Abstract
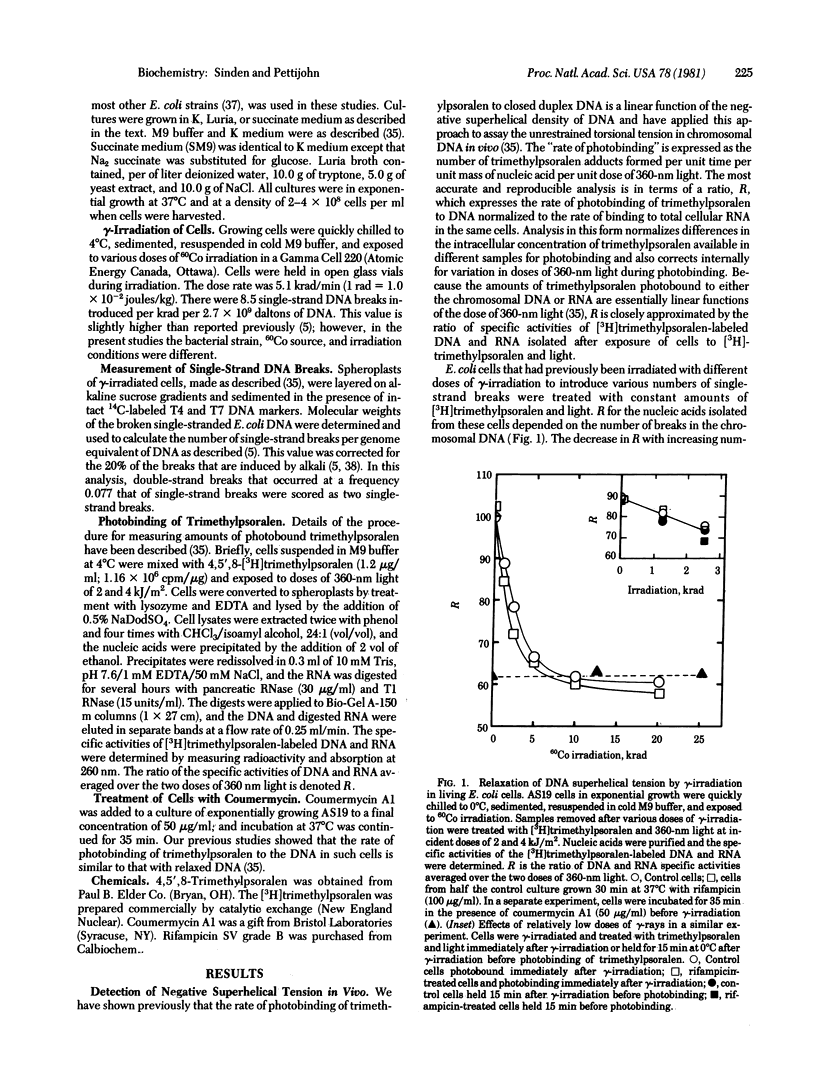
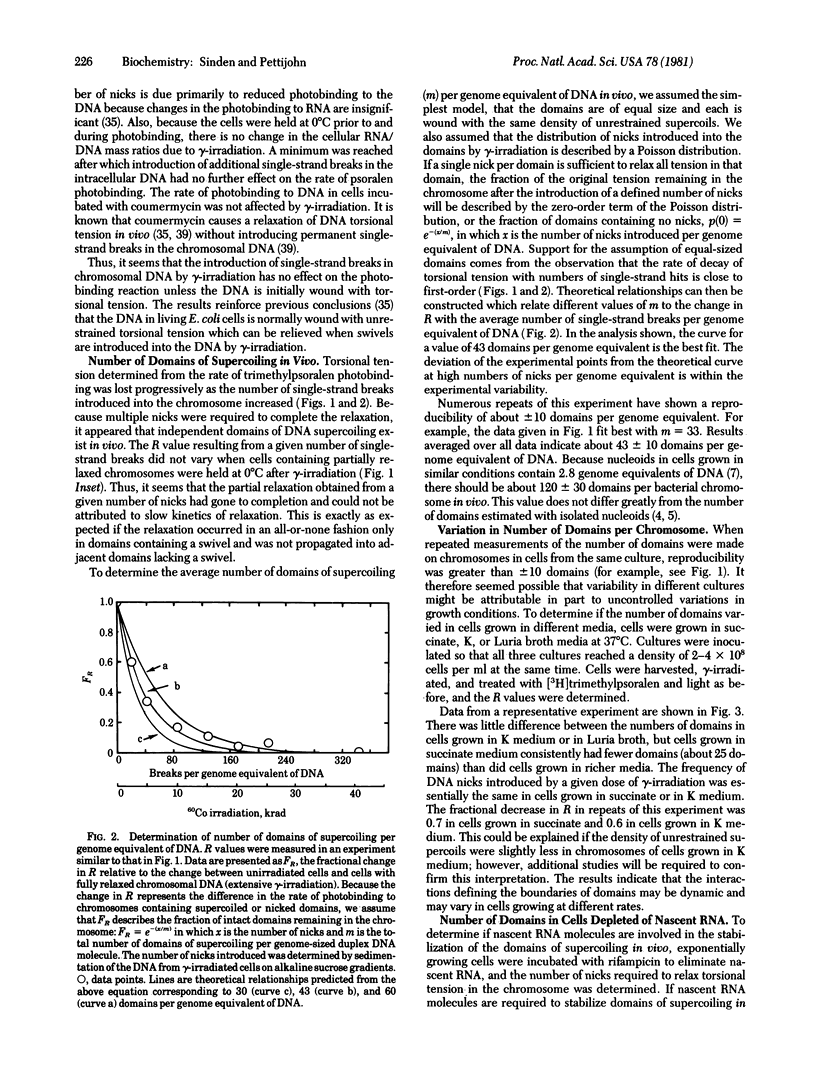
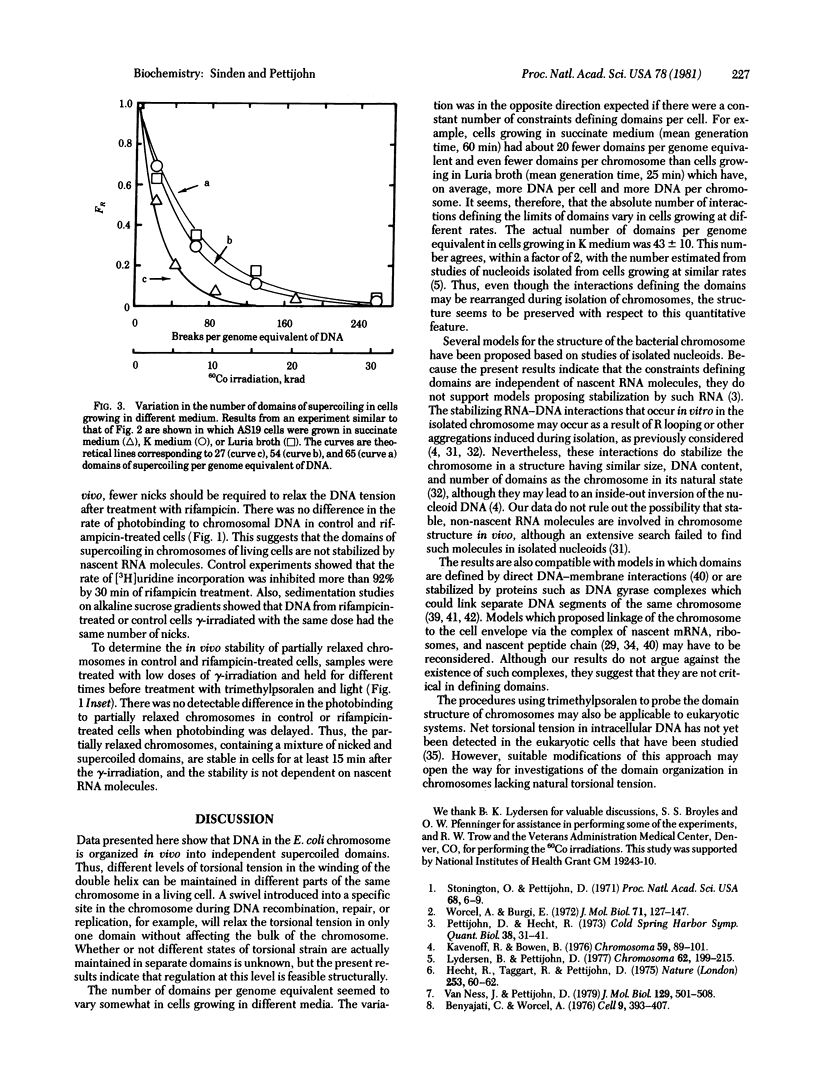
Torsional tension in the DNA double helix can be detected in living cells of Escherichia coli from measurements of the rate of trimethylpsoralen photobinding to the intracellular DNA. Here we show that this tension is relaxed in vivo when single-strand DNA breaks are introduced by gamma-irradiation and that approximately 160 nicks per genome equivalent of DNA are required to relax greater than 95% of the tension. Chromosomes containing less than 160 nicks per genome equivalent lose only a part of the tension, depending on the number of nicks. The remaining tension is maintained during incubations of cells at 0 degrees C. Chromosomes with tension relaxed by incubation of cells with inhibitors of DNA gyrase interact with the trimethylpsoralen probe independently of the number of nicks introduced by gamma-irradiation. The results fit a model in which the chromosome in growing E. coli cells (mean generation time, 30 min) is segregated into 43 +/- 10 domains of supercoiling per genome equivalent of DNA or 120 +/- 30 domains per nucleoid. The number of domains is unchanged in cells depleted of nascent RNA by growth with rifampicin, but varies somewhat in cells growing at different rates in different media.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bauer W. R., Ressner E. C., Kates J., Patzke J. V. A DNA nicking-closing enzyme encapsidated in vaccinia virus: partial purification and properties. Proc Natl Acad Sci U S A. 1977 May;74(5):1841–1845. doi: 10.1073/pnas.74.5.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benyajati C., Worcel A. Isolation, characterization, and structure of the folded interphase genome of Drosophila melanogaster. Cell. 1976 Nov;9(3):393–407. doi: 10.1016/0092-8674(76)90084-2. [DOI] [PubMed] [Google Scholar]
- Blumenthal A. B., Kriegstein H. J., Hogness D. S. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harb Symp Quant Biol. 1974;38:205–223. doi: 10.1101/sqb.1974.038.01.024. [DOI] [PubMed] [Google Scholar]
- Cook P. R., Brazell I. A. Supercoils in human DNA. J Cell Sci. 1975 Nov;19(2):261–279. doi: 10.1242/jcs.19.2.261. [DOI] [PubMed] [Google Scholar]
- Drlica K., Snyder M. Superhelical Escherichia coli DNA: relaxation by coumermycin. J Mol Biol. 1978 Apr 5;120(2):145–154. doi: 10.1016/0022-2836(78)90061-x. [DOI] [PubMed] [Google Scholar]
- Drlica K., Worcel A. Conformational transitions in the Escherichia coli chromosome: analysis by viscometry and sedimentation. J Mol Biol. 1975 Oct 25;98(2):393–411. doi: 10.1016/s0022-2836(75)80126-4. [DOI] [PubMed] [Google Scholar]
- Dworsky P., Schaechter M. Effect of rifampin on the structure and membrane attachment of the nucleoid of Escherichia coli. J Bacteriol. 1973 Dec;116(3):1364–1374. doi: 10.1128/jb.116.3.1364-1374.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hand R. Regulation of DNA replication on subchromosomal units of mammalian cells. J Cell Biol. 1975 Jan;64(1):89–97. doi: 10.1083/jcb.64.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi Y., Hayashi M. Template activities of the phi X-174 replicative allomorphic deoxyribonucleic acids. Biochemistry. 1971 Nov;10(23):4212–4218. doi: 10.1021/bi00799a009. [DOI] [PubMed] [Google Scholar]
- Hays J. B., Boehmer S. Antagonists of DNA gyrase inhibit repair and recombination of UV-irradiated phage lambda. Proc Natl Acad Sci U S A. 1978 Sep;75(9):4125–4129. doi: 10.1073/pnas.75.9.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht R. M., Pettijohn D. E. Studies of DNA bound RNA molecules isolated from nucleoids of Escherichia coli. Nucleic Acids Res. 1976 Mar;3(3):767–788. doi: 10.1093/nar/3.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht R. M., Taggart R. T., Pettijohn D. E. Size and DNA content of purfied E. coli nucleoids observed by fluorencence microscopy. Nature. 1975 Jan 3;253(5486):60–62. doi: 10.1038/253060a0. [DOI] [PubMed] [Google Scholar]
- Holloman W. K., Radding C. M. Recombination promoted by superhelical DNA and the recA gene of Escherichia coli. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3910–3914. doi: 10.1073/pnas.73.11.3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori T., Lark K. G. Effect of puromycin on DNA replication in Chinese hamster cells. J Mol Biol. 1973 Jul 5;77(3):391–404. doi: 10.1016/0022-2836(73)90446-4. [DOI] [PubMed] [Google Scholar]
- Ide T., Nakane M., Anzai K., Ando T. Supercoiled DNA folded by non-histone proteins in cultured mammalian cells. Nature. 1975 Dec 4;258(5534):445–447. doi: 10.1038/258445a0. [DOI] [PubMed] [Google Scholar]
- Kavenoff R., Bowen B. C. Electron microscopy of membrane-free folded chromosomes from Escherichia coli. Chromosoma. 1976 Dec 16;59(2):89–101. doi: 10.1007/BF00328479. [DOI] [PubMed] [Google Scholar]
- Kleppe K., Ovrebö S., Lossius I. The bacterial nucleoid. J Gen Microbiol. 1979 May;112(1):1–13. doi: 10.1099/00221287-112-1-1. [DOI] [PubMed] [Google Scholar]
- Kreuzer K. N., Cozzarelli N. R. Escherichia coli mutants thermosensitive for deoxyribonucleic acid gyrase subunit A: effects on deoxyribonucleic acid replication, transcription, and bacteriophage growth. J Bacteriol. 1979 Nov;140(2):424–435. doi: 10.1128/jb.140.2.424-435.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibowitz P. J., Schaechter M. The attachment of the bacterial chromosome to the cell membrane. Int Rev Cytol. 1975;41:1–28. doi: 10.1016/s0074-7696(08)60964-x. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Liu C. C., Alberts B. M. Type II DNA topoisomerases: enzymes that can unknot a topologically knotted DNA molecule via a reversible double-strand break. Cell. 1980 Mar;19(3):697–707. doi: 10.1016/s0092-8674(80)80046-8. [DOI] [PubMed] [Google Scholar]
- Lydersen B. K., Pettijohn D. E. Interactions stabilizing DNA tertiary structure in the Escherichia coli chromosome investigated with ionizing radiation. Chromosoma. 1977 Jul 8;62(3):199–215. doi: 10.1007/BF00286044. [DOI] [PubMed] [Google Scholar]
- Mattern M. R., Painter R. B. Dependence of mammalian DNA replication on DNA supercoiling. II. Effects of novobiocin on DNA synthesis in Chinese hamster ovary cells. Biochim Biophys Acta. 1979 Jul 26;563(2):306–312. doi: 10.1016/0005-2787(79)90049-2. [DOI] [PubMed] [Google Scholar]
- Meyer M., De Jong M. A., Woldringh C. L., Nanninga N. Factors affecting the release of folded chromosomes from Escherichia coli. Eur J Biochem. 1976 Apr 1;63(2):469–475. doi: 10.1111/j.1432-1033.1976.tb10249.x. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K., Nash H. A. Restriction assay for integrative recombination of bacteriophage lambda DNA in vitro: requirement for closed circular DNA substrate. Proc Natl Acad Sci U S A. 1976 Oct;73(10):3524–3528. doi: 10.1073/pnas.73.10.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson M. C., Roozen K. J., Setlow R. B. -Ray-induced chain breaks and alkali-labile bonds in lambda d upsilon DNA of Escherichia coli minicells irradiated under aerobic and anoxic conditions. Int J Radiat Biol Relat Stud Phys Chem Med. 1973 May;23(5):495–508. doi: 10.1080/09553007314550571. [DOI] [PubMed] [Google Scholar]
- Pettijohn D. E., Hecht R. RNA molecules bound to the folded bacterial genome stabilize DNA folds and segregate domains of supercoiling. Cold Spring Harb Symp Quant Biol. 1974;38:31–41. doi: 10.1101/sqb.1974.038.01.006. [DOI] [PubMed] [Google Scholar]
- Pettijohn D. E. Prokaryotic DNA in nucleoid structure. CRC Crit Rev Biochem. 1976 Nov;4(2):175–202. doi: 10.3109/10409237609105458. [DOI] [PubMed] [Google Scholar]
- Piñon R., Salts Y. Isolation of folded chromosomes from the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2850–2854. doi: 10.1073/pnas.74.7.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povirk L. F., Painter R. B. The effect of 313 nanometer light on initiation of replicons in mammalian cell DNA containing bromodeoxyuridine. Biochim Biophys Acta. 1976 May 19;432(3):267–272. doi: 10.1016/0005-2787(76)90135-0. [DOI] [PubMed] [Google Scholar]
- Ryan M. J. Coumermycin A1: A preferential inhibitor of replicative DNA synthesis in Escherichia coli. I. In vivo characterization. Biochemistry. 1976 Aug 24;15(17):3769–3777. doi: 10.1021/bi00662a020. [DOI] [PubMed] [Google Scholar]
- Sanzey B. Modulation of gene expression by drugs affecting deoxyribonucleic acid gyrase. J Bacteriol. 1979 Apr;138(1):40–47. doi: 10.1128/jb.138.1.40-47.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi M., Iida S. Mutants of Escherichia coli permeable to actinomycin. Proc Natl Acad Sci U S A. 1967 Dec;58(6):2315–2320. doi: 10.1073/pnas.58.6.2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J. A. Molecular model for the transposition and replication of bacteriophage Mu and other transposable elements. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1933–1937. doi: 10.1073/pnas.76.4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinden R. R., Carlson J. O., Pettijohn D. E. Torsional tension in the DNA double helix measured with trimethylpsoralen in living E. coli cells: analogous measurements in insect and human cells. Cell. 1980 Oct;21(3):773–783. doi: 10.1016/0092-8674(80)90440-7. [DOI] [PubMed] [Google Scholar]
- Smith C. L., Kubo M., Imamoto F. Promoter-specific inhibition of transcription by antibiotics which act on DNA gyrase. Nature. 1978 Oct 5;275(5679):420–423. doi: 10.1038/275420a0. [DOI] [PubMed] [Google Scholar]
- Snyder M., Drlica K. DNA gyrase on the bacterial chromosome: DNA cleavage induced by oxolinic acid. J Mol Biol. 1979 Jun 25;131(2):287–302. doi: 10.1016/0022-2836(79)90077-9. [DOI] [PubMed] [Google Scholar]
- Staudenbauer W. L. Letters to the editor: Novobiocin-a specific inhibitor of semiconservative DNA replication in permeabilized Escherichia coli cells. J Mol Biol. 1975 Jul 25;96(1):201–205. doi: 10.1016/0022-2836(75)90191-6. [DOI] [PubMed] [Google Scholar]
- Stonington O. G., Pettijohn D. E. The folded genome of Escherichia coli isolated in a protein-DNA-RNA complex. Proc Natl Acad Sci U S A. 1971 Jan;68(1):6–9. doi: 10.1073/pnas.68.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ness J., Pettijohn D. E. A simple autoradiographic method for investigating long range chromosome substructure: size and number of DNA molecules in isolated nucleoids of Escherichia coli. J Mol Biol. 1979 Apr 15;129(3):501–508. doi: 10.1016/0022-2836(79)90509-6. [DOI] [PubMed] [Google Scholar]
- Wang J. C. Interaction between DNA and an Escherichia coli protein omega. J Mol Biol. 1971 Feb 14;55(3):523–533. doi: 10.1016/0022-2836(71)90334-2. [DOI] [PubMed] [Google Scholar]
- Worcel A., Burgi E. On the structure of the folded chromosome of Escherichia coli. J Mol Biol. 1972 Nov 14;71(2):127–147. doi: 10.1016/0022-2836(72)90342-7. [DOI] [PubMed] [Google Scholar]


