479 |
| Macaca mulatta * | 2 | 81% | NM_001142940 | 2 | 80% | NM_001194657 |
| Canis familiaris * | 34 | 82% | XM_545216 | 23 | ENSCAFG0000000986 7 |
|
| Bos taurus * | 1 | 81% | NM_001105463 | 1 | 78% | NM_001163781 |
| Equus cabalus * | 19 | 80% | NM_001143799 | 16 | 80% | XM_001916428 |
| Ornithorhynchus anatinus * | 1 | 81% | XM_001506934 | 1 | AY112710 | |
| Sus scrofa | 13 | 82% | EU503117 | 13 | ENSSSCG0000001165 6 |
|
| Oryctolagus cuniculus | 14 | 82% | XM_002716451 | 14 | ENSOCUG000000016 86 |
|
| Loxodonta africana | NM | ENSLAFG0000000636 2 |
NM | ENSLAFG0000000344 7 |
||
| Rattus norvegicus * | 2 | 81% | NM_001109181 | 8 | NW047801.1 | |
| Mus musculus * | 3 | 81% | NM_011443 | 9 | 78% | NM_011440 |
| Monodelphis domestica * | 7 | 85% | XM_001368783 | 4 | ENSMODG000000249 83 |
|
| Cavia porcellus | NM | ENSCPOG0000000357 5 |
NM | ENSCPOG0000002627 2 |
||
| Echinops telfairi | NM | ENSETEG0000000512 2 |
NM | ENSETEG0000001857 6 |
||
| Birds | ||||||
| Gallus gallus * | 9 | 79% | D50603 | 9 | ENSGALG000000173 72 |
|
| Amphibians | ||||||
| Xenophus tropicalis | NM | 86% | BC159121 | NM | ENSXETG0000002268 9 |
|
| Fish | ||||||
| Danio rerio * | 22 | 82% | AB242329 | 6 | ENSDARG000000709 29 |
|
| Oryzias latipes | 4 | 93% | FJ895588 | NM | 91% | NM_001164872 |
| Tetraodon nigroviridis | 1 | ENSTNIG00000008596 | NM | 100% | AY612092 | |
| Gasterosteus aculeatus | 8 | ENSGACG0000002011 1 |
NA | NA | ||
Species whose Sox2 and Sox14 linkage was previously checked/revised in Popovic and Stevanovic 2009.
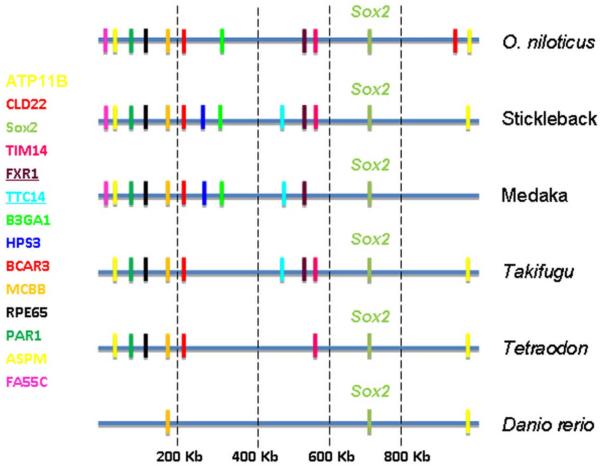
Analyses of the syntenic relationship between vertebrate chromosomal segments containing Sox2 and Sox14 genes were conducted using the Genomicus genome browser. The analyses of each gene separately using Homo sapiens as the reference species show that synteny is highly conserved in the chromosomal segments containing the Sox genes. A large genomic block containing several genes was conserved through vertebrates (Figure 3). At least 4 genes around Sox2 region are conserved even between more distant species like mammals and fishes: TTC14, FXR1, DNAJC19 and MCCC1 genes are present at least in three fish species and human (Figure 3). The Sox14 region seems to be more divergent between mammals and fishes, and sometimes the gene correspondence was not clear (see medaka and Tetraodon, for example, in Figure 3) or even the synteny correspond to a unique gene (gene CLDN18 in fugu and gene DZIP1L in zebrafish for example, Figure 3). Considering that O. niloticus gene content is not yet available in the Genomicus browser, the genomic block containing Sox2 was comparatively analyzed among several fish species using BouillaBase (Figure 4). In this new analysis several genes were detected conserved between O. niloticus, stickleback, medaka, Takifugu and Tetraodon (Figure 4). On the other hand, zebrafish presented few conserved genes with the other fish species. Furthermore, two genes (FXR1 and TTC14) were observed in most fish species (Figure 4) and also in several non-fish vertebrates (Figure 3). That analysis was not possible for Sox14 because the genome annotation for O. niloticus is not yet complete.
Fig. 3.
Chromosomal segments showing the conserved syntenic blocks containing Sox2 and Sox14 genes in diverse vertebrates and only in fish. Color squares indicate the same gene in the different vertebrate species (left) and its respective genomic position in relation to several other genes (right). The most conserved genes among fish and other vertebrates are underlined. See Supplementary Material S3 for more information on the gene abbreviations.
Fig. 4.
Chromosomal segments showing the conserved syntenic blocks containing Sox2 in several fish species, including the cichlid O. niloticus. Vertical color bars indicate the same gene in the different species (left) and its respective genomic position in relation to several other genes (right). The most conserved genes among fish and other vertebrates are underlined. The interrupted vertical lines indicate the genomic positions of the genes in reference to the scaffold 243 of O. niloticus genome that was used as reference. See Supplementary Material S4 for more information on the gene abbreviations.
Discussion
General aspects on the genome organization of Sox2 and Sox14 genes
The analysis of syntenic regions of the chromosomal locations that harbour the Sox genes using Genomicus have demonstrated that some genes which flank the Sox2 and Sox14 orthologs are conserved in their positions in some mammalian species. Sox14 is more conserved (only few rearrangements were detected) than Sox 2 (more rearrangements were observed) among mammals. However, when the Sox14 regions were compared using diverse groups (mammals, birds, fishes), they were not conserved as observed in mammals. Sox14 orthologs are highly diverged in non-mammal groups and Sox2 orthologs are more stable among all vertebrates.
The analysis of the genomic blocks containing Sox genes suggest the genes observed in the region are evolving as part of a large block of genes rather than individually. This is clearly observed among mammals but not much clear for fishes maybe because the (i) limited amount of genomic data available or (ii) the intense dynamism that rules the genome evolution in teleost fishes. The size of the syntenic blocks looks smaller in fishes, maybe because the low level of sequence similarities in distant comparisons has made it difficult to identify unambiguously orthologs, or the loss of Sox2 and Sox14 regions. Even using only fish species in the comparative analysis, the size of the syntenic blocks are still limited, but it is possible to detect some genes still present in mammals such as FXR1 and TTC14 (see Figures 3 and 4).
The analysis of Sox genes supports a model that at least four duplication events must have happened during vertebrate evolution, including a whole genome duplication that occurred before the radiation of teleost fishes (Kirby et al. 2002; Taylor et al. 2003; Guth and Wegner 2008). As a consequence, gene pairs can in theory exist in teleosts for every gene in the major Sox groups of other vertebrates. However, which of the pairs survived, differs among teleost species (Guth and Wegner 2008). In the pufferfish Takifugu rubripes, 25 Sox genes were identified occurring as duplicated paralogs with the mammalian Sox1, Sox4, Sox6, Sox8, Sox9, Sox10 and Sox14 (Koopman et al. 2004). In contrast, Sox8 and Sox10 are not duplicated in the zebrafish. However, zebrafish has three SoxB2 genes (Sox14, Sox21a and Sox21b) and six SoxB1 genes (Sox1a, Sox1b, Sox2, Sox3, Sox19a and Sox19b) (Okuda et al. 2006). The second copies of Sox2 and Sox3 might have been lost early in the teleost lineage, because the Takifugu genome also contains only one copy of Sox2 and Sox3 (Koopman et al. 2004). The same should be occurring with cichlids, where the second copies of Sox2 and Sox14 are absent and may have been lost very early in the teleost fish radiation. Interestingly, there is no direct SoxB ortholog for teleost Sox19a/b in other vertebrates, so these are fish specific genes. Instead, the highly divergent mammalian SoxG gene Sox15 and Xenopus SoxD appear to be the closest relatives to Sox19a/b (Okuda et al. 2006).
Cytogenetic mapping of Sox2 and Sox14 in cichlids and inferences on the chromosomal rearrangements involving both genes
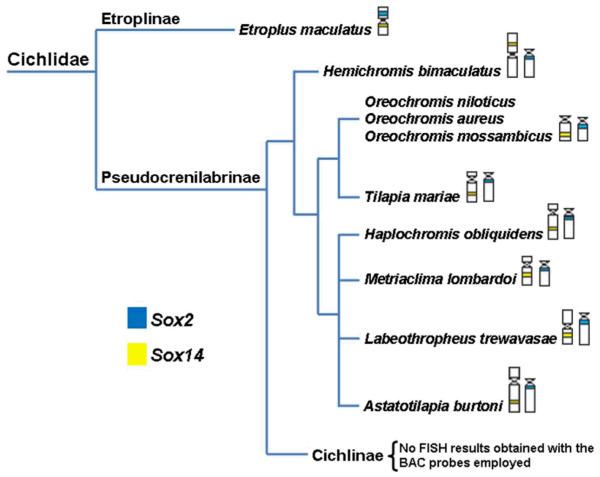
Etroplus maculatus contains the Sox2 and Sox14 genes preserved in the same chromosome (Figure 2 and 5), i.e. the ancestral vertebrate condition previously suggested (Kirby et al. 2002). The subfamily Etroplinae is considered the sister group of all remaining cichlids (Smith et al. 2008) and we could propose that these genes were syntenic in the ancestor of cichlids and were preserved linked on the same chromosome of Etroplinae species until now. On the other hand, cytogenetic mapping of Sox2 and Sox14 in Psedocrenilabrinae cichlids suggests that chromosomal rearrangements during the diversification of this group separated Sox2 and Sox14 genes onto different chromosomes (Figure 1 and 5). Although a higher number of species should be analyzed, the data obtained for the species here investigated suggest the chromosome that harbours Sox2 has been conserved during Pseudocrenilabrinae diversification. However, variations in the morphology of the chromosome carrying Sox14 among Pseudocrenilabrinae species were observed, suggesting that this chromosome has undergone more rearrangements during the evolution of the group (Figure 5).
Fig. 5.
Phylogenetic relationship of cichlids (adapted from Smith et al. 2008) showing the chromosomal distribution of Sox2 and Sox14 genes.
The chromosomal organization of these two genes among several vertebrates apparently does not follow a unique pattern. Human Sox2 and Sox14 are linked on chromosome 3 and map together in the platypus as well (Hope et al. 1990), demonstrating the synteny conservation of Sox2 and Sox14 over at least 170 million years since mammalian groups Prototheria and Theria diverged. Sox2-Sox14 maps together also in several other primates like, Gorilla gorilla, Macaca mulatta, Pan troglodytes and P. abelli, in Sus scrofa (pig), in Oryctolagus cuniculus (rabbit), and in Bos taurus (cattle) (Popovic and Stevanovic 2009, present work). On the other hand, no linkage was found for Sox2-Sox14 pair in dog, mouse, and in the primate Callitrix jacchus (Popovic and Stevanovic 2009, present work).
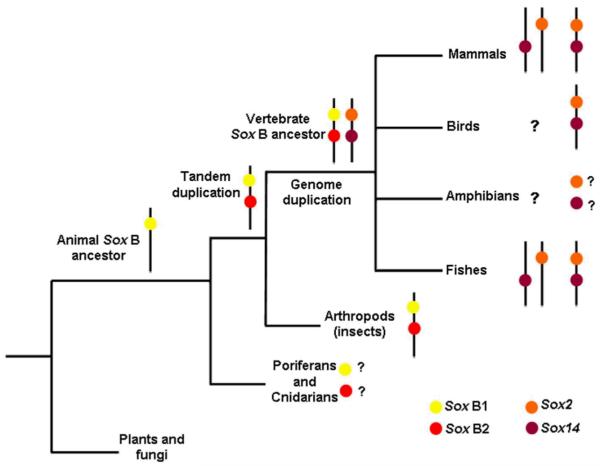
The present analysis suggests that the expected ancestral linkage for Sox2 and Sox14 is maintained in diverse vertebrate taxa and the genomic split of these genes to different chromosomes occurred independently in all major vertebrate groups (Figure 6) may be a consequence of particular karyotype rearrangements such as translocation or transposition, for example. The presence of linkage and non-linkage of Sox2 and Sox14 within Cichlidae suggests that the events of separation of both genes also occur in the terminal taxa level (recent evolutionary events) and are not only restricted to major vertebrate groups (ancient evolutionary events).
Fig. 6.
Evolutionary perspective for the distribution of Sox2 and Sox14 among vertebrates. Rounds of whole genomic duplications, rearrangement and divergence from ancestral Sox B genes have originated the actual scenario observed. There is no data concerning linkage of Sox2 and Sox14 in poriferans and cnidarians, as well as in amphibians. For birds, the linkage of both genes was confirmed only in Gallus gallus. Among fish species Sox2 and Sox14 genes are not linked in zebrafish and both conditions (linkage and non-linkage) were detected among cichlids (present work). Both linkage and non-linkage of Sox2 and Sox14 genes were detected in several mammals (see Table 2 for details).
The integration of cytogenetic mapping and comparative genomics of Sox genes in other vertebrates would further improve our understanding of the structure, organization and evolution of Sox genes. Unfortunately deep analysis integrating cytogenetics and genomic data were not possible for cichlids because there is no large scale genomic data available for the family yet. Although small chromosome variations were observed in the location of Sox2 among Pseudocrenilabrinae cichlids, it seems that Sox14 occupies a more dynamic genomic region resulting in variations in its chromosomal position among the species.
Supplementary Material
Acknowledgments
This study was supported by Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP), Coordenadoria de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) from Brazil.
Abbreviations
- BAC
bacterial artificial chromosome
- BLAST
Basic local alignment search tool
- CLDN18
claudin 18 gene
- CNS
central nervous system
- DAPI
4′,6-diamidino-2-phenyloindole
- DIG
digoxygenin
- DNAJC19
DnaJ (Hsp40) homolog gene, subfamily C, member 19
- DZIP1L
DAZ interacting protein 1-like gene
- FISH
fluorescence in situ hybridization
- FITC
fluoroscein isothiocyanate
- FR-LGI
Fish Room of the Laboratório Genômica Integrativa
- FXR1
autosomal homolog 1 gene of fragile X mental retardation
- HMG
high mobility group
- LG
linkage group
- MCCC1
methylcrotonoyl-CoA carboxylase 1 (alpha) gene
- NCBI
National Center for Biotechnology Information
- SSC
saline-sodium citrate
- TAF
Tropical Aquaculture Facility
- TTC14
tetratricopeptide repeat domain 14 gene
- UMD
University of Maryland
- UNESP
São Paulo State University
- WGA
whole genome amplification
References
- Bertollo LAC, Takahashi CS, Moreira-Filho O. Cytotaxonomic consideration on Hoplias lacerdae (Pisces, Erythrinidae) Braz J Genet. 1978;1:103–120. [Google Scholar]
- Bowles J, Schepers G, Koopman P. Phylogeny of the Sox family of developmental transcription factors based on sequence and structural indicators. Dev Biol. 2000;227:239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- Cnaani A, Lee BY, Ozouf-Costaz C, Bonillo C, Baroiller JF, D’Cotta H, Kocher TD. Mapping of Sox2 and Sox14 in Tilapia (Oreochromis spp.) Sex Dev. 2007;1:207–210. doi: 10.1159/000102109. [DOI] [PubMed] [Google Scholar]
- Collignon J, Sockanathan S, Hacker A, et al. A comparison of the properties of Sox-3 with Sry and two related genes, Sox-1 and Sox-2. Development. 1996;122:509–520. doi: 10.1242/dev.122.2.509. [DOI] [PubMed] [Google Scholar]
- Guo B, Tong C, He S. Sox genes evolution in closely related young tetraploid cyprinid fishes and their diploid relative. Gene. 2009;439:102–112. doi: 10.1016/j.gene.2009.02.016. [DOI] [PubMed] [Google Scholar]
- Guth SIE, Wegner M. Having it both ways: Sox protein function between conservation and innovation. Cell Mol Life Sci. 2008;65:3000–3018. doi: 10.1007/s00018-008-8138-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargrave M, James K, Nield K, et al. Fine mapping of the neurally expressed gene Sox14 to human 3q23, relative to three congenital diseases. Hum Genet. 2000;106:432–439. doi: 10.1007/s004390000266. [DOI] [PubMed] [Google Scholar]
- Hope RM, Cooper S, Wainwright B. Globin macromolecular sequence in marsupials and monotremes. In: Graves JAM, Hope RM, Cooper DW, editors. Mammals from pouches and eggs: genetic breeding and the evolution of marsupials and monotremes. CSIRO Press; Melbourne: 1990. pp. 147–171. [Google Scholar]
- Howard-Ashby M, Materna SC, Brown CT, Chen L, Cameron RA, Davidson EH. Gene families encoding transcription factors expressed in early development of Strongylocentrotus purpuratus. Dev Biol. 2006;300:90–107. doi: 10.1016/j.ydbio.2006.08.033. [DOI] [PubMed] [Google Scholar]
- Kiefer JC. Back to Basics: Sox Genes. Dev Dyn. 2007;236:2356–2366. doi: 10.1002/dvdy.21218. [DOI] [PubMed] [Google Scholar]
- Kirby PJ, Waters PD, Delbridge M, Svartman M, Stewart AN. Cloning and mapping of platypus Sox2 and Sox14: insights into Sox group B evolution. Cytogenet Genome Res. 2002;98:96–100. doi: 10.1159/000068539. [DOI] [PubMed] [Google Scholar]
- Kishi M, Mizuseki K, Sasai N, et al. Requirement of Sox2-mediated signaling for differentiation of early Xenopus neuroectoderm. Development. 2000;127:791–800. doi: 10.1242/dev.127.4.791. [DOI] [PubMed] [Google Scholar]
- Koopman P, Schepers G, Brenner S, Venkatesh B. Origin and diversity of the Sox transcription factor gene family: Genome-wide analysis in Fugu rubripes. Gene. 2004;328:177–186. doi: 10.1016/j.gene.2003.12.008. [DOI] [PubMed] [Google Scholar]
- Larroux C, Luke GN, Koopman P, Rokhsar DS, Shimeld SM, Degnan BM. Genesis and expansion of metazoan transcription factor gene classes. Mol Biol Evol. 2008;25:980–996. doi: 10.1093/molbev/msn047. [DOI] [PubMed] [Google Scholar]
- McKimmie C, Woerfel G, Russell S. Conserved genomic organisation of Group B Sox genes in insects. BMC Genet. 2005;19:6–26. doi: 10.1186/1471-2156-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meulemans D, Bronner-Fraser M. The amphioxus SoxB family: implications for the evolution of vertebrate placodes. Int J Biol Sci. 2007;3:356–364. doi: 10.7150/ijbs.3.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda Y, Yoda H, Uchikawa M, et al. Comparative genomic and expression analysis of group B1 Sox genes in zebrafish indicates their diversification during vertebrate evolution. Dev Dyn. 2006;235:811–825. doi: 10.1002/dvdy.20678. [DOI] [PubMed] [Google Scholar]
- Poletto AB, Ferreira IA, Martins C. The B chromosomes of the African cichlid fish Haplochromis obliquidens harbour 18S rRNA gene copies. BMC Genet. 2010a;11:1. doi: 10.1186/1471-2156-11-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poletto AB, Ferreira IA, Cabral-de-Mello DC, et al. Chromosome differentiation patterns during cichlid fish evolution. BMC Genet. 2010b;11:50. doi: 10.1186/1471-2156-11-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popovic J, Stevanovic M. Remarkable evolutionary conservation of Sox14 orthologues. J Genet. 2009;88:15–24. doi: 10.1007/s12041-009-0003-4. [DOI] [PubMed] [Google Scholar]
- Satou Y, Satoh N. Cataloging transcription factor and major signaling molecule genes for functional genomic studies in Ciona intestinalis. Dev Genes Evol. 2005;215:580–596. doi: 10.1007/s00427-005-0016-9. [DOI] [PubMed] [Google Scholar]
- Taylor JS, Braasch I, Frickey T, Meyer A, Van de Peer Y. Genome duplication, a trait shared by 22,000 species of ray-finned fish. Genome Res. 2003;13:382–390. doi: 10.1101/gr.640303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith WL, Chakrabarty P, Sparks JS. Phylogeny, taxonomy, and evolution of Neotropical cichlids (Teleostei: Cichlidae: Cichlinae) Cladistics. 2008;24:625–641. [Google Scholar]
- Uchikawa M, Kamachi Y, Kondoh H. Two distinct group B SoxSox genes for transcriptional activators and repressors: their expression during embryonic organogenesis of the chicken. Mech Dev. 1999;84:103–120. doi: 10.1016/s0925-4773(99)00083-0. [DOI] [PubMed] [Google Scholar]
- Wei L, Cheng D, Li D, et al. Identification and characterization of Sox genes in the silkworm, Bombyx mori. Mol Biol Rep. 2011;38:3573–3584. doi: 10.1007/s11033-010-0468-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.