Abstract
E2 enzymes catalyze the ATP-dependent polymerization of polyubiquitin chains which function as molecular signals in the regulation of numerous cellular processes. Here we present a method that uses genetically encoded unnatural amino acids to halt and re-start ubiquitin polymerization providing access to natural-linkage, precision-length ubiquitin chains that can be used for biochemical, structural, and dynamics studies.
Ubiquitin (Ub) is a small 76 amino acid protein that is central to the regulation of numerous cellular processes. The C-terminus of Ub can be attached to lysine residues of a target protein via an isopeptide bond by the combined actions of Ub-activating (E1) and conjugating (E2) enzymes and Ub ligases (E3). Monoubiquitinated proteins can then serve as substrates for further modification by E1 and E2 to generate proteins tagged with various polyubiquitins (poly-Ubs). Because each Ub monomer contains seven lysine residues (K6, K11, K27, K29, K33, K48, and K63), the extent and regiospecificity of ubiquitination can be highly varied, and as such can direct proteins to different fates within the cell.1–3 The specificity of this process is largely governed by E2 and E3 enzymes,4 many of which remain to be characterized.
Despite significant progress in elucidating various cellular processes regulated by ubiquitin-mediated signaling pathways, the mechanisms underlying specific recognition of poly-Ub signals by their receptors remain to be understood.2 Structural studies of these mechanisms require access to poly-Ub chains of defined length and linkage. Homogeneous K11-, K48-, or K63-linked poly-Ubs can be assembled using the conjugating enzymes Ube2S, E2-25K, or Ubc13/Mms2, respectively.5–7 While these enzymes are known to be linkage specific, the method is limited by the inability to halt an enzymatic assembly at defined lengths with the natural substrates. For example, the activity of E1/E2-25K will polymerize Ub to high molecular weight oligomers of various lengths. A partial solution to this problem can be achieved by employing “chain terminators”, for example, a mutant Ub such as K48C or K48R, which is not a substrate for further polymerization.6,8 In the case of the K48C mutation, the terminator-cysteine can be converted to a lysine mimic by reaction with ethyleneimine.8 This produces a surrogate linkage with different geometrical and electronic properties from a natural lysine residue. As the recognition of poly-Ubs may be mediated by the Ub–Ub linker itself, it is critical that poly-Ubs contain natural isopeptide linkages for biochemical and biophysical studies.9 Thus there is a significant interest in any methodologies that can produce milligram quantities of “natural” poly-Ubs of defined length and linkage, are compatible with isotopic labeling of individual monomers, and allow full control of where these individual monomers are incorporated in the chain.
Recently several groups have made impressive advances on generating synthetic Ub linkages using combinations of peptide synthesis, unnatural amino acid mutagenesis, and native chemical ligation.9–14 While powerful, many of these chemical methods produce surrogate linkages that are mimics of a natural ubiquitinated lysine residue. Furthermore, the aforementioned methods are not easily extendable to build poly-Ub chains longer than two units. In addition, total chemical synthesis is not conducive to isotopic labeling using biosynthetic precursors. With these studies in mind, our goal was to assemble a natural poly-Ub chain of defined length with high fidelity and with full control of which Ub variant is incorporated at each position in the chain.
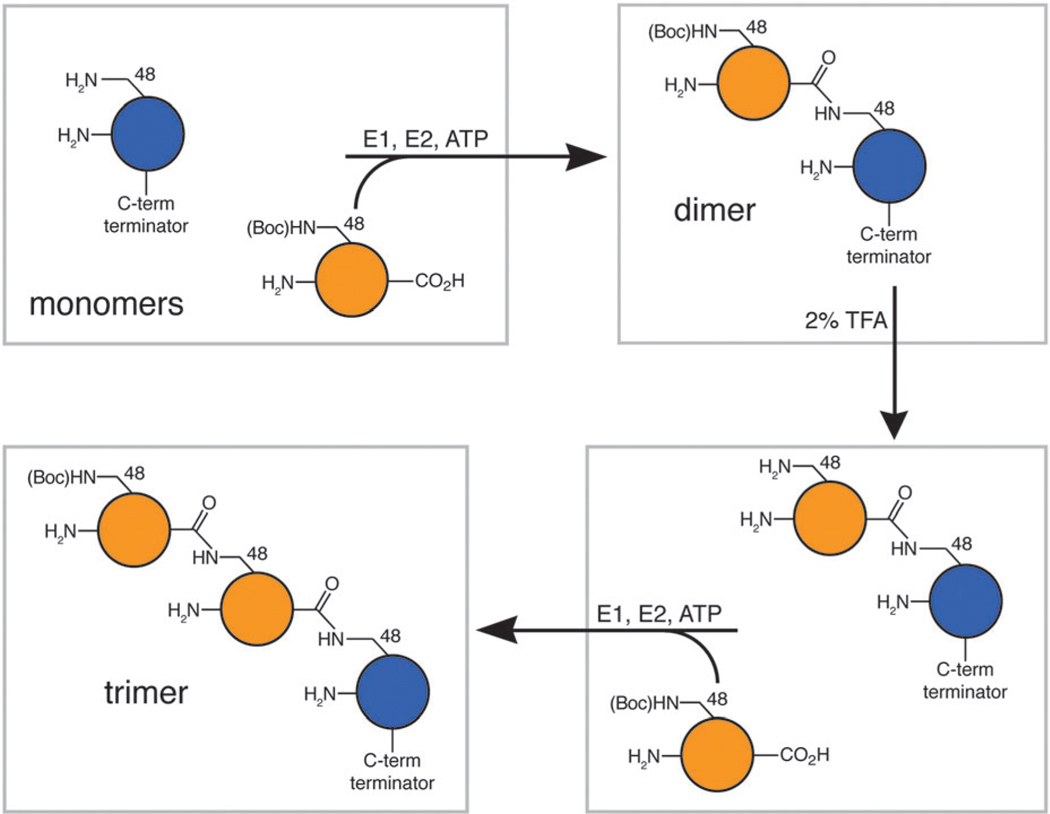
We developed a hybrid approach using modified lysines with removable protecting groups that are genetically encoded to direct enzymatic assembly of poly-Ub chains (Scheme 1). The lysine analogue Nε-(tert-butyloxycarbonyl)-l-lysine (Lys(Boc)) has been added to the genetic code of E. coli using the pyrrolysyl-tRNA-synthetase/tRNAPyl pair.15,16 This allows one to produce a homogeneous protein sample that contains a single Lys(Boc), encoded by a TAG stop codon, in the context of many other lysine residues. We produced full-length recombinant Ub containing a K48Lys(Boc) mutation (UbK48Lys(Boc)) in high yields and purity using a condensed protein expression culture.17 This Ub was then reacted in the presence of E1 and E2-25K with another (proximal) Ub containing a C-terminal modification that blocks polymerization to yield a di-ubiquitin (di-Ub) chain. As a proof of principle, we used a Ub variant (Ub1–74) lacking the two C-terminal glycines. (Another Ub variant (UbD77) can also be used as a C-terminal chain terminator which could subsequently be treated with C-terminal hydrolase to yield wild-type Ub.8) Under these conditions a new protein was produced consistent in size with di-Ub (Fig. 1). Here, Ub1–74 can only serve as the proximal unit in the chain and the Boc-protected Ub serves as the distal unit. In addition, further synthesis of longer chains is halted due to the blockage of the reactive lysine side chain, just as has been observed previously for a UbK48R variant.6,8 Importantly, neither UbK48Lys(Boc) (Fig. 1) nor Ub1–74 (not shown) alone can be polymerized by E2-25K.
Scheme 1.
Method to generate poly-Ub with natural linkages and defined length. Blue circle represents Ub with a C-terminal terminator (e.g. UbD77 or Ub1–74). Orange circle represents UbK48Lys(Boc) variant. The trimer can be then be treated with TFA and extended to longer lengths with more rounds of enzymatic assembly, as shown above.
Fig. 1.
SDS-PAGE gels showing the corresponding polymerization reactions with and without E1 as controls. (A) Full-length monomeric Ub substrates: K48Lys(Boc), K48R, K48Lys(Boc) treated with TFA, and wild-type. (B) Combinations of UbK48Lys(Boc) and Ub1–74 halt polymerization at di-Ub (Ub2) and can be iterated to produce tri-Ub (Ub3).
We performed another control experiment in which UbK48Lys(Boc) was de-protected by treatment with 2% TFA and exchanged into original buffer. As expected, when this Ub was used as a single substrate, the enzymatic reaction produced higher molecular weight oligomers, similar to those formed upon polymerization of wild type Ub (Fig. 1A).
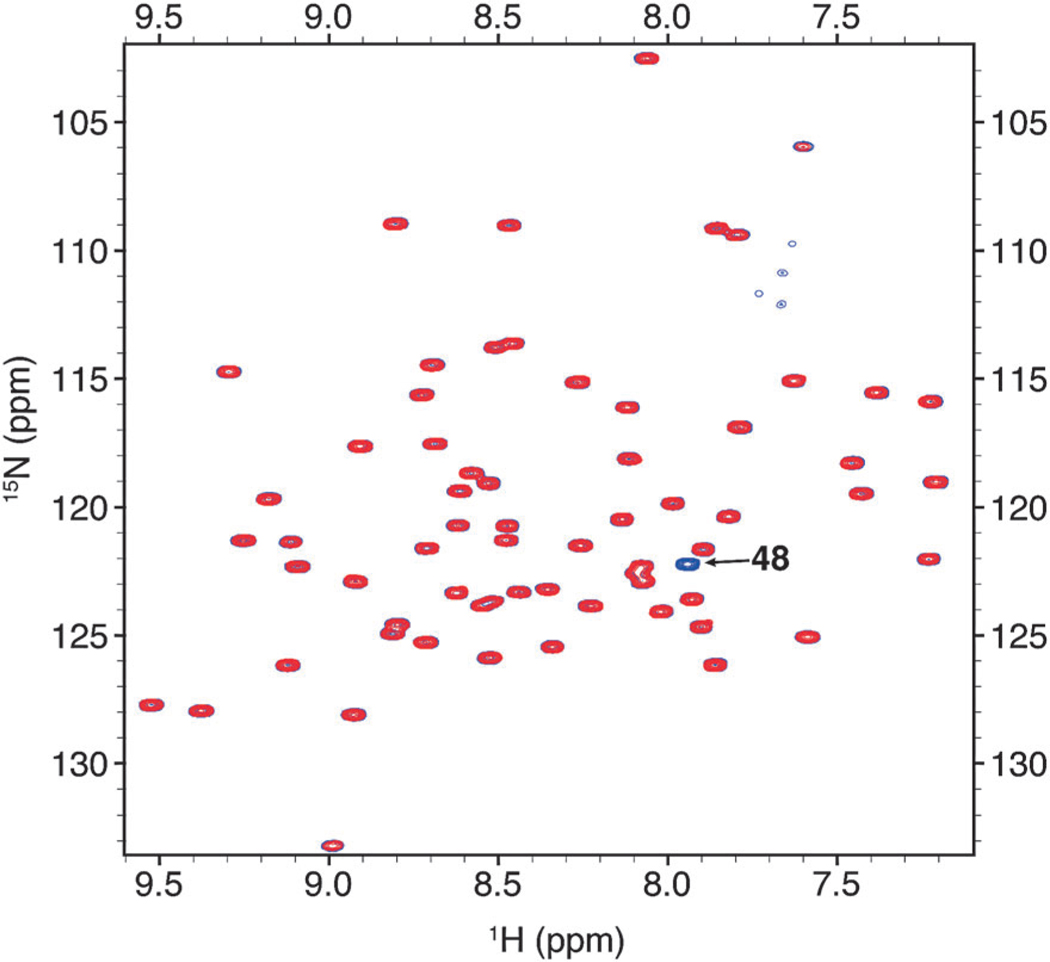
This indicates that after de-protection the protein behaves as wild-type Ub and is presumably in the native state. We then used NMR to verify that TFA-treated Ub is indeed in the native conformation. We produced Ub1–74K48Lys(Boc) that is uniformly 15N-enriched, removed the protecting group with TFA treatment (further verified by MS analysis, see ESI†), and compared its NMR spectra with those of separately prepared 15N-enriched Ub1–74. The NMR spectra (Fig. 2) of the two proteins are identical with the exception of the absent K48 signal in the spectrum of the TFA-treated sample, reflecting the site specific incorporation of unlabeled Lys(Boc).
Fig. 2.
Overlay of {15N–1H} TROSY-HSQC spectra of TFA-treated Ub1–74 K48Lys(Boc) monomer (red) and Ub1–74 monomer (blue).
As our goal is to make poly-Ub chains of various lengths, we needed to verify that the chain’s properties are not affected by TFA treatment. Therefore, we also produced a di-Ub sample using 15N-enriched Ub1–74 as the proximal Ub. NMR spectra were collected and compared with the corresponding spectra of monomeric Ub1–74 at both pH 4.5 and pH 6.8 (see Fig. S1, ESI†). The excellent spectral quality and chemical shift dispersion strongly suggest that TFA treatment did not irreversibly denature K48-linked di-Ub. Moreover, a detailed NMR analyis (see below) further confirms that the conformational properties of di-Ub are fully retained.
Encouraged by the fact that the TFA de-protection can be performed with the structure and function of both Ub and di-Ub intact, we de-protected a Boc-terminated di-Ub chain (as assembled above and shown in Scheme 1) and used it as a substrate to synthesize a tri-Ub. The K48-blocked di-Ub was purified by size-exclusion chromatography to homogeneity, subjected to TFA de-protection, and finally re-buffered to the original conditions. This sample should resemble a “native” di-Ub, and serve as a substrate for E2-25K. Indeed when the sample is combined with a second equivalent of UbK48Lys(Boc) in an enzymatic reaction, a trimeric species is formed, accompanied by a loss of di-Ub and UbK48Lys(Boc) substrates (Fig. 1B). This indicates that the de-protection reaction is quantitative (verified by MS analysis, see ESI†) and the deblocked lysine residue can subsequently be used as a reactive nucleophile in the enzymatic reaction. This also confirms that the TFA-treated di-Ub is of native conformation such that it can be recognized by E2-25K as a substrate. These results clearly demonstrate that our approach can be used iteratively to build higher order chains of precise length and composition.
Because poly-Ub chains are intrinsically flexible, and this flexibility is vital for their ability to recognize receptors in a linkage-specific manner, it is critical that the synthesized chains retain the conformational properties and inter-domain interactions characteristic of the native polyUb. Two defining properties of K48-linked di-Ub are (i) the existence of a hydrophobic Ub/Ub interface centered at residues L8, I44, and V7018,19 and (ii) the dynamic nature of K48-linked di-Ub which is in a pH-controlled equilibrium between a closed conformation (predominant at pH 6.8) and open conformation(s) (predominant at pH 4.5).18,20–22 These properties can be readily determined using NMR spectroscopy.18
Our NMR data (Fig. S1, ESI†) demonstrate that the synthesized di-Ub exhibits all of these properties. At pH 6.8, many residues in the proximal Ub show significant spectral perturbations with respect to monomeric Ub1–74 (Fig. S1B and S1D, ESI†). The perturbations center around the hydrophobic-interface residues L8, I44 and V70, in strong agreement with the closed conformation of K48-linked di-Ub.18 At pH 4.5 (Fig. S1A and S1C, ESI†), significant chemical shift perturbations are only observed for residues near the site of the K48 isopeptide linkage. The lack of perturbations for residues centered around L8 and V70 serves as the signature for the absence of the hydrophobic interface, in full agreement with previous observations for other K48-linked di-Ub constructs.18 Together, our NMR data prove that K48-linked di-Ub assembled from Ub K48Lys(Boc) and Ub1–74 exhibits the defining characteristics of K48-linked di-Ub constructed by other means (see ESI†).
In conclusion, we have developed a modified enzymatic protocol for the synthesis of poly-Ub chains. The enzymatic products are directed by genetically encoded protecting groups that can be removed with conditions compatible with biomacromolecules. Importantly, this method (i) produces poly-Ub chains of natural Ub linkages, (ii) allows precise control of chain length, (iii) provides precise control of the chain composition (for example, incorporation of isotope-labeled or mutated Ub at a desired position in the chain), and (iv) is easy to implement. Moreover, this method is capable of producing milligram quantities of products. We expect that the yield of product from our method is limited only by the efficiency of the E1 and E2 enzymes used.
Our method relies on the availability of a linkage-specific conjugating enzyme. Intriguingly, while many of the E2 enzymes remain uncharacterized, a growing number have been shown to be highly promiscuous and capable of targeting different lysine residues.4 Moreover it has been shown that the promiscuity can be amplified by using blocked substrates, such as arginine mutants.4 Using the strategy described here it might be possible to capitalize on promiscuous E2 enzymes and block multiple lysines simultaneously to direct the synthesis of atypical Ub linkages. We are currently exploring this possibility in our laboratory. With our approach and other recently published methods (see above) it might be easier than ever to study the structural basis for the complexity of Ub signaling.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (GM084396 to T.A.C and GM065334 to D.F.) and the National Science Foundation (CHE-0848398 to T.A.C and NSF Postdoctoral Fellowship DBI-0905967 to C.A.C.).
Footnotes
Electronic supplementary information (ESI) available: Supporting experimental procedures, figures showing chemical shift perturbations of K48-linked di-Ubs, and MS data.
Notes and references
- 1.Pickart CM. Annu. Rev. Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 2.Pickart CM, Fushman D. Curr. Opin. Chem. Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 3.Ye Y, Rape M. Nat. Rev. Mol. Cell Biol. 2009;10:755–764. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.David Y, Ziv T, Admon A, Navon A. J. Biol. Chem. 2010;285:8595–8604. doi: 10.1074/jbc.M109.089003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williamson A, Wickliffe KE, Mellone BG, Song L, Karpen GH, Rape M. Proc. Natl. Acad. Sci. U.S.A. 2009;106:18213–18218. doi: 10.1073/pnas.0907887106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piotrowski J, Beal R, Hoffman L, Wilkinson KD, Cohen RE, Pickart CM. J. Biol. Chem. 1997;272:23712–23721. doi: 10.1074/jbc.272.38.23712. [DOI] [PubMed] [Google Scholar]
- 7.Hofmann RM, Pickart CM. J. Biol. Chem. 2001;276:27936–27943. doi: 10.1074/jbc.M103378200. [DOI] [PubMed] [Google Scholar]
- 8.Pickart CM, Raasi S. Methods Enzymol. 2005;399:21–36. doi: 10.1016/S0076-6879(05)99002-2. [DOI] [PubMed] [Google Scholar]
- 9.Virdee S, Ye Y, Nguyen DP, Komander D, Chin JW. Nat. Chem. Biol. 2010;6:750–757. doi: 10.1038/nchembio.426. [DOI] [PubMed] [Google Scholar]
- 10.Li X, Fekner T, Ottesen JJ, Chan MK. Angew. Chem., Int. Ed. 2009;48:9184–9187. doi: 10.1002/anie.200904472. [DOI] [PubMed] [Google Scholar]
- 11.Yang R, Pasunooti KK, Li F, Liu XW, Liu CF. J. Am. Chem. Soc. 2009;131:13592–13593. doi: 10.1021/ja905491p. [DOI] [PubMed] [Google Scholar]
- 12.Kumar KS, Spasser L, Erlich LA, Bavikar SN, Brik A. Angew. Chem., Int. Ed. 2010;49:9126–9131. doi: 10.1002/anie.201003763. [DOI] [PubMed] [Google Scholar]
- 13.Ajish Kumar KS, Haj-Yahya M, Olschewski D, Lashuel HA, Brik A. Angew. Chem., Int. Ed. 2009;48:8090–8094. doi: 10.1002/anie.200902936. [DOI] [PubMed] [Google Scholar]
- 14.Chatterjee C, McGinty RK, Fierz B, Muir TW. Nat.Chem. Biol. 2010;6:267–269. doi: 10.1038/nchembio.315. [DOI] [PubMed] [Google Scholar]
- 15.Yanagisawa T, Ishii R, Fukunaga R, Kobayashi T, Sakamoto K, Yokoyama S. Chem. Biol. 2008;15:1187–1197. doi: 10.1016/j.chembiol.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 16.Wan W, Huang Y, Wang Z, Russell WK, Pai PJ, Russell DH, Liu WR. Angew. Chem., Int. Ed. 2010;49:3211–3214. doi: 10.1002/anie.201000465. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Castaneda CA, Wilkins BJ, Fushman D, Cropp TA. Bioorg. Med. Chem. Lett. 2010;20:5613–5616. doi: 10.1016/j.bmcl.2010.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Varadan R, Walker O, Pickart C, Fushman D. J. Mol. Biol. 2002;324:637–647. doi: 10.1016/s0022-2836(02)01198-1. [DOI] [PubMed] [Google Scholar]
- 19.Cook WJ, Jeffrey LC, Carson M, Chen Z, Pickart CM. J. Biol. Chem. 1992;267:16467–16471. doi: 10.2210/pdb1aar/pdb. [DOI] [PubMed] [Google Scholar]
- 20.Ryabov Y, Fushman D. Magn. Reson. Chem. 2006;44:S143–S151. doi: 10.1002/mrc.1822. [DOI] [PubMed] [Google Scholar]
- 21.Ryabov Y, Fushman D. Proteins: Struct., Funct., Bioinf. 2006;63:787–796. doi: 10.1002/prot.20917. [DOI] [PubMed] [Google Scholar]
- 22.Lam YA, DeMartino GN, Pickart CM, Cohen RE. J. Biol. Chem. 1997;272:28438–28446. doi: 10.1074/jbc.272.45.28438. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.