Abstract
Bacterial pathogens exhibit significant variation in their genomic content of virulence factors. This reflects the abundance of strategies pathogens evolved to infect host organisms by suppressing host immunity. Molecular arms-races have been a strong driving force for the evolution of pathogenicity, with pathogens often encoding overlapping or redundant functions, such as type III protein secretion effectors and hosts encoding ever more sophisticated immune systems. The pathogens' frequent exposure to other microbes, either in their host or in the environment, provides opportunities for the acquisition or interchange of mobile genetic elements. These DNA elements accessorize the core genome and can play major roles in shaping genome structure and altering the complement of virulence factors. Here, we review the different mobile genetic elements focusing on the more recent discoveries and highlighting their role in shaping bacterial pathogen evolution.
Key words: horizontal gene transfer, plasmid, transposon, integron, genomic island, insertion sequence, bacteriophage, virulence, animal pathogen, plant pathogen, pathogenicity islands
Introduction
Bacteria were the earliest forms of life on earth and the evolution of other organisms opened up a plethora of new niches for bacterial exploitation. Selection likely favored mutational variants that were able to infect a host and tap into the host's nutrients. In turn, this probably drove selection for hosts with enhanced resistance mechanisms. Over the course of millennia, many bacterial pathogens have evolved, and most, if not all, organisms have experienced bacterial infection and thus have been under selection pressure to evolve complex immune systems. Bacterial pathogens or pre-pathogens (strains that are close to gaining pathogenic potential) faced with evolving immune systems became under selection pressure themselves to rapidly adapt to—and break down—the defense barriers erected by their hosts. During this evolutionary arms race, many bacteria have accessorized their genomes with DNA from bacteria outside of their species or genus with the help of mobile genetic elements (MGE). The most common MGEs for horizontal transfer are plasmids, genomic islands and bacteriophages, most of which have strategies for enabling transfer between bacteria; consequently, these MGEs are relatively complex, usually encoding regulatory and structural mechanisms for replication and transfer. These MGEs can carry smaller and simpler insertion sequences (IS), transposons and integrons, which can facilitate genome rearrangements, gene duplications and deletions, and capture of new genes. The “accessory-metagenome” is therefore a massive resource for bacteria that provides unprecedented flexibility to improve their fitness and, potentially, pathogenicity and virulence. In this review, we illustrate the various MGEs that can influence changes in bacterial genomes with a focus on very recent discoveries on how MGEs influence bacterial pathogenicity. Finally, we outline the need to consider the wider ecology of bacterial pathogens in terms of other niches and alternative hosts that they inhabit, which leads to exposure to other MGEs that may contribute to bacterial pathogen evolution.
Mobile Genetic Elements Influence Bacterial Evolution
Plasmids: major gene movers between bacteria.
Plasmids are circular or linear DNA molecules defined by their ability to autonomously replicate in the host cell; they can appear in all domains of life, and usually carry genes encoding adaptive traits, such as resistance to antibiotics or heavy metals, degradation of aromatics, pathogenicity or the ability to exploit particular environmental niches.1,2 A network analysis of MGEs from complete genomes and environmental metagenomes indicates that plasmids, and not viruses, have been the main factors in horizontal gene transfer (HGT) between bacterial chromosomes,3 mostly because they can be transferred between distantly related organisms including transfer from bacteria to eukaryotes,4–6 they can accommodate large amounts of DNA, they code for important functions for survival, such as antibiotic resistance, and because they often impart the ability to interact with higher eukaryotes.7
Plasmids not only carry genes for selectable phenotypes, but are also efficient vehicles for the dissemination of other MGEs (also see below “Genomic islands” and “Integrons”). The availability of a growing number of plasmid sequences reveals that 5–20%, and up to 40% in extreme cases, of the DNA in plasmids larger than 20 kb correspond to IS elements, with no ISs in the majority of the smaller plasmids.8 Among other functions, IS elements can serve as mobile recombination regions, facilitating the exchange of DNA with other replicons.8,9 Mirroring the apparent preference of plasmids to propagate within a specific host clade,7 a given type of IS element is only rarely shared by distantly related prokaryotic clades.10 Plasmids can also induce the mobilization of other MGEs: in diverse Salmonella enterica serovars, which cause a diversity of human intestinal diseases, conjugative plasmids of the IncA/C incompatibility group often carry multidrug resistance (MDR) determinants and can mobilize in trans the antibiotic resistance pathogenicity island (PAI) SGI-1 at frequencies ranging from 10−3 to 10−6.11 Additionally, a new emerging genotype of S. enterica subsp. enterica serovar Typhimurium (S. Typhimurium, causing human gastroenteritis) in Mexico, genotype ST213, contains an IncA/C plasmid carrying a gene conferring resistance to extended-spectrum cephalosporins and a Class 1 integron carrying additional antibiotic resistance determinants. Although the plasmid is mobilized with a very low frequency, it is likely that the ecological success of this genotype is related to the carriage of the IncA/C plasmid.12
It is well established that plasmids in a variety of prokaryotes carry and distribute a plethora of genes conferring adaptation, such as resistance to antibacterial compounds against human, animal and plant pathogens, virulence genes, ultraviolet resistance genes, detoxifying enzymes, bacteriocins and enzymes for secondary metabolism.2,13–18 A remarkable aspect of plasmids as MGEs is that they can hold and transfer large amounts of DNA, allowing for quantum leap19,20 evolution and the acquisition of very complex phenotypes, including the transformation of non-pathogenic or low virulence bacteria into devastating pathogens of plants and animals. For example, the enterobacterium Pantoea agglomerans (syn. Enterobacter agglomerans) is a heterogeneous group including epiphytic (living on plant surfaces), commensal and opportunistic human pathogens.21 However, P. agglomerans pv. betae and pv. gypsophilae are plant pathogens that induce gall formation in several plant species due to the acquisition of a non-conjugative pathogenicity plasmid designated pPATH, which harbors most, if not all, of the genes required for tumorigenesis and host-specificity.22 The best studied pPATH, pPATHPag (ca. 135 kb) from P. agglomerans pv. gypsophilae 824-1, carries a loosely defined PAI of about 75 kb. This PAI includes a complete type III protein secretion system (T3SS) that is very similar to the one described in the plant pathogen Erwinia amylovora, with six T3SS effector (T3SE) genes and genes for the biosynthesis of the phytohormones-3-indoleacetic acid and cytokinins. The PAI also includes six IS elements, five of which were present in pathogenic but not in non-pathogenic strains of P. agglomerans, as well as remnants of known gene sequences from diverse bacteria that include Yersinia pestis and Xylella fastidiosa. This may indicate that the plasmid has been disseminated through a variety of different pathogens that typically inhabit different hosts. Another example is Vibrio, which includes species that inhabit various aquatic environments, including fish symbionts and invertebrate pathogens, and carry diverse plasmids that are essential for their pathogenesis.23 V. anguillarum serotype O1 is part of the natural microbiota of aquatic habitats, although it can cause fatal haemorrhagic septicaemia in freshwater and marine fish; these strains causing disease carry a virulence plasmid, pJM1, encoding the siderophore anguibactin biosynthesis and transport proteins, which are essential for virulence and survival in its naturally ironlimited habitat.24
Next generation sequencing is allowing the identification of the genetic changes behind plasmid adaptation to new hosts as well as the co-evolution of bacterial genomes with plasmids during the acquisition of new genes and capabilities. Concomitant with this genetic and potential phenotypic gain, the acquisition of a large amount of DNA with possibly a divergent base composition and an array of new genes can have a relevant impact in the cell metabolism, potentially imposing a hefty fitness cost that is solved by different evolutionary strategies to ensure stable plasmid maintenance. An interesting example is the conjugative plasmid pSf-R27 from S. Typhimurium.25 This plasmid has a 55% A + T content and titrates the cell global transcriptional repressor protein H-NS, which binds to A + T-rich sequences.25,26 This would normally cause a significant reduction in bacterial fitness, but this is avoided by the plasmid-encoded gene sfh, which is a paralogue of H-NS. Sfh binds to most of the H-NS targets in the chromosome, thus allowing pSf-R27 transfer with a minimal impact to the cell. In another case, controlled evolution and competition experiments showed that diverse IncP promiscuous plasmids can exhibit a shift in host range through changes in the replication protein gene, allowing the stable colonization of new bacterial hosts.27 Another study found that an IncQ plasmid has evolved to a lower copy number variant; this variant is more competitive within the bacterial population than a higher copy number variant because it places a lower metabolic load on the bacterial host.28 Likewise, transfer of the symbiotic plasmid from the β-rhizobium Cupriavidus taiwanensis to the taxonomically related plant pathogen Ralstonia solanacearum allowed the adaptive evolution of the latter, after the inactivation of the T3SS by a single or a double mutation, into a nodulating symbiont.29 Although these data resulted from artificial evolution settings, the experiments underscore the amazing plasticity of bacteriaplasmid associations and the potential for rapidly generating new phenotypes.
Bacteriophages.
Bacteriophages are viruses that infect bacteria. That bacteriophages contribute to virulence was first suggested in 1951, when Freeman30 found that avirulent strains of Corynebacterium diphtheriae, the causal agent of diphtheria, could be turned into virulent strains through infection with a bacteriophage. How a bacteriophage can turn a relatively harmless bacterium into a highly virulent pathogen became clear with the study of the cholera pathogen Vibrio cholerae, when it was found that the emergence of toxigenic strains of this species is the result of lysogenic conversion by a filamentous bacteriophage carrying the genes for cholera toxin.31 More recently, the emergence of the seventh pandemic clone of V. cholerae was probably due to the interaction with three filamentous bacteriophages and two helper bacteriophages.32 The most fascinating aspect of bacteriophages as MGEs is probably that they put their own pathogenicity on hold during lysogeny while contributing to the pathogenicity or virulence of their bacterial host, the lysogen. Brüssow33 reviews how bacteriophages might have evolved lysogeny because it allows them to lyse their hosts only when other susceptible host cells are present since otherwise lysis would lead to extinction of the host and the phage itself. Importantly, during lysogeny the prophage genome can be assumed to be under selection pressure to be eliminated since even its simple replication during cell division presents an energetic expense for the host. Therefore, Rankin et al.18 argue that only if the prophage provides a benefit to the lysogen, such as a virulence gene that enhances fitness, will selection favor prophage persistence in the host during lysogeny. This requires a tight integration of bacteriophage and lysogen gene regulation networks so that bacteriophage-encoded virulence genes are expressed only when needed during pathogenicity and bacteriophage lytic genes are only expressed when it is advantageous for the bacteriophage to lyse its host cell. For example, a previously unknown mechanism of sophisticated integration of lysogeny control with the host regulatory circuit was found in Staphylococcus aureus (a commensal and pathogen that can cause a variety of infections and diseases): the host sigma factor σH binds the promoter of the bacteriophage integrase gene of several prophages and contributes to the maintenance of their lysogenic state.34 On the other hand, prophages have been found to encode repressors of host genes, possibly contributing to the fitness of the lysogen. For example, the principal repressor, cI, of several prophages not only maintains lysogeny but also downregulates host growth rate, possibly by directly binding the promoter of the host pckA gene.35 In another example, a prophage repressor, RepR, of Clostridium difficile, a common causal agent of nosocomial infections of the intestine, has been found to bind the promoter region of the gene tcdR of the PaLoc pathogenicity island modulating toxin production.36 In a different strategy that avoids host death, many of the prophage genomes in enterohaemorrhagic E. coli (EHEC) strains and other bacteria are found to be missing genes or have mutations in genes known to be essential for a bacteriophage to enter the lytic cycle and to be horizontally transferred. These bacteriophage remnants were assumed to have in fact lost their ability to be transferred between cells. However, Asadulghani et al.37 experimentally determined that even bacteriophage remnants can be disseminated effectively to other cells, probably through various inter-prophage interactions. Moreover, the authors show that recombination between Stx1 and Stx2 genomes leads to new Stx1 bacteriophages. Another assumption that was recently proven wrong is that bacteriophages cannot be transferred between distantly related strains: Chen and Novick38 experimentally determined that bacteriophages can be horizontally transferred from S. aureus to Listeria monocytogenes (the causal agent of listeriosis).
In the last few years progress has been made in our understanding of many aspects of virulence capabilities provided to pathogens by prophages. In particular, E. coli EHEC O55:O157 has become a model for studying the role of bacteriophages in the evolution of virulence. Correlating virulence gene repertoires between very similar strains of EHEC O55:O157 with their phylogeny and the symptoms they cause, it was found that phylogeny based on single nucleotide polymorphisms (SNPs) is predictive of toxin repertoires and symptom severity.39 Moreover, the clade that is associated with more severe symptoms also had a higher ability to attach to epithelial cells and had higher virulence gene expression.40 This suggests that bacteriophage repertoires are relatively stably associated with individual lineages within EHEC O55:O157.
Genome comparisons of more distantly related E. coli strains also gave new insight into the role of bacteriophages in virulence evolution. Comparison of EHEC O55:O157 with its closest non-EHEC relative O55:H7 revealed an extreme difference in bacteriophage repertoires: while O55:H7 has 19 intact or degraded prophage genomes and O157:H7 has 23, only three are present in both.41 Importantly, O55:O157 and O55:H7 have close to 100% DNA identity in housekeeping genes revealing the unbelievable speed at which the strain's bacteriophage repertoires diverged. It is also important to point out that EHEC O55:O157 is not the only EHEC lineage within E. coli as at least 24 other E. coli lineages also acquired an enterohaemorrhagic lifestyle: the comparison of O55:O157 with three of these other lineages revealed that they independently acquired different bacteriophages that carry similar virulence gene repertoires.42 In conclusion, specific bacteriophage repertoires appear to be vertically inherited within specific bacterial lineages contributing to lineage-specific fitness and virulence39,40 while horizontal transfer of bacteriophages between phylogenetically quite distant groups allows for emergence of new pathogenic lineages with very similar phenotypes.42
Genomic islands—plasmid-bacteriophage hybrids.
Genomic islands (GIs) are a collection of large, potentially mobile regions of DNA that frequently carry virulence-related genes. There are a number of different types of GIs including PAIs and integrative and conjugative elements (ICE). In general, GIs are areas of the genome that are present only in certain strains of a bacterial species, which are often flanked by specific DNA sequences that contain direct repeats and are often inserted in highly conserved genes, e.g., tRNA genes. They also carry genes coding for genetic mobility such as plasmid and bacteriophage genes, IS elements, integrases and transposases; they may have evolved through bacteriophage and plasmid interchange leading to a hybrid structure.43–45 PAIs are present on the genomes of pathogenic strains, but absent from the genomes of nonpathogenic members of the same or related species.43 ICEs are similar to PAIs but a number of them have been demonstrated to facilitate their own conjugative transfer between bacteria.46 GIs have important implications in human health; for example, the GI OI-57 has been described as part of the ‘virulome’ of Shiga toxin-producing E. coli (STEC) which cause severe human disease, including STEC (also called EHEC) 0157, whereas this island is not present in less virulent strains.47 Additionally, ICEs have a suggested role in evolution of multidrug-resistant Streptococcus pneumoniae Spain 23F ST81 lineage.48 For example, the ICESpFST81 carries a tetM gene, responsible for the Spain 23F ST81 strain's tetracycline resistance. GIs are also important in the pathogenicity of plant pathogens: for example, Clavibacter michiganensis, a pathogen of tomato, contains a 129 kb island that is necessary for pathogenicity.49 Also, plant pathogenic Streptomyces species contain a large mobile PAI that encodes multiple virulence-associated genes, including the nitrated dipeptide phytotoxin thaxtomin biosynthetic genes and the virulence factor nec1, whose transfer to other Streptomyces species is responsible for the emergence of new pathogens in agricultural settings.50
A common PAI found in a number of animal and plant pathogenic bacteria is the T3SS PAI. The T3SS is used to translocate T3SE proteins from the bacterial cell into eukaryotic host cells to interfere with host defenses, change the metabolism of the host and cause disease. The T3SS is carried by the chromosome in the plant pathogen Pseudomonas syringae51 and can be carried on plasmids such as plasmid pPATH described above.22 Salmonella has two T3SS PAIs (SPI-1 and SPI-2) that are used to modify the host cells response. Analysis of SPI-2 has recently shown that the genes on the island are under tight regulatory control as overexpression of them can lead to attenuation of virulence in mammalian cells. It was found that this PAI has integrated into the host regulatory network and is expressed only at the critical times of infection, but switched off at other times to avoid toxicity or energy burdens.52
Investigations into GIs are helping to unravel host range restriction in a number of pathogens. Salmonella enterica Gallinarum is a pathogen with a host range specific to poultry, while S. enterica Enteritidis is a broad host range pathogen that is a leading cause of gastrointestinal salmonellosis in humans and many other species, but only colonizes poultry sub-clinically (i.e., is asymptomatic). The Gallinarum strain harbors a PAI (SPI-19) that carries a type VI secretion system (T6SS) which contributes to the colonization of the gastrointestinal tract and internal organs of chickens.53,54 SPI-19 appears to be degenerate in Enteritidis and it was postulated that the transfer of SPI-19 from Gallinarum to Enteritidis may have had a short initial positive effect on the ability of the bacterium to colonize chickens, but had a strong negative impact on its ability to colonize in the long term, leading to counter selection against components of SPI-19 and eventual degradation. This may reflect the different initial strategies the bacteria use to colonize their host and also the fact that, in the case of Gallinarum, having a fully functional SPI-19 is an advantage. It also implies a cooperative role of SPI-19: it is useful to the pathogen only when the pathogen has other particular virulence gene systems.
Streptococcus equi subspecies equi (S. equi) is a host-restricted pathogen of horses which appears to have evolved from the zoonotic pathogen S. equi subspecies zooepidemicus and shares 80% genome sequence identity with the human pathogen S. pyogenes. Comparative genomics of strains of S. equi and S. zooepidemicus revealed events that led to the emergence of S. equi.55 Amongst other changes, S. equi has gained an ICE (ICESe2) carrying a novel iron acquisition system with similarity to the high PAI of Yersinia pestis and it was postulated that this was a key speciation event in the evolution of S. equi.55 Conversely, loss of a GI can lead to the expansion of host range as demonstrated by the loss of the GI PPHGI-1 from P. syringae pv. phaseolicola (Pph) strain 1302A. When Pph 1302A infects a bean plant carrying the R3 resistance gene it triggers a resistance reaction in the plant known as hypersensitive response (HR) because of the plant's recognition of the T3SE AvrPphB (now called HopAR1) by R3:56 avrPphB is carried on PPHGI-1.57 In the stressful environment caused by the HR, PPHGI-1 can be lost from the genome of Pph 1302A. Evolved strains lacking PPHGI-1 are able to cause disease on R3-expressing bean plants and thus Pph has expanded its host range.58 The fact that PPHGI-1 is maintained in the population even at a very low frequency, suggests that carriage of this island has an advantage to the pathogen in other niches.59 In fact it has been demonstrated that PPHGI-1 is mobile and can be acquired by another Pph strain via transformation in planta.60
Bioinformatic analysis of whole genome sequences has revealed that a number of GIs are predicted to occur in any given strain and deciphering which ones are involved in virulence is a laborious task.61,62 Lloyd et al.61 investigated this by individually deleting 11 of the predicted 13 GIs in the uropathogenic E. coli strain CFT073. Three out of nine of these mutants were significantly outcompeted by the wild type following co-challenge in a mouse model for urinary tract infection, indicating a lower virulence. A focus on specific genes within these GIs showed that a number of them contributed to the fitness of the wild type. A simpler approach was taken by Diard et al.62 who characterised several mutants of the extraintestinal pathogenic E. coli strain 536 by either deletion of all seven PAIs individually or all together. They showed that although the PAIs where dispensable for growth in the absence of external stress, fitness was drastically reduced when the strain with all seven PAIs deleted was in competition with the wild-type in a mice intestine model. No reduction in virulence was observed with individual PAI deletions, suggesting that there is a redundancy of function between the PAIs. In summary, GIs can in some cases provide genes essential for pathogenicity or that enhance virulence and/or fitness, although they can have a detrimental effect if island products are recognised by the host immune system.
IS elements and transposons—construction and deconstruction agents.
Some of the simplest MGEs found in bacteria are IS elements and transposons. In their most basic form, IS elements consist of a single gene coding for a site-specific recombinase (called a transposase) and short terminal inverted repeat sequences that are recognized by the transposase for transposition. Transposons are more complex than IS elements since they carry additional genes, including virulence genes. Transposons and IS elements are usually only mobile within their host genome although conjugative transposons and/or ICE (described above) have the ability to promote their own transfer into other bacterial cells by conjugation since they carry their own conjugation genes.
In recent years, it has become clear that IS elements play an important role in pathogen evolution through genome rearrangements and in genome reduction. Salzberg et al.63 reported that a strain of the rice pathogen Xanthomonas oryzae pv. oryzae has an unusually long tandem duplication (each region is 212,087 bp long) with an IS element in between the two regions and two IS elements each flanking the other end of the two regions. The two regions are 100% identical besides one single mutation in one of the IS elements, suggesting a recent duplication involving homologous recombination likely mediated by the IS elements. Burkholderia mallei, causative agent of glanders, is an obligate intracellular pathogen of horses that evolved from the melioidosis pathogen, B. pseudomallei. The B. mallei genome is 20% smaller than the B. pseudomallei genome. Song et al.64 inferred from the comparison of multiple B. mallei and B. pseudomallei genomes that genome reduction largely occurred through homologous recombination via an IS element. A similar observation was made in Francisella tularensis, the causative agent of tularemia, where Larsson et al.65 found that IS elements provided the sites for genome rearrangements, duplications and deletions during evolution of F. tularensis subspecies, with one single IS element present up to 63 times in the same genome.
A second important role for IS elements and transposons is in disruption of virulence genes. This is important in bacterial plant pathogens where IS elements can disrupt a T3SE gene, whose product may be recognized by a plant disease resistance gene and trigger immunity.66–68 In these cases, IS elements can increase host range by abolishing T3SE-triggered immunity. Since T3SE genes are sometimes organized in operons, insertions within an operon can lead to inactivation of effectors downstream of the actual insertion. For example, an IS element in P. syringae pv. tomato, the causative agent of bacterial speck disease of tomato, inserted in the T3SE gene hopAG1, which also interfered with expression of the genes hopAH1 and hopAI1 located downstream of hopAG1 in the same operon.69
Integrons—gene capture systems.
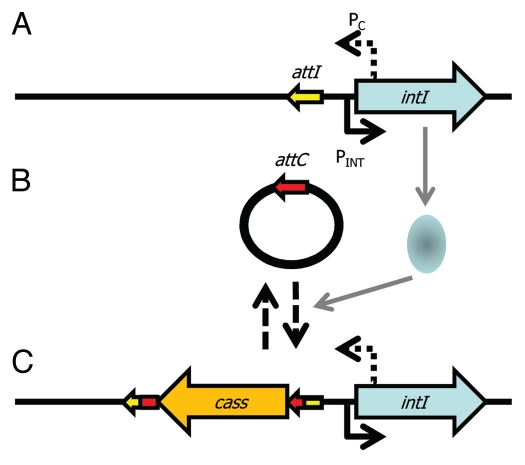
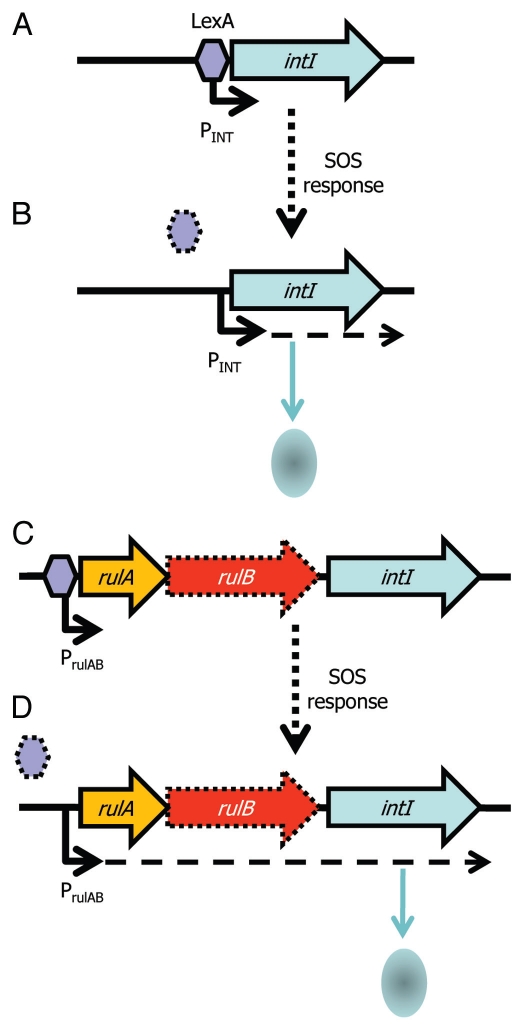
Integrons are gene capture systems, most famous for their rapid acquisition and spread of antibiotic resistance genes.70–72 Their common association with plasmids means they can be particularly promiscuous.73–79 Integrons are essentially composed of: (1) a core stable platform of a site-specific tyrosine recombinase (integrase) gene (intI) with its own promoter (Pint) and an outward facing promoter (PC) within the intI coding sequence that can express captured cassettes of gene(s),80,81 and (2) an adjacent attI recombination site (Fig. 1). The recombinase facilitates integration and excision of specific gene cassettes (DNA elements comprising one or more genes and a recombination site, attC) into the attI site. Multiple cassettes can be captured by the integron leading to the construction of large cassette arrays (reviewed in ref. 82). There can be significant co-assortment of cassettes within environmental bacteria83 highlighting the significant interchanging of genes between integrons. The gene cassette closest to intI may reflect the most recent adaptation and can be identified with a PCR based assay.84 Many of the cassettes are promoterless and integrons with small numbers of cassettes may rely on the PC promoter, while larger cassette arrays encode cassette-specific promoters that can respond to environmental signals independently of PC.81,85 A recent key discovery of the activity of integrons is that intI expression is regulated by the SOS response (Fig. 2A and B).86 The promoter of intI contains binding motifs for the transcriptional repressor LexA. LexA is derepressed under SOS conditions and thus leads to activation of intI and consequently to potential gene capture. Remarkably, plasmid conjugation triggers the SOS response and thus leads to activation of intI. This highlights an effective gene delivery and capture system for bacteria.87 Moreover, it raises the question of whether other recombinases (e.g., those of ICEs, GIs, IS elements and transposons) are also affected by conjugation and regulated by LexA and the SOS response—certainly, stress affects P. syringae PPHGI-1 integrase expression.58–60,88,89
Figure 1.
Capture of DNA by an integron. The integron core unit (A) is composed of an integrase (intI) gene, a promoter (PINT) to drive expression of intI, a promoter (PC) to drive expression of captured genes and a site for integration of genes (attI). (B) Expression of the integrase occurs during the SOS response and the integrase protein (pale blue oval) catalyses site-specific recombination of circularised gene cassettes with an attC site that matches attI so that (C) cassettes (cass) are incorporated into the integron. More cassettes can be integrated into attI and cassettes can also excise. PC can drive expression of the captured cassettes.
Figure 2.
Integron integrase expression is regulated by LexA and the SOS response. (A) The integrase (intI) gene of many integrons is preceded by a cis encoded LexA box, allowing the LexA repressor (purple hexagon) to bind upstream of intI and prevent expression of the gene. (B) Activation of the SOS response leads to derepression of intI by release and degradation of LexA (signified by dotted hexagon) and production of IntI and potential capture of gene cassettes. (C) An association of integrase genes with rulAB DNA repair genes (rulB is usually truncated, signified by dotted outline) may indicate that intI expression is controlled by the rulAB promoter (PrulAB), which is repressed by LexA and (D) relieved under SOS conditions.
Integrons have been found in a wide range of non-pathogenic and pathogenic bacteria in environments ranging from marine to terrestrial organisms and niches.73,77,83,90–101 Two types of integrons, the chromosomal integron (CI) and mobile integron (MI) have been identified:70 CIs appear to represent a “core” gene capture system in Gram negative bacteria to enable genome flexibility for adaptation to diverse environments. The five classes of MIs defined so far (based on intI sequence similarity) are associated with MGEs such as transposons and plasmids and thus provide a pool of elements for shuttling genes throughout microbial communities. MIs can rapidly transfer between members of a microbial community78 although the basis of transfer and selection are still unclear. For example, one study looking at the acquisition of antibiotic-resistance integrons by susceptible strains in the gut and intestines of humans and chickens showed that there may be a correlation in acquisition based on treatment with antibiotics102 although another study suggested this is not the case.78 Clearly, the host environment as well as the antibiotic treatment are likely to be influencing integron transfer on MGEs, a similar situation as seen with ICEs.60 An interesting observation by van der Veen et al.102 was that the prevalence of integrons within the intestinal microbial populations decreased despite continued antibiotic treatment. This may indicate that there is a cost to the bacteria of carrying either the integron or its shuttle element. It may also point to integron-based resistance playing a short term rescuer role, which is gradually superseded by the build up of antibiotic resistance through mutations elsewhere in the bacterial genome.
The impact of integrons on pathogen virulence evolution is less clear. For the purposes of this review, we do not consider antibiotic resistance to be a virulence trait, as antibiotic resistance only enhances bacterial fitness. Bona fide virulence factors such as toxins (e.g., heat-stable toxin (sto) and mannose-fucoseresistant hemagglutinin (mrhA)) are associated with Vibrio cholerae repeated sequence (VCRs) islands, which are integron-like structures.103 A VCR region encoding a gene essential for capsular polysaccharide (CPS) biosynthesis has been identified in V. vulnificus, which causes food poisoning and septicaemia, and requires CPS for protection from the mammalian immune system.104 The location of this gene within an integron implies that the gene has been captured independent of the core CPS biosynthesis system and it may indicate that the gene product controls the CPS gene expression or CPS secretion or that it modulates the CPS structure.
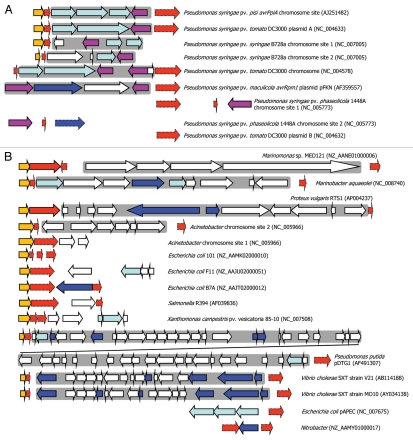
T3SE genes avrPpiA1 (avrRpm1) and avrPpiB1 (hopAM1-1) have also been identified in integron-like elements in plant pathogenic P. syringae.105,106 T3SEs play a key role in suppressing plant immunity and thus capture of a T3SE by an integron could potentially enable a plant pathogen to instantaneously evade host resistance. Two interesting observations arise from the analysis of T3SE integrons: firstly, the T3SE gene is often orientated so that transcription is towards the 3′ end of the integrase gene, suggesting that any integrase PC promoter would not be influencing the T3SE expression (Fig. 3A). This may be a mechanism to decouple the T3SE expression from the integrase (and LexA repression, see below) because the T3SE is only needed for overcoming the antimicrobial conditions of a plant host and expression from PC may lead to unwanted toxic effects. Secondly, the T3SE integrons appear to be inserted within the rulAB mutagenic DNA repair operon, which contains a LexA box within the promoter; a truncated 3′ end of rulB flanks the other end of the putative T3SE integrons. We have observed that the integrase appears to be less than 100 nt downstream of the 5′ part of rulB and that the integrase appears to lack an upstream LexA or RpoD binding site (unpublished results). We therefore postulate that the integrase is under the control of the rulAB promoter and, hence, regulated by LexA (Fig. 2C and D). In fact, this association appears to be much more broad ranging, with similar disruptions occurring in related DNA repair genes (rumAB, umuDC, impAB, mucAB, samAB, ruvAB) in many other bacteria although not all carry virulence factors107,108 (Fig. 3B). Hochut et al.107 found that an SXT conjugative element is embedded within rumB in V. cholera and suggested that a transposon had inserted into the DNA repair genes followed by insertion of the integron and the build up of gene cassettes. Evidence of this is clearly present within genome sequences, which can show substantial gene build up as well as what appears to be erosion, through loss of integron genes and loss of the DNA repair genes or parts thereof.
Figure 3.
Integron-like elements associated with DNA repair genes. (A) Type III protein secretion effector (T3SE) integron-like elements are associated with rulA (orange) and/or rulB (red) genes in various Pseudomonas syringae genomes; truncated rulB genes have a dotted edge. T3SE (purple) are found in many cases to be associated with an integrase gene (light blue) or sometimes transposases (dark blue). Other genes are shown in white. A “complete” integron insertion within rulAB is observed in Pseudomonas syringae pv. pisi (Ppi) and P. syringae pv. tomato (Pto), whereas there is evidence of erosion of the rulA and rulB genes in other strains. A grey background is used to highlight the more complete integron elements. The accession numbers refer to the source used for identifying these genes and for orientation, the locus tag for the first gene on the left of each diagram is: Ppi (ORFG); Pto DC3000 plasmid A (rulA); P. syringae pv. syringae B728a chromosomal site 1 (Psyr_0735) and site 2 (Psyr_1884); Pto DC3000 chromosome (intergenic rulB fragment between PSPTO_0585 and PSPTO_0586); P. syringae pv. maculicola (Orf2); P. syringae pv. phaseolicola chromosomal site 1 (PSPPH_0782) and 2 (avrB4-1). (B) Evidence of current or ancient integron associations with DNA repair genes such as umuDC and rumAB are seen in a wider range of bacterial genomes. The accession numbers refer to the source used for identifying these genes and for orientation, the locus tag or gene name of the first gene on the left of each diagram is: Marinomonas (MED121_22332); Marinobacter aquaeolei (1208); Proteus vulgaris (orf79); Acinetobacter chr. site 2 (umuD) and site 1 (ruvA); Escherichia coli 101 (samA), F11 (impA), B7A (EcB7A_1674); Salmonella R394 (mucA); Xanthomonas campestris (XCV3904); Pseudomonas putida (ruvA); Vibrio cholerae V21 (rumA) and MO10 (rumA); Escherichia coli pAPEC (O2ColV155); Nitrobacter (NB311A_19467). Note that differences in arrow lengths and a dotted edging for rulB-like genes (red) represent either truncated coding sequences or orphan non-coding sequences.
Taken together, integrons and gene cassettes are likely to be widely distributed within different environments and maintained in specific microbes through selection. They clearly represent a dynamic element for bacterial evolution and pose a major threat to animal and plant health through capture and proliferation of cassettes.
Alternative Hosts and Environments as MGE Reservoirs for DNA Exchange
We recently highlighted the diversity of environments in which bacteria can occur when not directly occupying their “host”.17 Plant and animal pathogens can be disseminated into the environment, which can potentially have an important influence on pathogen evolution (see also Morris et al.).109 For example, pathogens may be exposed to a variety of bacteriophages [e.g., vibriophages],52 and they frequently encounter other organisms that may act as alternative hosts and have resident microbial populations.110 Moreover, several virulence genes in human pathogens have been experimentally determined to provide resistance to grazing protozoa111,112 or to kill amoeba associated with them in biofilms.113 It has even been proposed that some virulence genes of human pathogens originally evolved as resistance mechanisms to protozoa111–113 and Brüssow33 goes so far as to propose that lysogeny itself evolved as an alliance between bacteria and phages “to fight grazing protists”.
The scope for HGT between bacteria is immense and plasmids provide an excellent example. Plasmids are most often transferred by conjugation and can be classified as: “conjugative”, when they contain all the necessary conjugation machinery; “mobilizable”, when they can be transferred by “piggybacking” and using the conjugation machinery of other plasmids and ICE; and “nonmobilizable”. The analysis of 1,730 plasmids indicates that, globally, half of the plasmids are either conjugative or mobilizable, whereas the other half is nonmobilizable.7 Mobilizable plasmids tend to be small (<30 kb), whereas conjugative plasmids are usually larger (15–500 kb);7 nonmobilizable plasmids come in all sizes, although most of the very large plasmids (>300 kb) are nonmobilizable, particularly those becoming a secondary chromosome and accumulating diverse essential genes, e.g., the 2.1 Mb plasmid of Ralstonia solanacearum,114 which causes bacterial wilt in many cultivated plants. Surprisingly, plasmids tend to preferentially persist in a given bacterial clade despite having a broader conjugative range,115 and the phylogeny of conjugation genes clearly show that mobility between distant clades, which implies transfer and stable maintenance of the plasmid—is sporadic.3,7 Nevertheless, exchange does occur among phylogenetically unrelated bacteria and in diverse habitats. As an example, resistance to antibiotics evolves in four main environments or genetic reactors: (1) human and animal microbiota, (2) places with crowds of susceptible individuals that favor cross-infection and bacterial gene exchange (e.g., hospitals, farms), (3) wastewater and biological residues originating from the second reactor and (4) soil and ground water environments, where bacteria from the previous reactors mix and interact with environmental microorganisms, with water environments being a major site for evolution and HGT.116 A network analysis integrating similarity data of resistance determinants and genes not prone to HGT, revealed that bacteria that are phylogenetically unrelated and/or inhabit distinct environments often shared common antibiotic resistance determinants.15 Importantly, we often have a narrow, human-centred view on the typical habitats colonized by microorganisms. For example, the plant pathogen Pseudomonas syringae is not restricted to agricultural contexts, and its life cycle is driven by the environmental cycle of water,117 where it can interact with a panoply of diverse microorganisms not linked to plants. Genome sequences from many pathogens highlight the presence of a wide range of genes that, on the basis of codon usage and low similarity to genes within the species, indicate they were probably horizontally acquired from other microbes. However, there needs to be more experimental studies that examine the source organism of the acquired genes and in which environment the genetic exchange occurred—only with this knowledge will we have a better grasp of the extent of HGT of MGEs within and without host organisms.
Concluding Comments
MGEs have played an important role in accessorising the genome of bacterial pathogens either by introducing new virulence factors into the genome or provisioning bacteria with new mechanisms for genome restructuring or gene capture. There clearly needs to be a greater consideration of the wider ecology of bacterial pathogens to understand the impact of the environments and the organisms and bacteriophages that reside in these environments in shaping pathogen evolution. While the primary host certainly shapes the direction of pathogen evolution through selection, the non-host environment can almost certainly shape the scope of it by provision of new genes and novel functions. A number of interesting questions are still outstanding:
In what environments can bacterial pathogens be found when away from the primary host and what is the microbial ecology and MGE pool of these environments?
What is the actual extent of MGE flux within and between bacterial genera—can modeling be used in conjunction with empirical studies to evaluate flux?
To what extent do MGEs impose a cost to the host and if so, what genes are providing trade-offs?
Does transduction and transformation trigger an SOS response as seen for conjugation and thus induce the movement of other MGEs within the cell?
Are GI and bacteriophage integrases controlled by LexA and the SOS response?
By addressing these questions, it will be possible to gain a much clearer insight into how pathogens evolve and, potentially, into their mechanisms for spread and dissemination. The results may also provide an understanding of the triggers of HGT and help us to reduce activities that might be pushing pathogen evolution towards increased virulence.
Useful Resources for MGEs
Several websites, databases and software resources have recently become available for storing and analyzing DNA sequences of MGEs. These include ACID (annotation of cassette and integron data118) and INTEGRALL119 for integrons; IS Finder120 (http://www-is.biotoul.fr/) for IS elements isolated from eubacteria and archaea, and ACLAME (a classification of mobile genetic elements;121 http://aclame.ulb.ac.be), comprising all known bacteriophage genomes, plasmids and transposons.
Acknowledgements
We thank José Manuel Palacios, Christopher Clarke and Sarah James and the two anonymous referees for critical review of the manuscript. J.M. thanks the Spanish Ministerio de Ciencia e Innovación (AGL2008-05311-C02-01) for financial support. B.A.V. acknowledges funding from the National Science Foundation (Award IOS 0746501). D.L.A. acknowledges support from the BBSRC for most of our work on the molecular genetics of the halo blight pathogen.
Abbreviations
- CI
chromosomal integron
- CPS
capsular polysaccharide
- EHEC
enterohaemorrhagic E. coli
- GI
genomic island
- HGT
horizontal gene transfer
- HR
hypersensitive response
- ICE
integrative and conjugative element
- IS
insertion sequence
- MDR
multidrug resistance
- MGE
mobile genetic element
- MI
mobile integron
- PAI
pathogenicity island
- Pph
Pseudomonas syringae pv. phaseolicola
- STEC
shiga-toxin producing E. coli
- T3SS
type III protein secretion system
- T3SE
type III secretion system effector
- T6SS
type VI protein secretion system
- VCR
Vibrio cholerae repeated sequence
References
- 1.Lipps HJ. Archaeal Plasmids. In: Lipps G, editor. Plasmids: Current Research and Future Trends. Norfolk: Caister Academic Press; 2008. [Google Scholar]
- 2.Sundin GW. Genomic insights into the contribution of phytopathogenic bacterial plasmids to the evolutionary history of their hosts. Annu Rev Phytopathol. 2007;45:129–151. doi: 10.1146/annurev.phyto.45.062806.094317. [DOI] [PubMed] [Google Scholar]
- 3.Halary S, Leigh JW, Cheaib B, Lopez P, Bapteste E. Network analyses structure genetic diversity in independent genetic worlds. Proc Natl Acad Sci USA. 2010;107:127–132. doi: 10.1073/pnas.0908978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Heinemann JA, Sprague GF. Bacterial conjugative plasmids mobilize DNA transfer between bacteria and yeast. Nature. 1989;340:205–209. doi: 10.1038/340205a0. [DOI] [PubMed] [Google Scholar]
- 5.Grillot-Courvalin C, Goussard S, Huetz F, Ojcius DM, Courvalin P. Functional gene transfer from intracellular bacteria to mammalian cells. Nat Biotech. 1998;16:862–866. doi: 10.1038/nbt0998-862. [DOI] [PubMed] [Google Scholar]
- 6.Zupan J, Ward D, Zambryski P. Inter-kingdom DNA transfer decoded. Nat Biotech. 2002;20:129–131. doi: 10.1038/nbt0202-129. [DOI] [PubMed] [Google Scholar]
- 7.Smillie C, Garcillan-Barcia MP, Francia MV, Rocha EPC, de la Cruz F. Mobility of Plasmids. Microbiol Mol Biol Rev. 2010;74:434–452. doi: 10.1128/MMBR.00020-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Siguier P, Filee J, Chandler M. Insertion sequences in prokaryotic genomes. Curr Opin Microbiol. 2006;9:526–531. doi: 10.1016/j.mib.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 9.Treangen TJ, Abraham AL, Touchon M, Rocha EPC. Genesis, effects and fates of repeats in prokaryotic genomes. FEMS Microbiol Rev. 2009;33:539–571. doi: 10.1111/j.1574-6976.2009.00169.x. [DOI] [PubMed] [Google Scholar]
- 10.Wagner A, de la Chaux N. Distant horizontal gene transfer is rare for multiple families of prokaryotic insertion sequences. Mol Genet Genomics. 2008;280:397–408. doi: 10.1007/s00438-008-0373-y. [DOI] [PubMed] [Google Scholar]
- 11.Douard G, Praud K, Cloeckaert A, Doublet B. The Salmonella genomic island 1 is specifically mobilized in trans by the IncA/C multidrug resistance plasmid family. PLoS One. 2010;5:15302. doi: 10.1371/journal.pone.0015302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wiesner M, Calva E, Fernández-Mora M, Cevallos MA, Campos F, Zaidi MB, et al. Salmonella typhimurium ST213 is associated with two types of IncA/C plasmids carrying multiple resistance determinants. BMC Microbiol. 2011;11:9. doi: 10.1186/1471-2180-11-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sundin GW, Murillo J. Gene traders: characteristics of native plasmids from plant pathogenic bacteria. In: Jackson RW, editor. Plant pathogenic bacteria: genomics and molecular biology. Cambs: Caister Academic Press; 2009. pp. 295–310. [Google Scholar]
- 14.Maurelli AT. Black holes, antivirulence genes and gene inactivation in the evolution of bacterial pathogens. FEMS Microbiol Lett. 2007;267:1–8. doi: 10.1111/j.1574-6968.2006.00526.x. [DOI] [PubMed] [Google Scholar]
- 15.Fondi M, Fani R. The horizontal flow of the plasmid resistome: clues from inter-generic similarity networks. Environ Microbiol. 2010;12:3228–3242. doi: 10.1111/j.1462-2920.2010.02295.x. [DOI] [PubMed] [Google Scholar]
- 16.Hegstad K, Mikalsen T, Coque TM, Werner G, Sundsfjord A. Mobile genetic elements and their contribution to the emergence of antimicrobial resistant Enterococcus faecalis and Enterococcus faecium. Clin Microbiol Infect. 2010;16:541–554. doi: 10.1111/j.1469-0691.2010.03226.x. [DOI] [PubMed] [Google Scholar]
- 17.Jackson RW, Johnson LJ, Clarke SR, Arnold DL. Bacterial pathogen evolution: breaking news. Trends Genet. 2010;27:32–40. doi: 10.1016/j.tig.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 18.Rankin DJ, Rocha EPC, Brown SP. What traits are carried on mobile genetic elements and why? Heredity. 2011;106:1–10. doi: 10.1038/hdy.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groisman EA, Ochman H. Pathogenicity islands: bacterial evolution in quantum leaps. Cell. 1996;87:791–794. doi: 10.1016/s0092-8674(00)81985-6. [DOI] [PubMed] [Google Scholar]
- 20.Paauw A, Leverstein-van Hall MA, Verhoef J, Fluit AC. Evolution in quantum leaps: Multiple combinatorial transfers of HPI and other genetic modules in Enterobacteriaceae. PLoS One. 5:14. doi: 10.1371/journal.pone.0008662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rezzonico F, Smits T, Montesinos E, Frey J, Duffy B. Genotypic comparison of Pantoea agglomerans plant and clinical strains. BMC Microbiol. 2009;9:204. doi: 10.1186/1471-2180-9-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barash I, Manulis-Sasson S. Recent evolution of bacterial pathogens: The gall-forming Pantoea agglomerans case. Annu Rev Phytopathol. 2009;47:133–152. doi: 10.1146/annurev-phyto-080508-081803. [DOI] [PubMed] [Google Scholar]
- 23.Hazen TH, Pan L, Gu JD, Sobecky PA. The contribution of mobile genetic elements to the evolution and ecology of Vibrios. FEMS Microbiol Ecol. 2010;74:485–499. doi: 10.1111/j.1574-6941.2010.00937.x. [DOI] [PubMed] [Google Scholar]
- 24.Naka H, Lopez CS, Crosa JH. Role of the pJM1 plasmid-encoded transport proteins FatB, C and D in ferric anguibactin uptake in the fish pathogen Vibrio anguillarum. Environ Microbiol Rep. 2010;2:104–111. doi: 10.1111/j.1758-2229.2009.00110.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doyle M, Fookes M, Ivens A, Mangan MW, Wain J, Dorman CJ. An H-NS-like stealth protein aids horizontal DNA transmission in bacteria. Science. 2007;315:251–252. doi: 10.1126/science.1137550. [DOI] [PubMed] [Google Scholar]
- 26.Dillon SC, Cameron ADS, Hokamp K, Lucchini S, Hinton JCD, Dorman CJ. Genome-wide analysis of the H-NS and Sfh regulatory networks in Salmonella typhimurium identifies a plasmid-encoded transcription silencing mechanism. Mol Microbiol. 2010;76:1250–1265. doi: 10.1111/j.1365-2958.2010.07173.x. [DOI] [PubMed] [Google Scholar]
- 27.Sota M, Yano H, Hughes JM, Daughdrill GW, Abdo Z, Forney LJ, et al. Shifts in the host range of a promiscuous plasmid through parallel evolution of its replication initiation protein. ISME J. 2010;4:1568–1580. doi: 10.1038/ismej.2010.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Loftie-Eaton W, Rawlings DE. Evolutionary competitiveness of two Natural variants of the IncQ-like plasmids, pRAS3.1 and pRAS3.2. J Bacteriol. 2010;192:6182–6190. doi: 10.1128/JB.00176-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marchetti M, Capela D, Glew M, Cruveiller S, Chane-Woon-Ming B, Gris C, et al. Experimental evolution of a plant pathogen into a legume symbiont. PLoS Biol. 2010;8:1000280. doi: 10.1371/journal.pbio.1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freeman VJ. Studies on the virulence of bacteriophage-infected strains of Corynebacterium diphtheriae. J Bacteriol. 1951;61:675–688. doi: 10.1128/jb.61.6.675-688.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 32.Hassan F, Kamruzzaman M, Mekalanos JJ, Faruque SM. Satellite phage TLCphi enables toxigenic conversion by CTX phage through dif site alteration. Nature. 2010;467:982–985. doi: 10.1038/nature09469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brüssow H. Bacteria between protists and phages: from antipredation strategies to the evolution of pathogenicity. Mol Microbiol. 2007;65:583–589. doi: 10.1111/j.1365-2958.2007.05826.x. [DOI] [PubMed] [Google Scholar]
- 34.Tao L, Wu X, Sun B. Alternative sigma factor sigmaH modulates prophage integration and excision in Staphylococcus aureus. PLoS Pathog. 2010;6:1000888. doi: 10.1371/journal.ppat.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y, Golding I, Sawai S, Guo L, Cox EC. Population fitness and the regulation of Escherichia coli genes by bacterial viruses. PLoS Biol. 2005;3:229. doi: 10.1371/journal.pbio.0030229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Govind R, Vediyappan G, Rolfe RD, Dupuy B, Fralick JA. Bacteriophage-mediated toxin gene regulation in Clostridium difficile. J Virol. 2009;83:12037–12045. doi: 10.1128/JVI.01256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Asadulghani M, Ogura Y, Ooka T, Itoh T, Sawaguchi A, Iguchi A, et al. The defective prophage pool of Escherichia coli O157: prophage-prophage interactions potentiate horizontal transfer of virulence determinants. PLoS Pathog. 2009;5:1000408. doi: 10.1371/journal.ppat.1000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen J, Novick RP. Phage-mediated intergeneric transfer of toxin genes. Science. 2009;323:139–141. doi: 10.1126/science.1164783. [DOI] [PubMed] [Google Scholar]
- 39.Manning SD, Motiwala AS, Springman AC, Qi W, Lacher DW, Ouellette LM, et al. Variation in virulence among clades of Escherichia coli O157:H7 associated with disease outbreaks. Proc Natl Acad Sci USA. 2008;105:4868–4873. doi: 10.1073/pnas.0710834105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abu-Ali GS, Ouellette LM, Henderson ST, Lacher DW, Riordan JT, Whittam TS, et al. Increased adherence and expression of virulence genes in a lineage of Escherichia coli O157:H7 commonly associated with human infections. PLoS One. 2010;5:10167. doi: 10.1371/journal.pone.0010167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Z, Li X, Liu B, Beutin L, Xu J, Ren Y, et al. Derivation of Escherichia coli O157:H7 from its O55:H7 precursor. PLoS One. 2010;5:8700. doi: 10.1371/journal.pone.0008700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ogura Y, Ooka T, Iguchi A, Toh H, Asadulghani M, Oshima K, et al. Comparative genomics reveal the mechanism of the parallel evolution of O157 and non-O157 enterohemorrhagic Escherichia coli. Proc Natl Acad Sci USA. 2009;106:17939–17944. doi: 10.1073/pnas.0903585106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hacker J, Kaper JB. Pathogenicity islands and the evolution of microbes. Annu Rev Microbiol. 2000;54:641–679. doi: 10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]
- 44.Hacker J, Carniel E. Ecological fitness, genomic islands and bacterial pathogenicity. A Darwinian view of the evolution of microbes. EMBO Rep. 2001;2:376–381. doi: 10.1093/embo-reports/kve097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van der Meer JR, Sentchilo V. Genomic islands and the evolution of catabolic pathways in bacteria. Curr Opin Biotechnol. 2003;14:248–254. doi: 10.1016/s0958-1669(03)00058-2. [DOI] [PubMed] [Google Scholar]
- 46.Seth-Smith H, Croucher NJ. Genome watch: breaking the ICE. Nat Rev Microbiol. 2009;7:328–329. doi: 10.1038/nrmicro2137. [DOI] [PubMed] [Google Scholar]
- 47.Imamovic L, Tozzoli R, Michelacci V, Minelli F, Marziano ML, Caprioli A, et al. OI-57, a genomic island of Escherichia coli O157, is present in other seropathotypes of Shiga toxin-producing E. coli associated with severe human disease. Infect Immun. 2010;78:4697–4704. doi: 10.1128/IAI.00512-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Croucher NJ, Walker D, Romero P, Lennard N, Paterson GK, Bason NC, et al. Role of conjugative elements in the evolution of the multidrug-resistant pandemic clone Streptococcus pneumoniaeSpain23F ST81. J Bacteriol. 2009;191:1480–1489. doi: 10.1128/JB.01343-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gartemann KH, Abt B, Bekel T, Burger A, Engemann J, Flügel M, et al. The genome sequence of the tomato-pathogenic actinomycete Clavibacter michiganensis subsp. michiganensis NCPPB382 reveals a large island involved in pathogenicity. J Bacteriol. 2008;190:2138–2149. doi: 10.1128/JB.01595-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kers JA, Cameron KD, Joshi MV, Bukhalid RA, Morello JE, Wach MJ, et al. A large, mobile pathogenicity island confers plant pathogenicity on Streptomyces species. Mol Microbiol. 2005;55:1025–1033. doi: 10.1111/j.1365-2958.2004.04461.x. [DOI] [PubMed] [Google Scholar]
- 51.Alfano JR, Charkowski AO, Deng WL, Badel JL, Petnicki-Ocwieja T, van Dijk K, et al. The Pseudomonas syringae Hrp pathogenicity island has a tripartite mosaic structure composed of a cluster of type III secretion genes bounded by exchangeable effector and conserved effector loci that contribute to parasitic fitness and pathogenicity in plants. Proc Natl Acad Sci USA. 2000;97:4856–4861. doi: 10.1073/pnas.97.9.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choi J, Shin D, Yoon H, Kim J, Lee CR, Kim M, et al. Salmonella pathogenicity island 2 expression negatively controlled by EIIANtr-SsrB interaction is required for Salmonella virulence. Proc Natl Acad Sci USA. 2010;107:20506–20511. doi: 10.1073/pnas.1000759107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blondel CJ, Jiménez JC, Contreras I, Santiviago CA. Comparative genomic analysis uncovers 3 novel loci encoding type six secretion systems differentially distributed in Salmonella serotypes. BMC Genomics. 2009;10:354. doi: 10.1186/1471-2164-10-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blondel CJ, Yang HJ, Castro B, Chiang S, Toro CS, Zaldivar M, et al. Contribution of the Type VI secretion system encoded in SPI-19 to chicken colonization by Salmonella enterica serotypes Gallinarum and Enteritidis. PLoS One. 2010;5 doi: 10.1371/journal.pone.0011724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holden MT, Heather Z, Paillot R, Steward KF, Webb K, Ainslie F, et al. Genomic evidence for the evolution of Streptococcus equi: host restriction, increased virulence and genetic exchange with human pathogens. PLoS Pathog. 2009;5:1000346. doi: 10.1371/journal.ppat.1000346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jenner C, Hitchin E, Mansfield J, Walters K, Betteridge P, Teverson D, et al. Gene-for-gene interactions between Pseudomonas syringae pv. phaseolicola and Phaseolus. Mol Plant-Microbe Interact. 1991;4:553–562. [PubMed] [Google Scholar]
- 57.Jackson RW, Mansfield JW, Arnold DL, Sesma A, Paynter CD, Murillo J, et al. Excision from tRNA genes of a large chromosomal region, carrying avrP-phB, associated with race change in the bean pathogen, Pseudomonas syringae pv. phaseolicola. Mol Microbiol. 2000;38:186–197. doi: 10.1046/j.1365-2958.2000.02133.x. [DOI] [PubMed] [Google Scholar]
- 58.Pitman A, Jackson R, Mansfield J, Kaitell V, Thwaites R, Arnold D. Exposure to host resistance mechanisms drives evolution of bacterial virulence in plants. Curr Biol. 2005;15:2230–2235. doi: 10.1016/j.cub.2005.10.074. [DOI] [PubMed] [Google Scholar]
- 59.Lovell HC, Jackson RW, Mansfield JW, Godfrey SAC, Hancock JT, Desikan R, et al. In planta conditions induce genomic changes in Pseudomonas syringae pv. phaseolicola. Mol Plant Pathol. 2011;12:167–176. doi: 10.1111/j.1364-3703.2010.00658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lovell HC, Mansfield JW, Godfrey SAC, Jackson RW, Hancock JT, Arnold DL. Bacterial evolution by genomic island transfer occurs via DNA transformation in planta. Curr Biol. 2009;19:1586–1590. doi: 10.1016/j.cub.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 61.Lloyd AL, Henderson TA, Vigil PD, Mobley HL. Genomic islands of uropathogenic Escherichia coli contribute to virulence. J Bacteriol. 2009;191:3469–3481. doi: 10.1128/JB.01717-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Diard M, Garry L, Selva M, Mosser T, Denamur E, Matic I. Pathogenicity-associated islands in extraintestinal pathogenic Escherichia coli are fitness elements involved in intestinal colonization. J Bacteriol. 2010;192:4885–4893. doi: 10.1128/JB.00804-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Salzberg SL, Sommer DD, Schatz MC, Phillippy AM, Rabinowicz PD, Tsuge S, et al. Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics. 2008;9:204. doi: 10.1186/1471-2164-9-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Song H, Hwang J, Yi H, Ulrich RL, Yu Y, Nierman WC, et al. The early stage of bacterial genomereductive evolution in the host. PLoS Pathog. 2010;6:1000922. doi: 10.1371/journal.ppat.1000922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Larsson P, Elfsmark D, Svensson K, Wikstrom P, Forsman M, Brettin T, et al. Molecular evolutionary consequences of niche restriction in Francisella tularensis, a facultative intracellular pathogen. PLoS Pathog. 2009;5:1000472. doi: 10.1371/journal.ppat.1000472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kearney B, Staskawicz BJ. Characterization of IS476 and its role in bacterial spot disease of tomato and pepper. J Bacteriol. 1990;172:143–148. doi: 10.1128/jb.172.1.143-148.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Landgraf A, Weingart H, Tsiamis G, Boch J. Different versions of Pseudomonas syringae pv. tomato DC3000 exist due to the activity of an effector transposon. Mol Plant Pathol. 2006;7:355–364. doi: 10.1111/j.1364-3703.2006.00343.x. [DOI] [PubMed] [Google Scholar]
- 68.Rivas LA, Mansfield J, Tsiamis G, Jackson RW, Murillo J. Changes in race-specific virulence in Pseudomonas syringae pv. phaseolicola are associated with a chimeric transposable element and rare deletion events in a plasmid-borne pathogenicity island. Appl Environ Microbiol. 2005;71:3778–3785. doi: 10.1128/AEM.71.7.3778-3785.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Greenberg JT, Vinatzer BA. Identifying type III effectors of plant pathogens and analyzing their interaction with plant cells. Curr Opin Microbiol. 2003;6:20–28. doi: 10.1016/s1369-5274(02)00004-8. [DOI] [PubMed] [Google Scholar]
- 70.Cambray G, Guerout AM, Mazel D. Integrons. Annu Rev Genet. 2010;44:141–166. doi: 10.1146/annurev-genet-102209-163504. [DOI] [PubMed] [Google Scholar]
- 71.Krauland MG, Marsh JW, Paterson DL, Harrison LH. Integron-mediated multidrug resistance in a global collection of nontyphoidal Salmonella enterica isolates. Emerg Infect Dis. 2009;15:388–396. doi: 10.3201/eid1503.081131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Partridge SR, Tsafnat G, Coiera E, Iredell JR. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol Rev. 2009;33:757–784. doi: 10.1111/j.1574-6976.2009.00175.x. [DOI] [PubMed] [Google Scholar]
- 73.Dawes FE, Kuzevski A, Bettelheim KA, Hornitzky MA, Djordjevic SP, Walker MJ. Distribution of class 1 integrons with IS26-mediated deletions in their 3′-conserved segments in Escherichia coli of human and animal origin. PLoS One. 2010;5:12754. doi: 10.1371/journal.pone.0012754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Poirel L, Carattoli A, Bernabeu S, Bruderer T, Frei R, Nordmann P. A novel IncQ plasmid type harbouring a class 3 integron from Escherichia coli. J Antimicrob Chemother. 2010;65:1594–1598. doi: 10.1093/jac/dkq166. [DOI] [PubMed] [Google Scholar]
- 75.Poirel L, Carrer A, Pitout JD, Nordmann P. Integron mobilization unit as a source of mobility of antibiotic resistance genes. Antimicrob Agents Chemother. 2009;53:2492–2498. doi: 10.1128/AAC.00033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rodriguez-Martinez JM, Nordmann P, Fortineau N, Poirel L. VIM-19, a metallo-beta-lactamase with increased carbapenemase activity from Escherichia coli and Klebsiella pneumoniae. Antimicrob Agents Chemother. 2010;54:471–476. doi: 10.1128/AAC.00458-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shaheen BW, Oyarzabal OA, Boothe DM. The role of class 1 and 2 integrons in mediating antimicrobial resistance among canine and feline clinical E. coli isolates from the US. Vet Microbiol. 2010;144:363–370. doi: 10.1016/j.vetmic.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 78.van Essen-Zandbergen A, Smith H, Veldman K, Mevius D. In vivo transfer of an incFIB plasmid harbouring a class 1 integron with gene cassettes dfrA1-aadA1. Vet Microbiol. 2009;137:402–407. doi: 10.1016/j.vetmic.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 79.Woodford N, Carattoli A, Karisik E, Underwood A, Ellington MJ, Livermore DM. Complete nucleotide sequences of plasmids pEK204, pEK499 and pEK516, encoding CTX-M enzymes in three major Escherichia coli lineages from the United Kingdom, all belonging to the international O25:H4-ST131 clone. Antimicrob Agents Chemother. 2009;53:4472–4482. doi: 10.1128/AAC.00688-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jové T, Da Re S, Denis F, Mazel D, Ploy MC. Inverse correlation between promoter strength and excision activity in class 1 integrons. PLoS Genet. 2010;6:1000793. doi: 10.1371/journal.pgen.1000793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.León G, Quiroga C, Centrón D, Roy PH. Diversity and strength of internal outward-oriented promoters in group IIC-attC introns. Nucleic Acids Res. 2010;38:8196–8207. doi: 10.1093/nar/gkq709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Naas T, Mikami Y, Imai T, Poirel L, Nordmann P. Characterization of In53, a class 1 plasmid- and composite transposon-located integron of Escherichia coli which carries an unusual array of gene cassettes. J Bacteriol. 2001;183:235–249. doi: 10.1128/JB.183.1.235-249.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Michael CA, Andrew NR. Co-assortment in integron-associated gene cassette assemblages in environmental DNA samples. BMC Genet. 2010;11:75. doi: 10.1186/1471-2156-11-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Huang L, Cagnon C, Caumette P, Duran R. First gene cassettes of integrons as targets in finding adaptive genes in metagenomes. Appl Environ Microbiol. 2009;75:3823–3825. doi: 10.1128/AEM.02394-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Michael CA, Labbate M. Gene cassette transcription in a large integron-associated array. BMC Genet. 2010;11:82. doi: 10.1186/1471-2156-11-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Guerin E, Cambray G, Sanchez-Alberola N, Campoy S, Erill I, Da Re S, et al. The SOS response controls integron recombination. Science. 2009;324:1034. doi: 10.1126/science.1172914. [DOI] [PubMed] [Google Scholar]
- 87.Baharoglu Z, Bikard D, Mazel D. Conjugative DNA transfer induces the bacterial SOS response and promotes antibiotic resistance development through integron activation. PLoS Genet. 2010;6:1001165. doi: 10.1371/journal.pgen.1001165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Godfrey SAC, Lovell HC, Mansfield JW, Corry DS, Jackson RW, Arnold DL. The stealth episome: suppression of gene expression on the excised genomic island PPHGI-1 from Pseudomonas syringae pv. phaseolicola. PLoS Pathog. 2011;7:1002010. doi: 10.1371/journal.ppat.1002010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arnold DL, Jackson RW, Waterfield NR, Mansfield JW. Evolution of microbial virulence: the benefits of stress. Trends Genet. 2007;23:293–300. doi: 10.1016/j.tig.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 90.Koenig JE, Bourne DG, Curtis B, Dlutek M, Stokes HW, Doolittle WF, et al. Coral-mucus-associated Vibrio integrons in the Great Barrier Reef: genomic hotspots for environmental adaptation. ISME J. 2011 doi: 10.1038/ismej.2010.193. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Elsaied H, Stokes HW, Kitamura K, Kurusu Y, Kamagata Y, Maruyama A. Marine integrons containing novel integrase genes, attachment sites, attI and associated gene cassettes in polluted sediments from Suez and Tokyo Bays. ISME J. 2011 doi: 10.1038/ismej.2010.208. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sajjad A, Holley MP, Labbate M, Stokes HW, Gillings MR. Preclinical class 1 integron with a complete Tn402-like transposition module. Appl Environ Microbiol. 2011;77:335–337. doi: 10.1128/AEM.02142-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yang H, Byelashov OA, Geornaras I, Goodridge LD, Nightingale KK, Belk KE, et al. Characterization and transferability of class 1 integrons in commensal bacteria isolated from farm and nonfarm environments. Foodborne Pathog Dis. 2010;7:1441–1451. doi: 10.1089/fpd.2010.0555. [DOI] [PubMed] [Google Scholar]
- 94.Heringa S, Kim J, Shepherd MW, Jr, Singh R, Jiang X. The presence of antibiotic resistance and integrons in Escherichia coli isolated from compost. Foodborne Pathog Dis. 2010;7:1297–1304. doi: 10.1089/fpd.2010.0544. [DOI] [PubMed] [Google Scholar]
- 95.Nawaz M, Khan SA, Khan AA, Sung K, Tran Q, Kerdahi K, et al. Detection and characterization of virulence genes and integrons in Aeromonas veronii isolated from catfish. Food Microbiol. 2010;27:327–331. doi: 10.1016/j.fm.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 96.Rosewarne CP, Pettigrove V, Stokes HW, Parsons YM. Class 1 integrons in benthic bacterial communities: abundance, association with Tn402-like transposition modules and evidence for coselection with heavy-metal resistance. FEMS Microbiol Ecol. 2010;72:35–46. doi: 10.1111/j.1574-6941.2009.00823.x. [DOI] [PubMed] [Google Scholar]
- 97.Colinon C, Jocktane D, Brothier E, Rossolini GM, Cournoyer B, Nazaret S. Genetic analyses of Pseudomonas aeruginosa isolated from healthy captive snakes: evidence of high inter- and intrasite dissemination and occurrence of antibiotic resistance genes. Environ Microbiol. 2010;12:716–729. doi: 10.1111/j.1462-2920.2009.02115.x. [DOI] [PubMed] [Google Scholar]
- 98.Gillings MR, Labbate M, Sajjad A, Giguere NJ, Holley MP, Stokes HW. Mobilization of a Tn402-like class 1 integron with a novel cassette array via flanking miniature inverted-repeat transposable element-like structures. Appl Environ Microbiol. 2009;75:6002–6004. doi: 10.1128/AEM.01033-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kiiru JN, Saidi SM, Goddeeris BM, Wamae NC, Butaye P, Kariuki SM. Molecular characterisation of Vibrio cholerae O1 strains carrying an SXT/R391-like element from cholera outbreaks in Kenya: 1994–2007. BMC Microbiol. 2009;9:275. doi: 10.1186/1471-2180-9-275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Koenig JE, Sharp C, Dlutek M, Curtis B, Joss M, Boucher Y, et al. Integron gene cassettes and degradation of compounds associated with industrial waste: the case of the Sydney Tar Ponds. PLoS One. 2009;4:5276. doi: 10.1371/journal.pone.0005276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ramirez MS, Pineiro S, Centron D. Novel insights about class 2 integrons from experimental and genomic epidemiology. Antimicrob Agents Chemother. 2010;54:699–706. doi: 10.1128/AAC.01392-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.van der Veen EL, Schilder AG, Timmers TK, Rovers MM, Fluit AC, Bonten MJ, et al. Effect of long-term trimethoprim/sulfamethoxazole treatment on resistance and integron prevalence in the intestinal flora: a randomized, double-blind, placebo-controlled trial in children. J Antimicrob Chemother. 2009;63:1011–1016. doi: 10.1093/jac/dkp050. [DOI] [PubMed] [Google Scholar]
- 103.Mazel D, Dychinco B, Webb VA, Davies J. A distinctive class of integron in the Vibrio cholerae genome. Science. 1998;280:605–608. doi: 10.1126/science.280.5363.605. [DOI] [PubMed] [Google Scholar]
- 104.Smith AB, Siebeling RJ. Identification of genetic loci required for capsular expression in Vibrio vulnificus. Infect Immun. 2003;71:1091–1097. doi: 10.1128/IAI.71.3.1091-1097.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Arnold DL, Jackson RW, Vivian A. Evidence for the mobility of an avirulence gene, avrPpiA1, between the chromosome and plasmids of races of Pseudomonas syringae pv. pisi. Mol Plant Pathol. 2000;1:195–199. doi: 10.1046/j.1364-3703.2000.00019.x. [DOI] [PubMed] [Google Scholar]
- 106.Arnold DL, Jackson RW, Fillingham AJ, Goss SC, Taylor JD, Mansfield JW, et al. Highly conserved sequences flank avirulence genes: isolation of novel avirulence genes from Pseudomonas syringae pv. pisi. Microbiology. 2001;147:1171–1182. doi: 10.1099/00221287-147-5-1171. [DOI] [PubMed] [Google Scholar]
- 107.Hochhut B, Lotfi Y, Mazel D, Faruque SM, Woodgate R, Waldor MK. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT constins. Antimicrob Agents Chemother. 2001;45:2991–3000. doi: 10.1128/AAC.45.11.2991-3000.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rohmer L, Kjemtrup S, Marchesini P, Dangl JL. Nucleotide sequence, functional characterization and evolution of pFKN, a virulence plasmid in Pseudomonas syringae pathovar maculicola. Mol Microbiol. 2003;47:1545–1562. doi: 10.1046/j.1365-2958.2003.03402.x. [DOI] [PubMed] [Google Scholar]
- 109.Morris CE, Bardin M, Kinkel LL, Moury B, Nicot PC, Sands DC. Expanding the paradigms of plant pathogen life history and evolution of parasitic fitness beyond agricultural boundaries. PLoS Pathog. 2009;5:1000693. doi: 10.1371/journal.ppat.1000693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Nadarasah G, Stavrinides J. Insects as alternative hosts for phytopathogenic bacteria. FEMS Microbiol Rev. 2011 doi: 10.1111/j.1574-6976.2011.00264.x. In press. [DOI] [PubMed] [Google Scholar]
- 111.Steinberg KM, Levin BR. Grazing protozoa and the evolution of the Escherichia coli O157:H7 Shiga toxinencoding prophage. Proc R Soc B. 2007;274:1921–1929. doi: 10.1098/rspb.2007.0245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Adiba S, Nizak Cm, van Baalen M, Denamur E, Depaulis F. From grazing resistance to pathogenesis: The coincidental evolution of virulence factors. PLoS One. 2010;5:11882. doi: 10.1371/journal.pone.0011882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Matz C, Moreno AM, Alhede M, Manefield M, Hauser AR, Givskov M, et al. Pseudomonas aeruginosa uses type III secretion system to kill biofilm-associated amoebae. ISME J. 2008;2:843–852. doi: 10.1038/ismej.2008.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Salanoubat M, Genin S, Artiguenave F, Gouzy J, Mangenot S, Arlat M, et al. Genome sequence of the plant pathogen Ralstonia solanacearum. Nature. 2002;415:497–502. doi: 10.1038/415497a. [DOI] [PubMed] [Google Scholar]
- 115.De Gelder L, Ponciano JM, Joyce P, Top EM. Stability of a promiscuous plasmid in different hosts: no guarantee for a long-term relationship. Microbiology. 2007;153:452–463. doi: 10.1099/mic.0.2006/001784-0. [DOI] [PubMed] [Google Scholar]
- 116.Baquero F, Martinez JL, Canton R. Antibiotics and antibiotic resistance in water environments. Curr Opin Biotechnol. 2008;19:260–265. doi: 10.1016/j.copbio.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 117.Morris CE, Sands DC, Vanneste JL, Montarry J, Oakley B, Guilbaud C, et al. Inferring the evolutionary history of the plant pathogen Pseudomonas syringae from its biogeography in headwaters of rivers in North America, Europe and New Zealand. mBio. 2010;1:107–110. doi: 10.1128/mBio.00107-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Joss M, Koenig J, Labbate M, Polz M, Gillings M, Stokes H, et al. ACID: annotation of cassette and integron data. BMC Bioinformatics. 2009;10:118. doi: 10.1186/1471-2105-10-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Moura A, Soares M, Pereira C, Leitao N, Henriques I, Correia A. INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics. 2009;25:1096–1098. doi: 10.1093/bioinformatics/btp105. [DOI] [PubMed] [Google Scholar]
- 120.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:32–36. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Leplae Rl, Lima-Mendez G, Toussaint A. ACLAME: A CLAssification of Mobile genetic Elements, update 2010. Nucleic Acids Res. 2010;38:57–61. doi: 10.1093/nar/gkp938. [DOI] [PMC free article] [PubMed] [Google Scholar]