Abstract
RmInt1 is a mobile group II intron from Sinorhizobium meliloti that is exceptionally abundant in this bacterial species. We compared the presence of RmInt1 and two of its insertion sequence homing sites (ISRm2011-2 and ISRm10-2) in two phylogenetic clusters (I and II) identified by AFLP analysis in a collection of S. meliloti field isolates from Italy. Both clusters contained several copies of the ISRm2011-2 element, which is present at high copy number in almost all S. meliloti isolates. By contrast, isolates from cluster I harbored no copies of ISRm10-2 and only a truncated copy of RmInt1, despite the absence of constraints on intron mobility in this genetic background, whereas cluster II strains harbored several copies of this intron. The absence of ISRm10-2 from one of the strains of this cluster suggests that this element was acquired more recently than the other two elements. Furthermore, studies of insertional polymorphisms in cluster II strains revealed the acquisition of ISRm10-2 and subsequent retrohoming of RmInt1 to this homing site. These results highlight the role of intron homing sites (ISs) in facilitating intron dispersal and the dynamic and ongoing nature of the spread of the group II intron RmInt1 in S. meliloti.
Key words: group II introns, IS elements, lateral gene transfer, retroelements, Sinorhizobium meliloti, Medicago sativa
Introduction
Mobile genetic elements drive bacterial evolution and adaptation via recombination and horizontal transfer events, and may be responsible for some of the genetic and phenotypic variability of bacteria.1 These mobile elements include bacteriophages, transposons, integrons, insertion sequences (ISs) and group II introns.
ISs are small genetic elements, usually less than 2.5 kb in size. They generally encode no functions other than those involved in their mobility. These include factors required in cis, such as the DNA sequences active in recombination that define the ends of the element (inverted repeats), together with an enzyme, the transposase, that recognizes and processes these ends. This enzyme is generally encoded by one or two open reading frames covering almost the entire length of the element.2 The transposition process can be divided into several steps, generally comprising the binding of the transposase to the ends of the element, the formation of a complex involving the enzyme, and possibly some accessory proteins, together with the two ends of the transposon, cleavage and strand transfer of the ends of the transposon into the target, followed by final processing of the strand transfer complex to generate a final product.3
Group II introns are catalytic RNAs and self-splicing mobile retroelements that are believed to have been the progenitors of nuclear pre-mRNA introns4 and the ancestors of non-LTR retrotransposons.5 A group II intron consists of a large catalytic RNA molecule displaying a conserved secondary structure with six double-helical domains (dI to dVI), one of which (dIV) may encode a multifunctional reverse transcriptase protein (the intronencoded protein or IEP).4 This IEP facilitates intron splicing and intron mobility in vivo. Group II introns can move in a sitespecific manner to homologous intron-less genes, in a process known as retrohoming; they may also move at a much lower frequency to new, ectopic sites, in a process known as retrotransposition.6–11 These properties are of interest because they have been used in the development of a new type of gene-targeting tool.12,13 The basic retrohoming process involves a target DNA-primed reverse transcription (TPRT) mechanism mediated by a ribonucleoprotein complex containing the IEP and the excised intron lariat RNA. This mechanism has been studied for a limited number of these mobile genetic elements, all within the IIA subclass of group II introns (reviewed in ref. 14). The IEPs of introns from this subclass have several conserved domains, including an N-terminal RT domain, domain X, a putative RNA-binding domain associated with RNA splicing or maturase activity and C-terminal DNA-binding (D) and DNA-endonuclease (En) domains for target DNA cleavage. However, many bacterial group II intron IEPs lack the endonuclease domain.15,16 One of the best studied introns of this type is the RmInt1 intron, which belongs to the IIB3/D subclass.10 It was found in Sinorhizobium meliloti, the nitrogen-fixing symbiotic bacterium that establish symbiosis in the roots of leguminous plants of the genus Medicago.17 This intron has been shown to be highly mobile in vivo.18–20 This mobility is characterized by a bias in the orientation of replication of the DNA target, indicating that cDNA synthesis is primed by the 3′ end of the DNA at the replication fork.21
Detailed information about group II intron content is currently available for only four bacterial taxa: Escherichia coli strains,22 the Bacillus cereus group,23 Wolbachia bacterial endosymbionts24 and Sinorhizobium meliloti and related rhizobiales.25 Studies on these introns have reported considerable variability in intron copy number between strains. In particular, RmInt1 is usually very abundant in S. meliloti strains, which may contain up to 11 copies. It is widespread and has been detected in 90% of the S. meliloti strains tested.26,27 Within the genome of S. meliloti, this intron is found mostly within copies of ISRm2011-2, an IS element present in almost all S. meliloti strains.25,26,28 Other copies of the intron have been found in genes such as oxi1, and other IS elements closely related to ISRm2011-2 have been identified (ISRm10-1 and ISRm10-2).9 ISRm10-1 has been detected in several isolates (but only in one or two copies), whereas ISRm10-2 was initially found as a single copy in one isolate from Uruguayan soils.9 Interestingly, ISRm10-2 was found to be more abundant, with several copies per genome, in 11 of 36 field isolates from an Italian soil collection. This IS element was originally detected in the intergenic region between nodQ1 and nodJ.29
Recent experimental data have shown that RmInt1 propagation within the S. meliloti genome occurs principally by retrohoming into the ISRm2011-2 element.30 Other Rhizobium and Sinorhizobium species were recently shown to have acquired the RmInt1 intron by vertical inheritance and independent horizontal transfer events.25 It has been suggested that RmInt1 location, together with the inefficiency of the splicing, is consistent with a role for this intron in preventing the spread of other potentially harmful mobile elements in these bacteria.31 However, the dynamics of bacterial group II introns in natural conditions and the factors influencing their gain or loss from some strains remain to be elucidated.32
In this study, we investigated the presence and distribution of RmInt1 and its IS homing sites in two genomic clusters from a collection of Italian field isolates of S. meliloti. Our results suggest that RmInt1 is probably still spreading and that the presence of intron homing sites (ISs) has facilitated intron dispersion in S. meliloti, partly accounting for the exceptionally high abundance of this element in this rhizobial species.
Results and Discussion
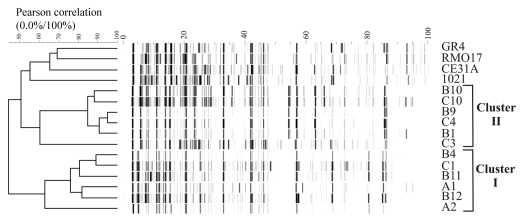
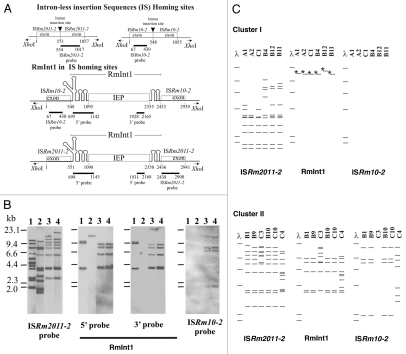
We investigated the genomic structure of S. meliloti isolates from an Italian collection of alfalfa-nodulating field isolates (Table 1),33 by AFLP (amplified fragment length polymorphism) analysis.34 The dendogram obtained identified two main clusters (I and II), with a Pearson correlation index value in the range of 75 to 95% (Fig. 1). These AFLP differences are significant because the isolates can be differentiated from the reference strain 1021 and other strains from different sources. Cluster I comprises the isolates of types B4, C1, B11, A1 and B12, and the more distantly related A2 (correlation index of only 62%). Cluster II comprises isolates of types B1, C10, B9, C4 and B10, and the more distantly related C3 (correlation index of only 60%). This clustering pattern was further supported by IS/intron fingerprint data (Fig. 2). Clusters I and II were clearly distinguished by the ISRm2011-2 fingerprint. Thus, all the strains of cluster I shared at least five hybridizing bands and differed in terms of the number of additional copies (copies 7–11; Fig. 2B and C). The cluster II fingerprint was also defined by five common bands, but these bands differed in size from those of cluster I. The number of additional copies (9–12 bands in total) differentiated between the isolates within this cluster. This genetic variation was even more pronounced in C4, which had six additional bands absent from the other isolates of cluster II (Fig. 2C). By contrast, ISRm10-2 fingerprinting showed that only five isolates, all belonging to cluster II, harbored this IS element (Fig. 2C and Table 1). C3 from cluster II was devoid of this IS element; four isolates contained four identical copies (B1, B9, B10 and C10), and C4 harbored seven copies with only the highest molecular weight band in common (Fig. 2C). Thus, cluster II isolates account for the high relative abundance of the ISRm10-2 element previously reported for this Italian collection (32% of the isolates).29 These findings further support the hypothesis that ISRm2011-2 is ancestral in the evolution of S. meliloti,35 whereas ISRm10-2 is probably a recent acquisition.
Table 1.
Presence of RmInt1 and host elements (ISRm2011-2 and ISRm10-2) in the genome of S. meliloti isolates
| Cluster | S. meliloti isolate | Location | ISRm2011-2* | ISRm10-2* |
| - | 1021 | Toulouse | 12/2† | 0/0† |
| - | GR4 | Granada | 12/9† | 0/0† |
| - | CE31A | Uruguay | 4/2† | 1/1† |
| - | RMO17 | Salamanca | 13/0‡ | ND |
| I | A1 | Lodi | 9/0 | 0/0 |
| I | A2 | Lodi | 7/0 | 0/0 |
| I | C1 | Mix§ | 7/0 | 0/0 |
| I | B4 | Rome | 10/0 | 0/0 |
| I | B12 | Rome | 10/0 | 0/0 |
| I | B11 | Rome | 11/0 | 0/0 |
| II | C3 | Mix§ | 12/7 | 0/0 |
| II | B9 | Rome | 9/5 | 4/0 |
| II | B1 | Rome | 9/5 | 4/0 |
| II | B10 | Rome | 9/5 | 4/0 |
| II | C10 | Mix§ | 9/5 | 4/0 |
| II | C4 | Mix | 12/7 | 7/3 |
Figure 1.
AFLP analysis of the S. meliloti isolates. AFLP patterns were normalized and transformed to horizontal electrophoretic gel format by the software package GelCompar 4.1, with the program Abicon. The dendrogram, based on ABI310 data obtained from AFLP fingerprints, was generated with the UPGMA algorithm, and shows two differentiated genomic clusters (I and II). Other S. meliloti strains (GR4, RMO17, CE31A and 1021) were included in the analysis as a reference.
Figure 2.
RFLP analysis of the S. meliloti isolates. (A) Schematic diagrams of intron-less and intron-invaded DNA sites. The DNA probes used for DNA hybridization are indicated below each diagram. Numbers indicate relevant nucleotide positions within the exons and intron sequences. (B) Examples of RFLP analysis depicted in (C) for XhoI-digested total DNA from S. meliloti 1021 (lane 1) and representative isolates from cluster I (B4, lane 2) and cluster II (B10, lane 3 and B1, lane 4), with probes for the mobile elements indicated under each part and represented in (A). DNA molecular size markers are indicated on the left of the first part. (C) Schematic diagrams of Southern blot hybridizations of XhoI-digested total DNA from isolates of clusters I (above) and II (below). The mobile elements used as probes are indicated at the bottom. DNA molecular size markers (λ) are also shown. Asterisks (*) indicate that the band hybridizes only with the probe for the 5′-end of RmInt1.
The presence and abundance of ISs in cluster I (up to 11 copies of ISRm2011-2; Table 1) and cluster II (up to 19 copies of ISRm2011-2 plus ISRm10-2), corresponding to potential DNA target sites (homing sites) for RmInt1, suggest that this intron should be well represented in both clusters of isolates. However, the distribution of RmInt1 in the Italian isolates showed unexpected differences between the two clusters. In cluster I, only one hybridizing band was detected for RmInt1, corresponding to a 3′-truncated remnant of RmInt1 (Fig. 2B and lane 2). This fragmented form of the intron can reflect a tendency of RmInt1 to evolve toward an inactive form by fragmentation, with loss of the intron-encoded protein ORF, similar to those previously described in rhizobia other than S. meliloti,25 and to other truncated forms of group II introns described elsewhere in reference 36. By contrast, cluster II strains contained five to nine full-length copies of RmInt1 (Fig. 2B and C), consistent with previous findings for S. meliloti strains from different collections (Table 1).25–27 Genetic variation based on this retroelement was more pronounced for isolates C3 and C4, consistent with the considerable genomic diversity of these isolates revealed by both IS fingerprint analyses. Thus, S. meliloti isolates may lack active RmInt1 despite the presence of IS homing sites. Furthermore, the absence of full-length active copies of RmInt1 in the genome of S. meliloti cluster I isolates is not associated with particular restrictions on the mobility of this element. Mobility assays with an intron donor plasmid and target recipient plasmid,19 performed with representative isolates from the two clusters showed similar homing efficiencies between isolates and for the positive control S. meliloti strain RMO17 (data not shown). The most plausible explanation for these findings is that RmInt1 is still spreading in the S. meliloti species. The isolates of cluster I thus provide an illustration of the cycle of gains and losses of RmInt1.
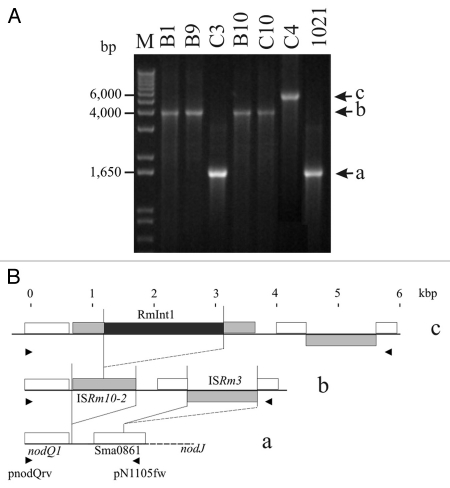
The ISRm10-2 element was initially found in these Italian isolates in studies of insertional polymorphisms in the nodQ1 and nodJ intergenic region.29 In cluster II strains, three different fragments were amplified from this intergenic region with primers pnodQrv and pN6313fw (Fig. 3A). An analysis of the sequence of the smallest of these fragments (1,614 bp), obtained from isolate C3, showed it to contain a fragmented ISRm8 element (identical to the Sma0861 ORF; Fig. 3B). The structure of the intergenic region between nodJ and nodQ1 was identical to that of strain 1021.37 Isolates B1, B9, B10 and C10 gave an amplified fragment of 3,971 bp. This fragment was larger than that obtained from C3, due to the insertion of elements ISRm10-2 and ISRm3,38 82 and 825 bp downstream from the nodQ1 gene (GenBank accession number: AY570924; see also Fig. 3B), respectively. A third band, 5,855 bp in size, was amplified from isolate C4. This fragment contained the same elements as obtained from isolates B1, B9, B10 and C10, but also carried a copy of RmInt1 inserted into the corresponding intron insertion site of ISRm10-2 (identical to the published sequence, accession number: Y11597). These results reveal the occurrence of genetic variation caused by successive transposition events in the nodQ1 and nodJ intergenic region and involving different IS elements, including ISRm10-2, with subsequent RmInt1 retrohoming to this homing site. The sequences of these insertional polymorphisms suggest that ISRm10-2 is an active transposable element, colonizing the intergenic region between nodQ1 and nodJ genes in particular. The ISRm10-2 copy in the former intergenic region seems to have become a target site for RmInt1, providing further evidence that the spread of RmInt1 in the genome is dependent on previous transposition events of its IS target sites.27
Figure 3.
Independent invasion of ISRm10-2 copies by RmInt1. (A) Ethidium bromide-stained agarose gel of amplified fragments of the intergenic region between nodJ and nodQ1, from total DNA of the cluster II isolates and the control strain 1021 (see Materials and Methods). Various amplified fragments were obtained: 1,614 a, 3,971 b and 5,855 c bp. (B) Structure of each amplified fragment. White boxes indicate genes present on pSymA in 1021 and the C3 isolate; gray boxes indicate the acquisition of IS transposable elements in the B1, B9, B10 and C10 isolates and the black box indicates the insertion of the RmInt1 group II intron into ISRm10-2 in isolate C4. Arrowheads indicate the positions of the primers used.
Our data also suggest that RmInt1 is continuing to spread in S. meliloti. RmInt1 requires a DNA target site for the initial invasion of the S. meliloti genome; this site may be provided by a conservative transposition event involving an existing (e.g., ISRm2011-2 ancestral in S. meliloti) or newly acquired ISRm2011-2-type homing site (e.g., transposition of ISRm10-2 to the intergenic region between the nodQ1 and nodJ genes). RmInt1 then moves from its presumed natural homing site (ISRm2011-2) into the new target site and spreads throughout the genome, providing a confirmation in the natural environment of the findings of experimental studies.30 Thus, the successful spread of RmInt1 in S. meliloti, as a retroelement, is based principally on a strategy of targeting alternative insertion sequences as homing sites.
Materials and Methods
S. meliloti isolates and reference strains.
All the Italian field isolates used in this work were obtained from a collection described in a previous study in reference 29 and 33. The other strains used were S. meliloti 1021,38 CE31A, GR4,9 RMO17.39
AFLP analysis.
We used a modified version of the experimental protocol described in the Gibco BRL AFLP manual and published by Biondi et al.34 The PCR products obtained were analyzed with a Perkin-Elmer ABI 310 analyzer.
Electrophoretic data were collected with ABI Genescan software (PE Applied Biosystems). After normalization, the levels of genetic similarity between the AFLP patterns were calculated with the Pearson product-moment correlation coefficient (r). For cluster analysis of AFLP banding patterns, we used the unweighted pair group method using arithmetic averages (UPGMA).40 We considered only band patterns that were reproducible in three independent amplification reactions.
IS/intron-fingerprint.
Fingerprints were obtained by DNA hybridization with various IS- and intron-derived DNA probes, as described elsewhere in reference 9 and 25. DNA hybridization analysis was carried out on the same filters, with the various probes. DNA probes for RmInt1 and the insertion elements ISRm2011-2 and ISRm10-2 were obtained by PCR amplification with the following oligonucleotides: for ISRm2011-2, 2011B1 (5′-TGG ACG AAG ACG AAC ATG G-3′) and 2011B2 (5′-TTG AAG TAG GCT GCG CAT T-3′); for ISRm10-2, ISRm10-67f (5′-ACG TCC GCC GTG TGG AGG-3′) and ISRm10-430r (5′-CGC GTG ATG TTG TGC CGC-3′); for the 5′ end of RmInt1, Epsilon (5′-GTG AGC GTC GGA TGA AAC-3′) and C18R0 (5′-ACG TTT CTC AAT TCG AAA CG-3′) and for the 3′ end of RmInt1, Int1 (5′-GTA TCC GAA TGT CAC GTT CG-3′) and Int2 (5′-CCG TCC ATA GTA GGC AAT CC-3′).
PCR amplification and DNA sequencing.
The intergenic region between the nodQ1 and nodJ genes (from nt 475,031 to 476,645 in pSymA; 3) of cluster II strains was obtained by PCR, with the High Fidelity PCR System (Roche), using primers pnodQrv (5′-AAT CAG CTC CCT GCC GTT CTC TGG TTC ACC-3′) and pN1105fw (5′-GGTA GCC ATC CGA GCA GGG-3′). The amplified DNA fragment was purified and used as a template for sequencing. We used pN6313fw, pnodQrv and pN61105fw for the sequencing of ISRm10-2 and ISRm10-2-RmInt1 in the C4, B1, B9, B10 and C10 isolates; pN61143fw (5′-GGC CGC GCT CCT GCC ACG-3′) and pN61055rv (5′-GCT GAC TCA GCC TCG GTG CAG G-3′) were used for the sequencing of ISRm3 in these isolates. The accession number of the sequence obtained from the B1 isolate is AY570924. Sequence similarity was evaluated against the BLAST database at the National Center for Biotechnology Information (NCBI).
Acknowledgements
We thank Mrs. Ascensión Martos for technical assistance. This work was supported by an Acción Integrada con Italia. Ref: HI 2000-0171 grant from the EU and by the Spanish Ministerio de Ciencia e Innovación [BIO2008-00740 and CSD 2009-0006 of Programme Consolider-Ingenio 2010] and Junta de Andalucía [CVI-01522] including ERDF (European Regional Development Fund) funds.
References
- 1.Kazazian HH., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–1632. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 2.Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mizuuchi K. Transpositional recombination: mechanistic insights from studies of mu and other elements. Annu Rev Biochem. 1992;61:1011–1051. doi: 10.1146/annurev.bi.61.070192.005051. [DOI] [PubMed] [Google Scholar]
- 4.Michel F, Ferat JL. Structure and activities of group II introns. Annu Rev Biochem. 1995;64:435–461. doi: 10.1146/annurev.bi.64.070195.002251. [DOI] [PubMed] [Google Scholar]
- 5.Toro N, Jiménez-Zurdo JI, García-Rodríguez FM. Bacterial group II introns: not just splicing. FEMS Microbiol Rev. 2007;31:342–358. doi: 10.1111/j.1574-6976.2007.00068.x. [DOI] [PubMed] [Google Scholar]
- 6.Belfort M, Derbyshire V, Parker MM, Cousineau B, Lambowitz AM. Mobile introns: pathways and proteins. In: Craig N, Craigie R, Gellert M, Lambowitz AM, editors. Mobile DNA. 2nd Edition. Washington DC: American Society for Microbiology; 2002. pp. 761–783. [Google Scholar]
- 7.Lambowitz AM, Caprara MG, Zimmerly S, Perlman PS. Group I and group II ribozymes as RNPs: clues to the past and guides to the future. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 2nd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1999. pp. 451–485. [Google Scholar]
- 8.Lambowitz AM, Zimmerly S. Mobile group II introns. Annu Rev Genet. 2004;38:1–35. doi: 10.1146/annurev.genet.38.072902.091600. [DOI] [PubMed] [Google Scholar]
- 9.Martínez-Abarca F, Toro N. RecA-independent ectopic transposition in vivo of a bacterial group II intron. Nucleic Acids Res. 2000;28:4397–4402. doi: 10.1093/nar/28.21.4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toro N. Bacteria and Archaea Group II introns: additional mobile genetic elements in the environment. Environ Microbiol. 2003;5:143–151. doi: 10.1046/j.1462-2920.2003.00398.x. [DOI] [PubMed] [Google Scholar]
- 11.Beauregard A, Curcio MJ, Belfort M. The take and give between retrotransposable elements and their hosts. Annu Rev Genet. 2008;42:587–617. doi: 10.1146/annurev.genet.42.110807.091549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guo H, Karberg M, Long M, Jones JP, 3rd, Sullenger B, Lambowitz AM. Group II introns designed to insert into therapeutically relevant DNA target sites in human cells. Science. 2000;289:452–457. doi: 10.1126/science.289.5478.452. [DOI] [PubMed] [Google Scholar]
- 13.Karberg M, Guo H, Zhong J, Coon R, Perutka J, Lambowitz AM. Group II introns as controllable gene targeting vectors for genetic manipulation of bacteria. Nat Biotechnol. 2001;19:1162–1167. doi: 10.1038/nbt1201-1162. [DOI] [PubMed] [Google Scholar]
- 14.Lambowitz AM, Mohr G, Zimmerly S. Group II intron homing endonucleases: ribonucleoprotein complexes with programmable target specificity. In: Belfort M, editor. The Homing endonucleases and Inteins. Heidelberg: Springer-Verlag; 2005. pp. 121–145. [Google Scholar]
- 15.Martínez-Abarca F, Toro N. Group II introns in the bacterial world. Mol Microbiol. 2000;38:917–926. doi: 10.1046/j.1365-2958.2000.02197.x. [DOI] [PubMed] [Google Scholar]
- 16.Robart AR, Zimmerly S. Group II intron retroelements: function and diversity. Cytogenet Genome Res. 2005;110:589–597. doi: 10.1159/000084992. [DOI] [PubMed] [Google Scholar]
- 17.Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC. How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat Rev Microbiol. 2007;5:619–633. doi: 10.1038/nrmicro1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiménez-Zurdo JI, García-Rodríguez FM, Barrientos-Durán A, Toro N. DNA target site requirements for homing in vivo of a bacterial group II intron encoding a protein lacking the DNA endonuclease domain. J Mol Biol. 2003;326:413–423. doi: 10.1016/s0022-2836(02)01380-3. [DOI] [PubMed] [Google Scholar]
- 19.Martínez-Abarca F, García-Rodríguez FM, Toro N. Homing of a bacterial group II intron with an intron-encoded protein lacking a recognizable endonuclease domain. Mol Microbiol. 2000;35:1405–1412. doi: 10.1046/j.1365-2958.2000.01804.x. [DOI] [PubMed] [Google Scholar]
- 20.Muñoz-Adelantado E, San Filippo J, Martínez-Abarca F, García-Rodríguez FM, Lambowitz AM, Toro N. Mobility of the Sinorhizobium meliloti group II intron RmInt1 occurs by reverse splicing into DNA, but requires an unknown reverse transcriptase priming mechanism. J Mol Biol. 2003;327:931–943. doi: 10.1016/s0022-2836(03)00208-0. [DOI] [PubMed] [Google Scholar]
- 21.Martínez-Abarca F, Barrientos-Durán A, Fernández-López M, Toro N. The RmInt1 group II intron has two different retrohoming pathways for mobility using predominantly the nascent lagging strand at DNA replication forks for priming. Nucleic Acids Res. 2004;32:2880–2888. doi: 10.1093/nar/gkh616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dai L, Zimmerly S. The dispersal of five group II introns among natural populations of Escherichia coli. RNA. 2002;8:1294–1307. doi: 10.1017/s1355838202023014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tourasse NJ, Kolstø AB. Survey of group I and group II introns in 29 sequenced genomes of the Bacillus cereus group: insights into their spread and evolution. Nucleic Acids Res. 2008;36:4529–4548. doi: 10.1093/nar/gkn372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leclercq S, Giraud I, Cordaux R. Remarkable Abundance and evolution of mobile group II introns in Wolbachia bacterial endosymbionts. Mol Biol Evol. 2010 doi: 10.1093/molbev/msq238. [DOI] [PubMed] [Google Scholar]
- 25.Fernández-López M, Muñoz-Adelantado E, Gillis M, Willems A, Toro N. Dispersal and evolution of the Sinorhizobium meliloti group II RmInt1 intron in bacteria that interact with plants. Mol Biol Evol. 2005;22:1518–1528. doi: 10.1093/molbev/msi144. [DOI] [PubMed] [Google Scholar]
- 26.Martínez-Abarca F, Zekri S, Toro N. Characterization and splicing in vivo of a Sinorhizobium meliloti group II intron associated with particular insertion sequences of the IS630-Tc1/IS3 retroposon superfamily. Mol Microbiol. 1998;28:1295–1306. doi: 10.1046/j.1365-2958.1998.00894.x. [DOI] [PubMed] [Google Scholar]
- 27.Muñoz E, Villadas PJ, Toro N. Ectopic transposition of a group II intron in natural bacterial populations. Mol Microbiol. 2001;41:645–652. doi: 10.1046/j.1365-2958.2001.02540.x. [DOI] [PubMed] [Google Scholar]
- 28.Selbitschka W, Arnold W, Jording D, Kosier B, Toro N, Pühler A. The insertion sequence element ISRm2011-2 belongs to the IS630-Tc1 family of transposable elements and is abundant in Rhizobium meliloti. Gene. 1995;163:59–64. doi: 10.1016/0378-1119(95)00371-c. [DOI] [PubMed] [Google Scholar]
- 29.Biondi EG, Fancelli S, Bazzicalupo M. ISRm10: a new insertion sequence of Sinorhizobium meliloti: nucleotide sequence and geographic distribution. FEMS Microbiol Lett. 1999;181:171–176. doi: 10.1111/j.1574-6968.1999.tb08841.x. [DOI] [PubMed] [Google Scholar]
- 30.Nisa-Martínez R, Jiménez-Zurdo JI, Martínez-Abarca F, Muñoz-Adelantado E, Toro N. Dispersion of the RmInt1 group II intron in the Sinorhizobium meliloti genome upon acquisition by conjugative transfer. Nucleic Acids Res. 2007;35:214–222. doi: 10.1093/nar/gkl1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chillón I, Martínez-Abarca F, Toro N. Splicing of the Sinorhizobium meliloti RmInt1 group II intron provides evidence of retroelement behavior. Nucleic Acids Res. 2011;39:1095–1104. doi: 10.1093/nar/gkq847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edgell DR, Belfort M, Shub DA. Barriers to intron promiscuity in bacteria. J Bacteriol. 2000;182:5281–5289. doi: 10.1128/jb.182.19.5281-5289.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paffetti D, Daguin F, Fancelli S, Gnocchi S, Lippi F, Scotti C, et al. Influence of plant genotype on the selection of nodulating Sinorhizobium meliloti strains by Medicago sativa. Antonie Van Leeuwenhoek. 1998;73:3–8. doi: 10.1023/a:1000591719287. [DOI] [PubMed] [Google Scholar]
- 34.Biondi EG, Pilli E, Giuntini E, Roumiantseva ML, Andronov EE, Onichtchouk OP, et al. Genetic relationship of Sinorhizobium meliloti and Sinorhizobium medicae strains isolated from Caucasian region. FEMS Microbiol Lett. 2003;220:207–213. doi: 10.1016/S0378-1097(03)00098-3. [DOI] [PubMed] [Google Scholar]
- 35.Toro N, Martínez-Abarca F, Fernández-López M, Muñoz-Adelantado E. Diversity of group II introns in the genome of Sinorhizobium meliloti strain 1021: splicing and mobility of RmInt1. Mol Genet Genomics. 2003;268:628–636. doi: 10.1007/s00438-002-0778-y. [DOI] [PubMed] [Google Scholar]
- 36.Dai L, Zimmerly S. Compilation and analysis of group II intron insertions in bacterial genomes: evidence for retroelement behavior. Nucleic Acids Res. 20;30:1091–1102. doi: 10.1093/nar/30.5.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wheatcroft R, Laberge S. Identification and nucleotide sequence of Rhizobium meliloti insertion sequence ISRm3: similarity between the putative transposase encoded by ISRm3 and those encoded by Staphylococcus aureus IS256 and Thiobacillus ferrooxidans IST2. J Bacteriol. 1991;173:2530–2538. doi: 10.1128/jb.173.8.2530-2538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Galibert F, Finan TM, Long SR, Puhler A, Abola P, Ampe F, et al. The composite genome of the legume symbiont Sinorhizobium meliloti. Science. 2001;293:668–672. doi: 10.1126/science.1060966. [DOI] [PubMed] [Google Scholar]
- 39.Villadas PJ, Velázquez E, Martínez-Molina E, Toro N. Identification of nodule dominant Rhizobium meliloti strains carrying pRmeGR4b-type plasmid within indigenous soil populations by PCR using primers derived from specific DNA sequences. FEMS Microbiol Ecol. 1995;17:161–168. [Google Scholar]
- 40.Vauterin L, Vauterin P. Computer-aided objective comparison of electrophoretic patterns for grouping and identification of microorganisms. Eur Microbiol. 1992;1:37–41. [Google Scholar]