Abstract
Malignant transformation often leads to both loss of normal proliferation control and inhibition of cell differentiation. Some tumor cells can be stimulated to reenter their differentiation program and to undergo terminal growth arrest. The in vitro differentiation of mouse erythroleukemia (MEL) cells is an important example of tumor cell reprogramming. MEL cells are malignant erythroblasts that are blocked from differentiating into mature RBC due to dysregulated expression of the transcription factor PU.1, which binds to and represses GATA-1, the major transcriptional regulator of erythropoiesis. We used RNA interference to ask whether inhibiting PU.1 synthesis was sufficient to cause MEL cells to lose their malignant properties. We report here that transfection of MEL cells with a PU.1-specific short interfering RNA oligonucleotide causes the cells to resume erythroid differentiation, accumulate hemoglobin, and undergo terminal growth arrest. RNA interference directed at specific, aberrantly expressed transcription factors may hold promise for the development of potent antitumor therapies in other hematologic malignancies.
Introduction
Most cancers are characterized by both loss of normal cell division controls and features of immature cells owing to a block in completing their normal differentiation program. Identifying ways to reestablish proliferation controls and differentiation in such malignant cells may lead to new approaches to cancer therapy. Even though tumor cells are usually blocked at an early or intermediate stage of differentiation, treating them with certain agents can sometimes cause them to reenter their normal differentiation program. Resumption of differentiation by such cells can lead to terminal cell division and loss of tumorigenicity (1).
Friend virus–induced erythroleukemias of mice have served as important model-for understanding the multistep aspects of leukemia development (2–4). The permanent murine erythroleukemia (MEL) cell lines that can be established from such mice (5, 6) have also stimulated interest in approaches to cancer therapy based on forced differentiation of tumor cells. MEL cells are blocked at about the proerythroblast stage of RBC development. However, treating MEL cells with various chemical agents causes the cells to resume erythroid differentiation, accumulate hemoglobin, and undergo terminal cell divisions and growth arrest (7). To date, three events have been associated with the generation of malignant erythroid cells in Friend virus–infected mice (3). The early stage of the disease is characterized by polyclonal proliferation of nonleukemic erythroid progenitors. Proliferation of the cells is caused by interaction of a virus-encoded 55-kDa fusion glycoprotein (gp55) with the erythropoietin (Epo) receptor, triggering constitutive activation of mitogenic signaling. gp55 and the Epo receptor can form a complex with the short form of the tyrosine kinase STK (sf-STK), resulting in phosphorylation of sf-STK and its association with signal-transducing molecules (8). Because erythroid cells from spleen focus-forming virus–susceptible (but not resistant) mice express sf-STK (8), this kinase may mediate Epo-independent proliferation of Friend virus–infected cells. Subsequently, clonal or oligoclonal malignant cells emerge. This stage is associated with two additional genetic events: inactivation of p53 (9–11) and proviral insertions at the Spi-1 locus that encodes the PU.1 transcription factor (12–15). Integration of the provirus does not disrupt PU.1 expression. Instead, it activates or deregulates PU.1 synthesis in erythroid precursors, in which PU.1 is ordinarily expressed at a low level. PU.1 is an Ets family transcription factor that plays key roles in the development of monocytic, granulocytic, and B lymphoid lineages (16, 17). However, it does not have an established role in the normal development of the erythroid lineage. The causative oncogenic role of PU.1 in erythroleukemogenesis is further supported by the finding that expressing an exogenous PU.1 gene in erythroid cells in transgenic mice leads to erythroleukemia (18–20) and by experiments showing that a PU.1-encoding retrovirus can immortalize erythroblasts in long-term bone marrow cultures (20).
Recently, the role of PU.1 in blocking erythroid differentiation in MEL tumor cells has been elucidated. One of the early events occurring during chemically induced differentiation of MEL cells is a marked decline in the level of PU.1 (21 –24). This decline is necessary for the reentry of the cells into the differentiation program because transfecting the cells with expression vectors encoding exogenous PU.1 blocks induced differentiation (23). The molecular mechanism by which PU.1 blocks erythroid differentiation is now fairly well understood. MEL blasts contain substantial levels of GATA-1 as well as other transcription factors involved in erythropoiesis, and yet, they are blocked from differentiating. PU.1 binds directly to and inhibits the transcriptional activity of GATA-1, the major transcription factor required for erythroid differentiation (25, 26). Recent evidence indicates that GATA-1 is bound to its target genes in the MEL blasts but that PU.1 binds to it on DNA and recruits a protein complex, including a histone methyltransferase, that creates a repressive chromatin structure in the vicinity of the GATA-1 target genes (27, 28).
Thus, the current view of the status of the malignant MEL tumor cell is that it is poised for differentiation, but because of the presence of abnormally high levels of PU.1, it is blocked from undergoing terminal differentiation and is instead locked into a program of uncontrolled proliferation. Based on this view, we recently described a strategy for overcoming PU.1-mediated inhibition of GATA-1 by providing the cells with additional GATA-1 in a conditionally active form. Activation of the exogenous GATA-1 causes the cells to resume erythroid differentiation and undergo terminal cell division (29). These results suggest that simply changing the relative stoichiometry of PU.1 to GATA-1 in the cells in favor of GATA-1 might lead to the reversal of their malignant phenotype.
The advent of RNA interference techniques provides a much more practical approach to cancer therapy compared with expressing exogenous transcription factors in tumor cells. In this report, we used RNA interference to ask whether inhibiting PU.1 synthesis in MEL tumor cells is sufficient to cause the cells to lose their malignant properties. We found that treating the cells with a PU.1-specific short interfering RNA (siRNA) oligonucleotide causes them to reenter the erythroid differentiation program and undergo terminal arrest. Because hematologic malignancies in humans are often associated with alterations in the structure or regulation of a specific transcription factor, our results suggest that RNA interference directed at inhibiting the synthesis of such factors might lead to the development of potent antitumor therapy.
Results
Transient Inhibition of PU.1 Synthesis Induces Hemoglobin Accumulation in Erythroleukemia Cells
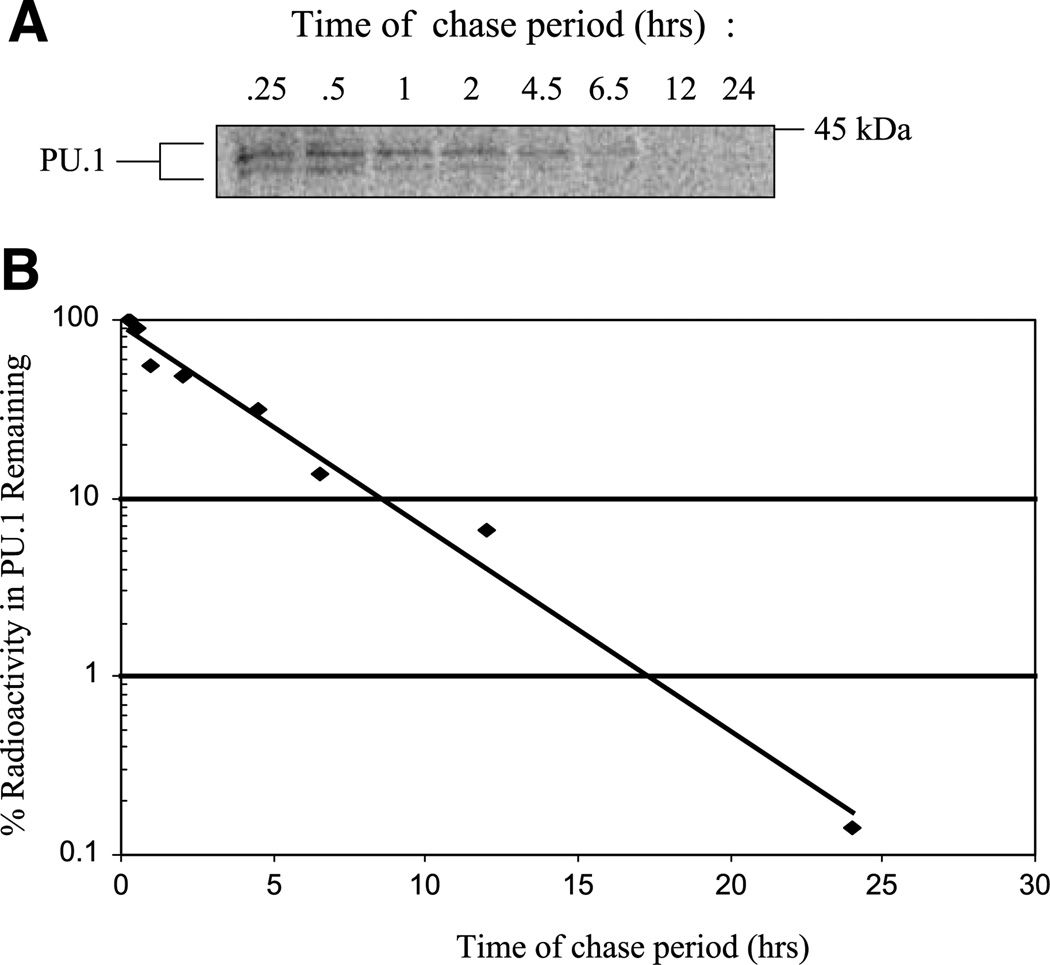
To determine whether blocking PU.1 synthesis is sufficient to induce erythroid differentiation in MEL cells, we sought to establish conditions for reducing PU.1 levels in the cells by transfecting them with a siRNA oligonucleotide directed against PU.1 mRNA. The effectiveness of a single siRNA treatment of cells in reducing target protein levels depends on many factors, of which the half-life of the protein of interest is one of the most critical. The treatment will be more effective if the half-life of the target protein is short in relation to the time required for maximal siRNA-induced degradation of the target mRNA sequence. To determine the half-life of PU.1 in MEL cells, we carried out a pulse-chase experiment. MEL cells were labeled with 35S-methionine for 15 min, and the cells were then transferred to medium containing excess unlabeled methionine. At various times thereafter, cell extracts were prepared, and the relative amounts of radioactivity in PU.1 were measured by immunoprecipitation with anti-PU.1 antibody followed by denaturing PAGE and quantitation of radioactivity in the PU.1 band by phosphorimaging (Fig. 1). The rate of loss of radioactivity in PU.1 during the chase period indicates that the half-life of PU.1 in MEL cells is ~2.4 h, suggesting that the protein will disappear fairly rapidly once PU.1 mRNA is degraded by siRNA treatment.
FIGURE 1.
Determination of PU.1 half-life in MEL cells. A. MEL cells were pulse-labeled with 100 µCi/mL of 35S-methionine for 15 min and chased in the presence of excess unlabeled methionine for the times indicated. Extracts of labeled cells were prepared with radioimmunoprecipitation assay buffer and immunoprecipitated with 1 µg of anti-PU.1. Immunoprecipitated complexes were separated by SDS-PAGE, and the dried gel was analyzed with a phosphorimager. B. Quantitation of radioactivity remaining in PU.1-specific immunoprecipitated complexes.
To determine whether a siRNA oligonucleotide directed against PU.1 mRNA can reduce the PU.1 protein level in MEL cells, we designed a PU.1-specific siRNA sequence using guidelines recommended by Reynolds et al. (30). These criteria, which include a lower internal stability at the 5′ antisense end, theoretically facilitate siRNA duplex unwinding and maximize strand retention by the RNA-induced silencing complex. Experiments were carried out in both MEL cells and in a MEL-transfected cell line (GATA-1-ER MEL) that expresses an exogenous GATA-1-ER fusion protein in which GATA-1 is fused to the ligand-binding domain of the estrogen receptor. We showed previously that in the absence of estrogen, these cells behave like untransfected MEL cells (29). However, in the presence of 10−7 mol/L of 17β-estradiol, the GATA-1-ER protein is activated, stimulating the transcription of GATA-1 target genes and inducing terminal erythroid differentiation. We also showed that estrogen treatment of GATA-1-ER MEL cells causes a rapid reduction of PU.1 (29), probably because the increase in active GATA-1 represses PU.1’s ability to stimulate its own gene transcription (31, 32). These results indicate that increasing the concentration of active GATA-1 relative to PU.1 in MEL cells strongly favors erythroid differentiation. The question we wished to address is whether the same result can be achieved by simply decreasing the concentration of PU.1 by RNA interference.
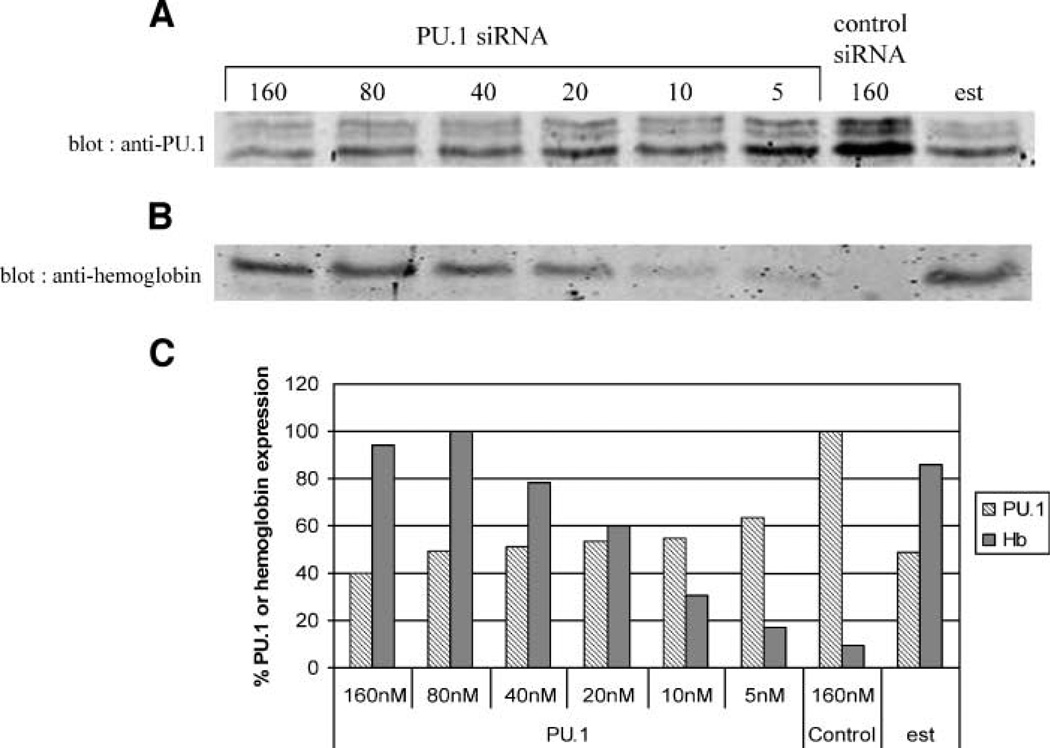
Cells were treated for 26 h with various concentrations of a PU.1-specific siRNA or a control siRNA, and the levels of PU.1 were determined at the end of the treatment by immunoblotting. Increasing concentrations of PU.1 siRNA up to 160 nmol/L causes a progressive decrease in PU.1 protein to a maximum 60% reduction (Fig. 2). The maximum reduction caused by the PU.1 siRNA is similar to that induced by estrogen treatment for the same time. To determine whether this reduction in PU.1 leads to derepression of hemoglobin synthesis, cell extracts were analyzed for hemoglobin by immunoblotting. The progressive decrease of PU.1 protein achieved with increasing concentrations of PU.1 siRNA leads to the accumulation of increasing amounts of hemoglobin (Fig. 2). The amounts of hemoglobin formed at the most effective concentrations of PU.1 siRNA (80 and 160 nmol/L) are slightly greater than that achieved with estrogen treatment. Careful comparison of the amounts of reduction in PU.1 level with the amounts of hemoglobin formed at various concentrations of PU.1 siRNA suggests that there is a threshold reduction in PU.1 level necessary to allow derepression of hemoglobin synthesis. For example, 80 nmol/L of PU.1 siRNA causes only a slightly greater reduction in PU.1 level than 10 nmol/L of PU.1 siRNA, but it allows much more hemoglobin to be formed. This effect may reflect the high degree of sensitivity of the proliferation versus differentiation decision in MEL cells to the relative stoichiometry of PU.1 and GATA-1 in the cells.
FIGURE 2.
PU.1 siRNA treatment suppresses PU.1 protein levels and induces hemoglobin expression. GATA-1-ER MEL cells were treated with 10−7 mol/L of β-estradiol or transiently transfected with a double-stranded PU.1 siRNA or control siRNA at the concentrations indicated for 26 h. Cell extracts were prepared by sonication, separated by SDS-PAGE, and immunoblotted for PU.1 (20 µg/lane; A) or hemoglobin (9 µg/lane; B). C. Quantitation of PU.1 and hemoglobin immunoblot band intensities.
Optimizing siRNA-Mediated Reduction in PU.1 and Induction of Hemoglobin Synthesis
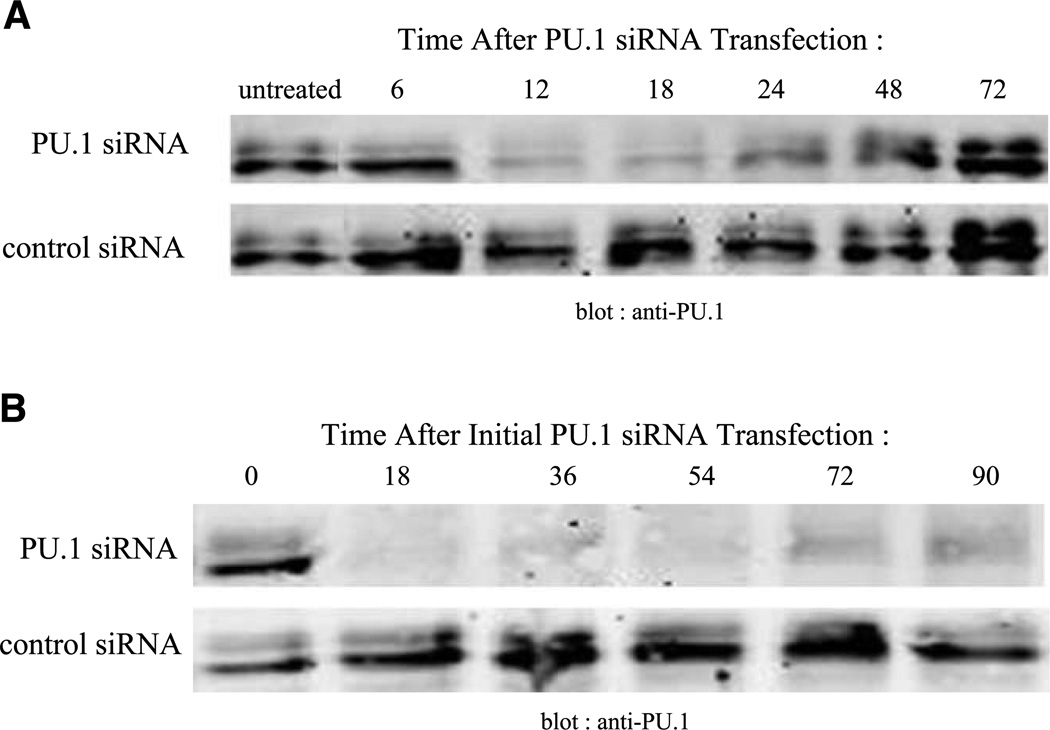
MEL cells typically are induced to differentiate by continuous treatment with a chemical inducing agent such as DMSO (7) or in the case of GATA-1-ER MEL cells, by continuous treatment with estrogen (29). These inducing agents presumably remain active during the entire differentiation period which usually occurs over 4 to 5 days. During this process, the level of PU.1 declines significantly by 24 h and remains low thereafter (21–24). To ascertain if a single siRNA transfection suppresses PU.1 for an extended period of time, PU.1 protein levels were measured at various times after transfection. PU.1-siRNA treatment induces a rapid decline in PU.1 protein, with the greatest reduction at 18 h (Fig. 3A). However, PU.1 protein subsequently increases, reaching the level of untreated cells by 72 h. Thus, a single treatment with PU.1-siRNA does not cause an extended decline in PU.1 levels typical of other differentiation induction protocols.
FIGURE 3.
PU.1 siRNA treatment induces a transient suppression of PU.1 protein levels. A. GATA-1-ER MEL cells were transfected once with 80 nmol/L of PU.1 siRNA. Cell extracts were prepared by sonication at the indicated times, separated by SDS-PAGE (20 µg/lane), and immunoblotted for PU.1. B. GATA-1-ER MEL cells were repeatedly transfected with PU.1 siRNA or control siRNA as described in (A) every 18 h. Extracts of the cells were prepared by sonication at the indicated times, separated by SDS-PAGE and immunoblotted for PU.1.
In an attempt to produce an extended reduction in PU.1 expression, we transfected GATA-1-ER MEL cells with siRNA multiple times. An 18-h schedule for repeated transfections was chosen because 18 h is the point of maximum reduction after a single siRNA transfection (Fig. 3A). This retransfection strategy produces an extended reduction in PU.1 levels that lasts for at least 90 h (Fig. 3B). The effect is specific because multiple transfections with the control siRNA do not cause PU.1 proteins to decline.
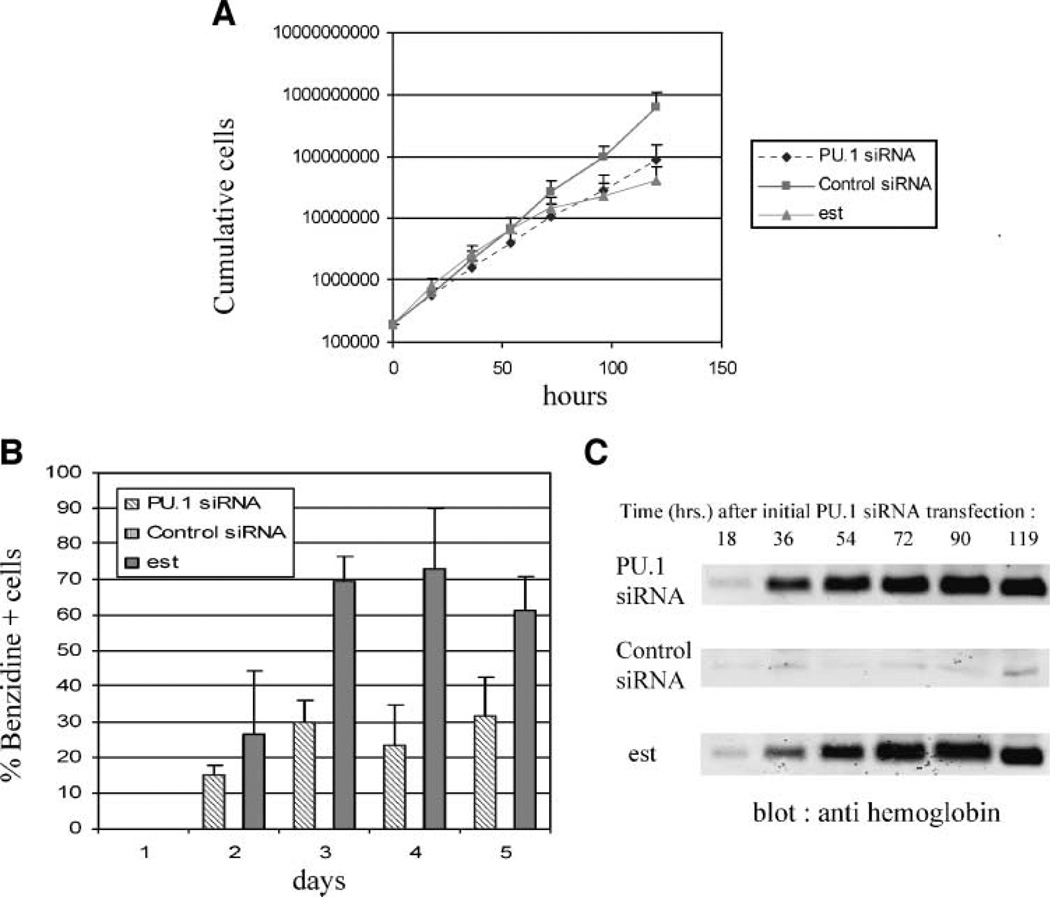
We determined the effect of multiple PU.1-siRNA transfections on proliferation by cell counting and the effect on differentiation by benzidine staining of the cells. Benzidine will positively stain cells that express hemoglobin. For comparison, measurements were also made on the GATA-1-ER MEL cells treated with 17β-estradiol. PU.1 siRNA treatment causes a marked reduction in cell proliferation. (Fig. 4A). Compared with control siRNA-treated cells that grow exponentially for 5 days with a doubling time of 9.9 h, PU.1 siRNA-treated cells grow initially with a doubling time of 10.9 h. Their growth rate begins to significantly decline at ~72 h, and by 5 days, they accumulate only 22% of the cell number of the control siRNA-treated culture. Furthermore, ~15% of PU.1 siRNA-treated cells are benzidine positive after 2 days and ~30% of the cells are positive at 3 to 5 days. In contrast, control siRNA treatment produces very few benzidine-positive cells (Fig. 4B). The rapid induction of differentiated cells by PU.1-siRNA treatment was confirmed by immunoblotting cell extracts for hemoglobin (Fig. 4C). By comparison, estrogen treatment of the GATA-1-ER MEL cells produces a more gradual accumulation of benzidine-positive cells and hemoglobin (Fig. 4B and C). However, estrogen treatment causes substantially more cells to differentiate and a concomitant greater reduction in cell proliferation (Fig. 4A). These results indicate that reducing PU.1 proteins by specific siRNA treatment is sufficient to induce substantial cell differentiation and hemoglobin accumulation.
FIGURE 4.
PU.1 siRNA treatment inhibits cell growth and induces sustained hemoglobin expression. GATA-1-ER MEL cells were treated once with 10−7 mol/L of β-estradiol or repeatedly transfected with 80 nmol/L of PU.1-siRNA or control siRNA every 18 h. Cumulative cell numbers (A) and benzidine-positive cells (B) were measured at 24-h intervals during the treatment. For control siRNA, benzidinepositive cells are <1% at each time point and therefore do not register on the graph. C. Cell extracts from estradiol-, PU.1 siRNA-, or control siRNA – treated cells were prepared by sonication at the indicated times, separated by SDS-PAGE (5 µg/lane) and, immunoblotted for hemoglobin.
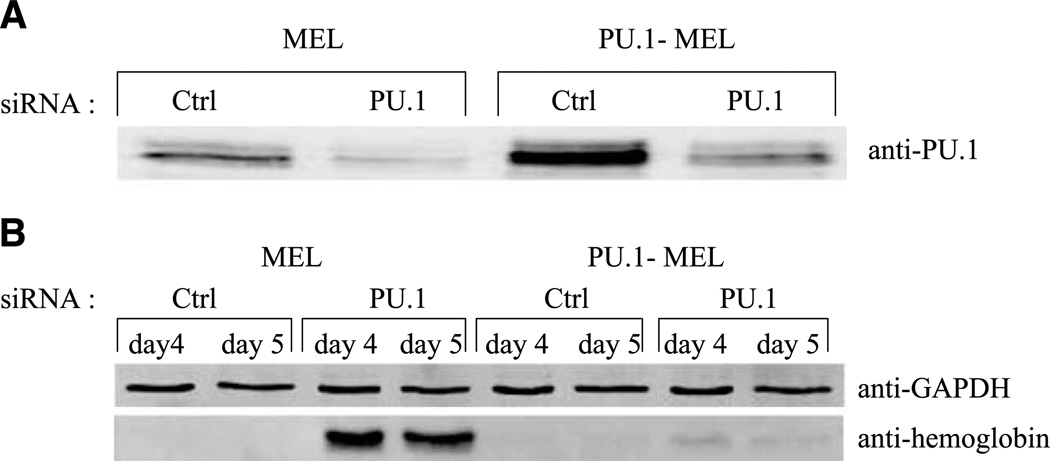
As a further check on the specificity of the PU.1 siRNA treatment, we sought to determine if expression of excess PU.1 would counteract the effects of the PU.1 siRNA oligonucleotide. For this purpose, we used a transfected MEL cell line that stably expresses an exogenous PU.1 protein (PU.1-MEL cells), in addition to endogenous PU.1 (23). The total level of PU.1 in these cells is several times that in MEL cells (Fig. 5A). PU.1 siRNA treatment of these cells does reduce the level of PU.1, but the resulting level is considerably higher than that in siRNA-treated MEL cells (Fig. 5A). Moreover, the siRNA treatment of these cells fails to induce significant levels of hemoglobin. These results strongly support the view that induction of differentiation by PU.1-siRNA treatment is due specifically to a reduction in the level of PU.1 in the cells and not due to off-target effects.
FIGURE 5.
PU.1 siRNA-induced hemoglobin expression is blocked by exogenous PU.1. A. Parental MEL cells or MEL stable transfectants constitutively overexpressing PU.1 (PU.1-MEL) were transfected once with 80 nmol/L PU.1 siRNA or control siRNA. Eighteen hours after transfection, cell extracts were prepared by sonication, separated by SDS-PAGE (20 µg/lane), and immunoblotted for PU.1. B. Four and 5 days after the single transfection with PU.1 or control siRNA, cell extracts were prepared by sonication, separated by SDS-PAGE (5 µg/lane), and immunoblotted for hemoglobin or glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Inhibition of PU.1 Synthesis Leads to Terminal Cell Division
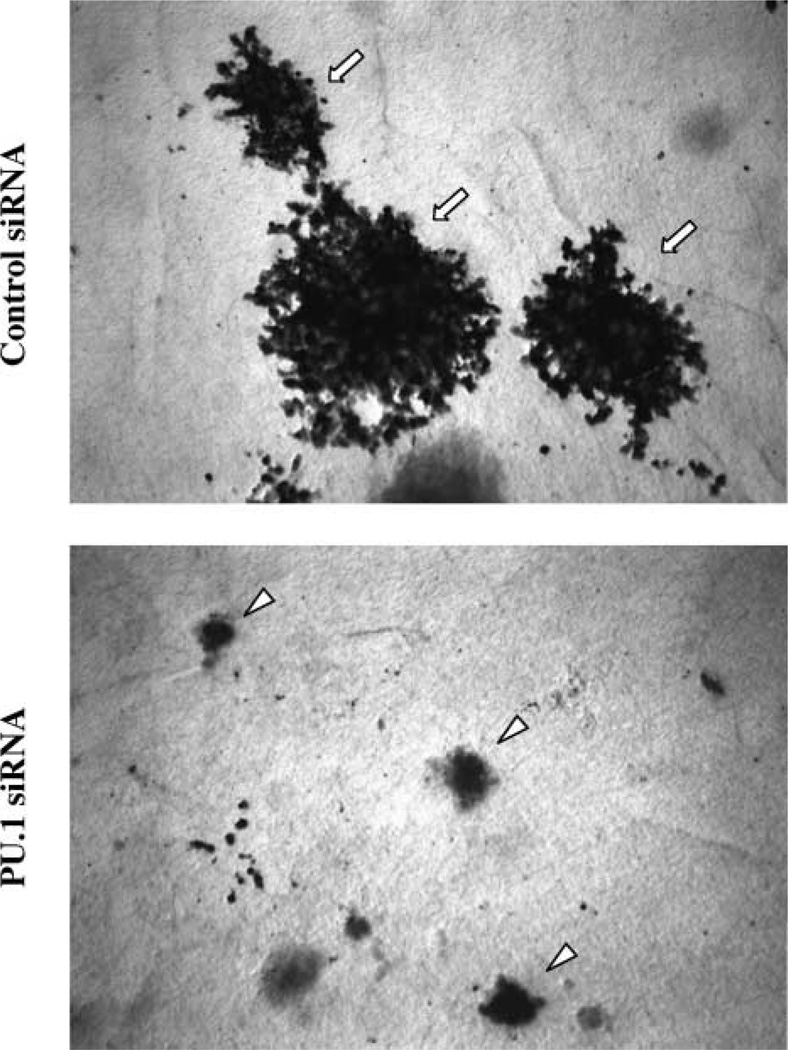
An important feature of the in vitro differentiation of MEL cells is that it initially leads to the production of cells that are not overtly differentiated but are irreversibly committed to differentiate (33). These committed cells no longer require the presence of the inducing stimulus to execute the terminal differentiation program. In contrast to untreated MEL cells that have unlimited proliferative potential, the committed cells can undergo a maximum of five to six cell divisions, similar to normal erythroid precursors. To determine if PU.1 siRNA treatment causes MEL cells to undergo irreversible commitment to terminal differentiation, we treated cells with PU.1 siRNA for various periods of time and then plated them in plasma clots in the absence of siRNA. PU.1 siRNA treatment causes up to 55% of the cells to form small, hemoglobinized colonies in plasma clots (Fig. 6). Importantly, all cells positive for hemoglobin form small colonies, indicating that hemoglobin production and terminal growth arrest are coupled in cells treated with PU.1 siRNA. In contrast, cells treated with the control siRNA form only very large, nonhemoglobinized colonies, typical of uncommitted MEL cells. These results indicate that PU.1 siRNA treatment induces not only hemoglobin synthesis, but also causes the cells to switch irreversibly into the terminal differentiation program.
FIGURE 6.
Inhibition of PU.1 synthesis leads to terminal cell division. GATA-1-ER MEL cells were repeatedly transfected with 80 nmol/L of PU.1 or control siRNA every 18 h. At 24-h intervals they were plated as single cells in plasma clots in the absence of siRNA and incubated for an additional 4 days at 37°C as described in Materials and Methods. The clots were then transferred to glass slides and stained for hemoglobin expression with benzidine reagent and counterstained with hematoxylin. Small benzidine-positive colonies represent committed cells undergoing terminal cell division whereas large benzidine-negative colonies represent uncommitted cells. Representative colonies in plasma clots from cells treated for 3 days with the indicated siRNAs. Arrows, uncommitted colonies; arrowheads, colonies committed to differentiation.
Reprogramming by PU.1 siRNA Occurs Preferentially in G1 Phase of the Cell Cycle
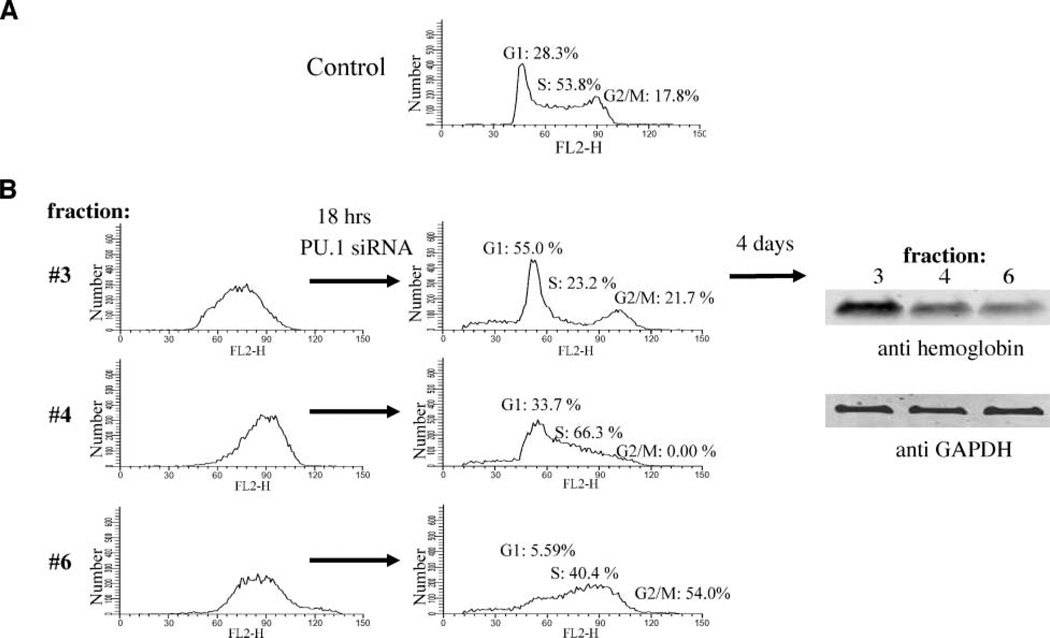
We have shown previously that MEL cells can be induced to differentiate by inhibiting specific cyclin-dependent kinases and that the sensitive period for induction is restricted to the G1 phase of the cell cycle. To determine whether cell cycle phase plays a role in the differentiation triggered by PU.1 siRNA treatment, we prepared cells at various stages of the cell cycle by centrifugal elutriation, a method that separates cells by their size, which correlates with their position in the cell cycle (34, 35). Fractions containing high percentages of cells at particular stages of the cell cycle were collected. For example, almost 100% of the cells in fraction 3 were originally in S phase, whereas fractions 4 and 6 were initially composed of some cells in S and most in G2-M phase. These samples were treated with the PU.1 siRNA for 18 h, a period of time during which the PU.1 level reaches a minimum value (Fig. 3A). The cell cycle distribution of each of the fractions was then determined by FACScan. Samples containing a high percentage of cells in G1 (55%), S (66%) or G2-M (54%) were chosen for further analysis (Fig. 7B). The amount of hemoglobin formed by each of these three cell samples was determined after 4 days of further incubation in the absence of PU.1 siRNA. Cell fractions enriched in G1 after PU.1 siRNA treatment were able to produce larger amounts of hemoglobin than cell fractions enriched in other cell cycle phases (Fig. 7B). Similar results were obtained by first treating cells with PU.1 siRNA for 18 h, then sorting them into G1, S and G2-M populations and examining them 4 days later for hemoglobin content (data not shown). The greater ability of cells in G1 to produce hemoglobin upon PU.1 siRNA treatment is not due to different efficiencies of PU.1 siRNA-induced knockdown of PU.1 in G1 versus S and G2-M phase cells because PU.1 siRNA suppresses PU.1 protein to very similar levels (9–10% of control) in different phases of the cell cycle (data not shown). These results indicate that upon inhibition of PU.1 by specific siRNA treatment, cells lying in G1 phase have a higher probability of differentiating, similar to our earlier observations with the induction of differentiation by cell cycle inhibitors (34).
FIGURE 7.
Reprogramming by PU.1 siRNA occurs preferentially in the G1 phase of the cell cycle. A. FACScan analysis of unfractionated, exponentially growing GATA-1-ER MEL cells. B. GATA-1-ER MEL cells were separated by size using centrifugal elutriation. Fractions containing cells enriched in different stages of the cell cycle (fractions 3, 4, and 6; left) were collected and transfected once with 80 nmol/L of PU.1-siRNA. Eighteen hours later, the cells were analyzed by FACScan, and fractions containing a high percentage of cells in G1 (55%), S (66%), or G2-M (54%) phase were incubated at 37°C for an additional 4 days (right). Cells were lysed with radioimmunoprecipitation assay buffer and extracts(7.5µg/lane) were separated by SDS-PAGE and immunoblotted for hemoglobin or glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Cell cycle modeling was done using ModFit LT software (Verity Software House).
Discussion
There is now an increasing appreciation that many transcription factors interact with one another through direct protein-protein contacts, the consequences of which can positively or negatively affect the activity of the factors depending on the specific proteins involved and the cellular context. PU.1 and GATA-1 are two key hematopoietic transcription factors whose interaction results in the repression of each factor’s transcriptional activity (25, 26). Thus, in cells that express both factors simultaneously, the stoichiometry of the two factors is expected to determine which transcriptional program, the myeloid program directed by PU.1 or the erythroid program directed by GATA-1, will predominate. Indeed, Nerlov and Graf showed that the stoichiometry of PU.1 and GATA-1 is important in determining the differentiation status of avian multipotential precursor cells (36), and we showed that the stoichiometry of the two factors is important in erythropoiesis in Xenopus embryos (25). Increasing evidence suggests that multipotential hematopoietic progenitors express a “promiscuous” gene expression program encompassing subsets of genes expressed by their numerous lineage-restricted progeny (37, 38). The ability of PU.1 to repress GATA-1 seems to be important in extinguishing the low-level erythroid gene expression that is present in multipotential progenitors as they differentiate into macrophages (27).
Perturbing the balance between PU.1 and GATA-1 can also contribute to leukemia development. Proviral insertions at the PU.1 locus, leading to deregulation of PU.1 synthesis, occur in at least 95% of erythroleukemias in Friend virus–infected mice (12). Therefore, MEL cells express a high level of PU.1, relative to GATA-1, that inhibits their further differentiation into more mature erythroid cells. A wide variety of chemical agents have been discovered that can induce the cells to overcome this block, but in most cases, their mechanisms of action are not known (7, 39). Nevertheless, we recently showed that stable transfection of MEL cells with an expression vector encoding a conditionally active form of GATA-1 (GA TA-1-ER) causes the cells to resume erythroid differentiation and undergo terminal cell division (29). These results suggest that changing the stoichiometry of PU.1 and GATA-1 in MEL cells in favor of GATA-1 is sufficient to cause them to lose their malignant properties. However, additional exogenous GATA-1 may have many actions in MEL cells beyond counteracting PU.1. Furthermore, as a cancer therapy, expression of exogenous transcription factors in tumor cells has numerous obstacles. Therefore, we turned to RNA interference to ask whether inhibiting PU.1 synthesis in MEL cells is sufficient to cause them to undergo terminal erythroid differentiation, including growth arrest.
We found that careful optimization of siRNA concentration and frequency of siRNA delivery is necessary to induce a large percentage of the cells to undergo erythroid differentiation. This is likely due to the fact that after a single treatment with PU.1 siRNA, the PU.1 protein level begins to increase after ~18 h (Fig. 3A). Previous studies have shown that induction of MEL cell differentiation is a stochastic process in which an increasing number of cells are committed to differentiate depending on inducer type, inducer concentration, and time of exposure (33). In most induction protocols, cells are continually exposed to the inducer. Therefore, we reasoned that a retransfection strategy might be useful, and we chose an 18-h schedule, because after 18 h of a single PU.1-siRNA transfection, the PU.1 level begins to increase. The retransfection strategy produced an extended reduction in PU.1 protein that lasts at least 90 h (Fig. 3B). It also leads to substantial hemoglobin accumulation in the cells, ~30% of which stain positive with benzidine, and a marked reduction in cell proliferation (Fig. 4). By comparison, activation of the GATA-1-ER fusion protein by estrogen treatment of these cells produces a somewhat slower accumulation of hemoglobin, but ultimately, more benzidine-positive cells and a concomitant greater reduction in cell proliferation. The reason for the difference in the percentage of cells undergoing differentiation with the two treatments is not clear. It is possible that the discrepancy is due to a difference in the GATA-1/PU.1 ratio produced in the cells by the two methods. Another possibility is that the kinetics of PU.1 suppression may differ between the two treatments. Perhaps stable transfection of the cells with a PU.1-siRNA expression vector would be more effective than treatment with exogenous siRNA oligonucleotides. It is also possible that GATA-1 activation results in a greater transient prolongation in the G1 phase of the cell cycle than that mediated by PU.1 siRNA. This prolongation occurs upon chemical induction of MEL cell differentiation and has been suggested to allow the progression of events critical for the commitment to differentiation (40, 41). Indeed, we showed that the cyclin-dependent kinase inhibitor p21 is only effective at inducing MEL cell differentiation during G1 phase (34). Therefore, GATA-1 activation may be more effective than PU.1 siRNA treatment at inducing other events, in addition to PU.1 suppression, that are necessary for differentiation.
An important goal of the current work is to determine whether specific reduction of the PU.1 level in MEL cells is sufficient to cause the cells to reenter their terminal differentiation program including growth arrest. Previous attempts to inhibit PU.1 synthesis in MEL cells have produced variable results. Delgado et al. found that reducing the PU.1 level by treating MEL cells with three doses of PU.1-specific antisense RNA (at 0, 5, and 15 h) leads to a decrease in proliferation but does not cause the accumulation of hemoglobin as measured by benzidine staining (24). Atar and Levi showed that MEL cells transfected with a retroviral DNA construct encoding a PU.1 siRNA accumulate hemoglobin and exhibit reduced growth (42). The antisense and siRNA constructs used in both studies each targets a region of PU.1 coding sequence distinct from that targeted by the PU.1 siRNA used in the current work. Neither study determined whether any of the PU.1-suppressed cells had actually undergone terminal cell division. Likewise, we recently reported that the same PU.1 siRNA used in the current work can induce the acquisition of an active chromatin structure and derepression of transcription of GATA-1 target genes previously repressed by PU.1 (27); however, our studies did not address the issue of terminal differentiation and growth arrest. The only definitive way to ascertain these properties is to determine the fate of individual cells in colony formation assays carried out in the absence of the siRNA following treatment of the cells with the siRNA. To this end, we used the plasma clot assay originally devised to characterize the differentiation potential of hematopoietic progenitors (43). The advantage of this assay is that it allows a determination of both the differentiation state and proliferative capacity of the progeny of single cells. Typically, MEL cells that are irreversibly committed to differentiate give rise to small (32–64 cells) colonies in which all cells stain positive with benzidine. The colonies are similar in size to those produced by committed erythroid progenitors, CFU-E, in the plasma clot assay. On the other hand, uncommitted MEL cells, exhibiting properties of the original tumor line, produce very large colonies with unlimited proliferative potential. Using the plasma clot assay, we found that up to 55% of PU.1-siRNA–treated MEL cells produce small, hemoglobinized colonies (Fig. 6), indicating that the cells had resumed their terminal differentiation program. Note that the cells giving rise to these small colonies are not in terminal arrest at the time of their plating in plasma clot, but their proliferative potential, like that of normal CFU-E, is limited to five to six cell divisions. We conclude that merely depleting the level of PU.1 in MEL cells is sufficient to cause the cells to switch their fate and to lose their malignant properties. This commitment to differentiation, including both the expression of differentiation-specific phenotypic markers as well as terminal growth arrest, is a key issue in tumor therapy. Induction of death or differentiation, not merely growth suppression, in tumors would ensure the irreversibility of the conversion to a noncancerous state.
Whereas overexpression of PU.1 in erythroid cells blocks differentiation and promotes erythroleukemic transformation, reduced expression of PU.1 in mice has been associated with the development of leukemia in myeloid cells (44, 45), and heterozygous mutations in PU.1 have been reported in some patients with acute myeloid leukemia (46). In myeloid leukemia cells that express an AML1-ETO fusion protein, PU.1 activity is reduced, and overexpression of exogenous PU.1 in such cells induces monocytic differentiation (47). Likewise, the expression of the PML-RARα fusion protein in acute promyelocytic leukemia reduces PU.1 expression, and restoration of PU.1 expression in acute promyelocytic leukemia cells induces neutrophil differentiation. Furthermore, treatment of acute promyelocytic leukemia cells with all-trans retinoic acid increases PU.1 levels and induces neutrophil differentiation, whereas reduction of PU.1 by siRNA treatment blocks all-trans retinoic acid–induced differentiation (48). Therefore, in both erythroid and myeloid lineages, abnormally high or low levels of PU.1, respectively, could contribute to the development of leukemia, and restoration of normal PU.1 expression drives differentiation.
The studies reported here strongly support the concept that the levels of certain transcription factors are an important determinant of the proliferation versus differentiation decision in leukemia and other types of malignant cells. Thus, siRNA inhibition of the synthesis of specific transcription factors may be a viable strategy for differentiation therapy in hematologic malignancies. In this context, we also wish to emphasize that a combination of therapeutic siRNAs may be useful. The effect of PU.1 siRNA in MEL cells is greatest in the G1 phase of the cell cycle (Fig. 7). This result is consistent with our earlier finding that MEL cells can be reprogrammed to terminal differentiation by inhibiting specific cyclin-dependent kinases in G1 phase (34). These results suggest that perhaps a combination of siRNAs, some directed at appropriate transcription factors and others directed at certain pro-proliferative G1 cyclin-dependent kinases, may synergize to cause tumor cells to undergo terminal growth arrest.
Materials and Methods
Cell Culture
Clone DS19 MEL cells were grown at 37°C, 10% CO2 in DMEM supplemented with 10% fetal bovine serum, 2 mmol/L of l-glutamine, 100 units/mL of penicillin, and 100 µg/mL of streptomycin. MEL cells stably expressing GATA-1 fused to the ligand-binding domain of the estrogen receptor (GATA-1/ER MEL cells) were generated, cultured, and induced to differentiate as described previously (29). Cell growth was measured by counting aliquots of the cultures with a model Zf Coulter counter (Coulter Electronics). Benzidine staining for hemoglobinized cells was done as described previously (49). Briefly, 4 µL of 30% hydrogen peroxide was added to 200 µL of 0.2% benzidine dihydrochloride/3% glacial acetic acid. This mixture was added to 200 µL of a cell suspension in a 24-well plate. After 10 min, the percentage of benzidine-positive (blue) cells was determined by examining at least 100 cells by bright-field light microscopy.
Determination of PU.1 Half-life
MEL cells (2 × 107) were washed twice and incubated in 4 mL of labeling medium (methionine-deficient DMEM supplemented with 10% fetal bovine serum) for 15 min at 37°C to deplete intracellular methionine pools. Cells were centrifuged, resuspended in 2 mL of labeling medium + 100 µCi/mL of 35S-methionine (New England Nuclear) and incubated for 15 min at 37°C. To stop the incorporation of labeled methionine, 4 mL of chase medium (methionine-deficient DMEM supplemented with 15 mg/L methionine + 10% fetal bovine serum) was added. After centrifugation and washing with 10 mL of chase medium, the cells were resuspended in 40 mL of chase medium and incubated at 37°C. Four-milliliter aliquots were removed after 15 and 30 min, and 1, 2, 4.5, 6.5, 12, and 24 h, and cell pellets were frozen at −80°C. Frozen cells were lysed by resuspending in 1 mL of radioimmunoprecipitation assay buffer [150 mmol/L NaCl, 30 mmol/L Tris (pH 8.0), 0.1% SDS, 0.5% sodium deoxycholate, 1% NP40] + protease inhibitors (pepstatin, leupeptin, aprotinin, phenylmethylsulfonyl fluoride) and rotating at 4°C for 1 h. For immunoprecipitation of PU.1, 200 µL of each lysate was precleared with 10 µL protein A/agarose (Repligen Corp.) for 1 h at 4°C. Meanwhile, 200 µL of protein A/agarose was incubated with 100 µL of anti-PU.1 (20 µg, clone T-21; Santa Cruz Biotechnology) for 1 h at room temperature and washed once with radioimmunoprecipitation assay buffer. Ten microliters of antibody/agarose complexes (containing 1 µg of anti-PU.1) were incubated with each precleared lysate at 4°C for 16 h, washed four times with 0.5 mL/wash of radio-immunoprecipitation assay buffer and washed once with 0.5 mL of 30 mmol/L Tris (pH 8.0), and 50 mmol/L of NaCl. Immunocomplexes were resuspended in 50 µL of loading buffer (4% SDS, 20% glycerol, 100 mmol/L Tris (pH 7.5), and 200 mmol/L DTT) and separated by SDS-PAGE. The gel was fixed for 1 h in 50% methanol/10% acetic acid, rinsed 15 min in distilled water, incubated 1 h in 1 mol/L sodium salicylate, dried, and exposed to a phosphorimaging screen (Molecular Dynamics, Sunnyvale, CA) for 3 days. The screen was read by a Storm 860 Phosphorimager (Molecular Dynamics), and radioactivity in the PU.1 bands was quantitated with ImageQuant software. The time at which 50% of the original radioactivity in PU.1 remained was designated the half-life of PU.1.
Plasma Clot Assay
Plasma clot assays were done essentially as described (43). Briefly, GATA-1-ER cells treated with either PU.1 or control siRNA for various periods of time were washed once in DMEM and plated in plasma clots consisting of the following assembled in 96-well microtiter plates: 100 µL of 0.2 mg/mL L-asparagine; 100 µL of 10 units/mL thrombin (Amersham); 200 µL of heat inactivated fetal bovine serum (Gemini Bioproducts); 100 µL of 10% bovine serum albumin (Fraction V, Sigma); 300 µL of DMEM, 100 µL of treated cells (200–500 cells); 100 µL of citrated bovine plasma (Animal Technologies). The clots were allowed to form for 5 to 10 min at room temperature resulting in a single-cell suspension. Plasma clots were incubated at 37°C, 10% CO2 for 4 days, then transferred to glass slides as follows: each clot was incubated with 8 drops of 5% glutaraldehyde (Electron Microscopy Sciences)/PBS at room temperature for 20 min. The clots were then scooped from the wells and placed onto glass slides. Approximately two more drops of 5% glutaraldehyde/PBS were added to each clot, and a piece of Whatman no. 1 filter paper was carefully placed on top of the clots. Another piece of Whatman no. 1 filter paper was placed on top of the first and removed just as it became saturated with glutaraldehyde. The clots were allowed to dehydrate at room temperature for 12 to 15 min and then the remaining piece of Whatman filter paper was carefully removed. The slides were rinsed with distilled water for 8 min and then completely dried with a stream of air. The slides were stained with 1% O-dianisidine/100% methanol for 5 min followed by 2.5% hydrogen peroxide in 70% ethanol for 5 min, rinsed with distilled water for 1 min, and counterstained with Harris’ modified hematoxylin for 2 min. Cells committed to differentiation formed small orange colonies of 8 to 50 cells whereas uncommitted cells formed large blue colonies containing hundreds of cells.
Western Blotting
Cells were centrifuged, and the pellets were frozen at −80°C. Cell pellets were resuspended in sonication buffer [50 mmol/L HEPES (pH 7.5), 150 mmol/L NaCl, 1 mmol/L EDTA (pH 8.0), 2.5 mmol/L EGTA (pH 7.2), 1 mmol/L DTT, 0.1% Tween 20, 10% gylcerol; 10 mmol/L β-glycerophosphate] + complete protease inhibitor cocktail (Roche) at ~106 cells per 300 µL and sonicated on ice with three pulses of 20 s at 15% to 20% output with intervening pauses of 30 s on a Model 500 Sonic Dismembrator (Fisher Scientific) equipped with a microtip (Branson). Cell extracts were centrifuged at 20,817 rcf at 4°C, and proteins in the supernatants were separated by SDS-PAGE and transferred to nitrocellulose with a Trans-Blot SD Semi-Dry Transfer Cell (Bio-Rad Laboratories) for 1 h at 15 V. Membranes were blocked for 1 h at room temperature or overnight at 4°C in Odyssey blocking buffer (LiCor Biosciences). Immunodetection was done by incubating blocked membranes with primary antibody for 1.5 to 2 h at room temperature followed by incubation with secondary antibody (1:10,000 goat anti-rabbit IgG-IR Dye 800; Rockland Immunochemicals) for 1 h at room temperature. Primary antibodies used were as follows: 1:1,000 rabbit anti-hemoglobin (ICN), 1:1,000 rabbit anti–glyceraldehyde-3-phosphate dehydrogenase (Abcam), and 1:500 rabbit anti-PU.1 (NH2-terminal epitope, a generous gift from Dr. Francoise Moreau-Gachelin, Institut Curie, INSERM U528, Paris, France). Membranes were scanned by an IR imaging system (LiCor) with Odyssey software (LiCor).
siRNA Transfection
A double-stranded RNA oligo targeting PU.1 (5′-GGAGGUGUCUGAUGGAGAA-3′) was designed using GeneScript1 and Dharmacon2 siRNA design tools. An oligo consisting of a mutated Snf2h sequence (5′-GAGGAUAGGGAAGAGCUAU-3′; ref. 50) was used as a control. All siRNA oligos were synthesized by Qiagen and contained 3′ dTdT overhangs. Cells were transfected as follows (per well of a six-well plate): 105 cells were plated in 800 µL of antibiotic-free DMEM supplemented with 10% fetal bovine serum. Four microliters of a 20 µmol/L siRNA oligo stock diluted to 185 µL with DMEM and 4 µL of Oligofectamine (Invitrogen) diluted to 15 µL with DMEM were incubated separately at room temperature for 5 min. The two mixtures were combined and incubated at room temperature for an additional 20 min. The siRNA-Oligofectamine mixtures were then added dropwise to cells in the well. Different concentrations of oligos were used for optimization experiments, but for subsequent studies, the optimal final concentration of 80 nmol/L was used. For retransfection experiments, cells were first incubated with siRNA-Oligofectamine for 18 h. The cells were counted, and 105 cells were centrifuged, resuspended in 800 µL of DMEM supplemented with 10% fetal bovine serum, reseeded in a new well of a six-well plate, and transfected with newly prepared siRNA-Oligofectamine mixtures as described above. The retransfection procedure was repeated every 18 h up to 3 to 5 days.
Acknowledgments
We thank Emily Cook and Carl Schildkraut for assistance with centrifugal elutriation, as well as William King and Clarissa Santos-Peña for help with FACS.
Grant support: NIH grants HL78381 (A.I. Skoultchi) and 1F32 HL077242-0102 (M. Papetti), and National Cancer Institute Cancer Center grant 2P30CA13330 (A.I. Skoultchi).
Footnotes
References
- 1.Leszczyniecka M, Roberts T, Dent P, Grant S, Fisher PB. Differentiation therapy of human cancer: basic science and clinical applications. Pharmacol Ther. 2001;90:105–156. doi: 10.1016/s0163-7258(01)00132-2. [DOI] [PubMed] [Google Scholar]
- 2.Lee CR, Cervi D, Truong AH, Li YJ, Sarkar A, Ben-David Y. Friend virus-induced erythroleukemias: a unique and well-defined mouse model for the development of leukemia. Anticancer Res. 2003;23:2159–2166. [PubMed] [Google Scholar]
- 3.Ben-David Y, Bernstein A. Friend virus-induced erythroleukemia and the multistage nature of cancer. Cell. 1991;66:831–834. doi: 10.1016/0092-8674(91)90428-2. [DOI] [PubMed] [Google Scholar]
- 4.Kabat D. Molecular biology of Friend viral erythroleukemia. Curr Top Microbiol Immunol. 1989;148:1–42. doi: 10.1007/978-3-642-74700-7_1. [DOI] [PubMed] [Google Scholar]
- 5.Friend C, Patuleia MC, De Harven E. Erythrocytic maturation in vitro of murine (Friend) virus-induced leukemic cells. J Natl Cancer Inst Monogr. 1966;22:505–522. [PubMed] [Google Scholar]
- 6.Ikawa Y, Sugano H. An ascites tumor derived from early splenic lesion of Friend’s disease: a preliminary report. Gann. 1966;57:641–643. [PubMed] [Google Scholar]
- 7.Marks PA, Rifkind RA. Erythroleukemic differentiation. Annu Rev Biochem. 1978;47:419–448. doi: 10.1146/annurev.bi.47.070178.002223. [DOI] [PubMed] [Google Scholar]
- 8.Nishigaki K, Thompson D, Hanson C, Yugawa T, Ruscetti S. The envelope glycoprotein of friend spleen focus-forming virus covalently interacts with and constitutively activates a truncated form of the receptor tyrosine kinase Stk. J Virol. 2001;75:7893–7903. doi: 10.1128/JVI.75.17.7893-7903.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rovinski B, Munroe D, Peacock J, Mowat M, Bernstein A, Benchimol S. Deletion of 5′-coding sequences of the cellular p53 gene in mouse erythroleukemia: a novel mechanism of oncogene regulation. Mol Cell Biol. 1987;7:847–853. doi: 10.1128/mcb.7.2.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ben-David Y, Lavigueur A, Cheong GY, Bernstein A. Insertional inactivation of the p53 gene during friend leukemia: a new strategy for identifying tumor suppressor genes. New Biol. 1990;2:1015–1023. [PubMed] [Google Scholar]
- 11.Munroe DG, Peacock JW, Benchimol S. Inactivation of the cellular p53 gene is a common feature of Friend virus-induced erythroleukemia: relationship of inactivation to dominant transforming alleles. Mol Cell Biol. 1990;10:3307–3313. doi: 10.1128/mcb.10.7.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moreau-Gachelin F, Tavitian A, Tambourin P. Spi-1 is a putative oncogene in virally induced murine erythroleukaemias. Nature. 1988;331:277–280. doi: 10.1038/331277a0. [DOI] [PubMed] [Google Scholar]
- 13.Paul R, Schuetze S, Kozak SL, Kabat D. A common site for immortalizing proviral integrations in Friend erythroleukemia: molecular cloning and characterization. J Virol. 1989;63:4958–4961. doi: 10.1128/jvi.63.11.4958-4961.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paul R, Schuetze S, Kozak SL, Kozak CA, Kabat D. The Sfpi-1 proviral integration site of Friend erythroleukemia encodes the ets-related transcription factor Pu.1. J Virol. 1991;65:464–467. doi: 10.1128/jvi.65.1.464-467.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goebl MK. The PU.1 transcription factor is the product of the putative oncogene Spi-1. Cell. 1990;61:1165–1166. doi: 10.1016/0092-8674(90)90676-6. [DOI] [PubMed] [Google Scholar]
- 16.Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 17.McKercher SR, Torbett BE, Anderson KL, et al. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996;15:5647–5658. [PMC free article] [PubMed] [Google Scholar]
- 18.Moreau-Gachelin F, Wendling F, Molina T, et al. Spi-1/PU.1 transgenic mice develop multistep erythroleukemias. Mol Cell Biol. 1996;16:2453–2463. doi: 10.1128/mcb.16.5.2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barnache S, Wendling F, Lacombe C, et al. Spi-1 transgenic mice develop a clonal erythroleukemia which does not depend on p53 mutation. Oncogene. 1998;16:2989–2995. doi: 10.1038/sj.onc.1202095. [DOI] [PubMed] [Google Scholar]
- 20.Schuetze S, Stenberg PE, Kabat D. The Ets-related transcription factor PU.1 immortalizes erythroblasts. Mol Cell Biol. 1993;13:5670–5678. doi: 10.1128/mcb.13.9.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schuetze S, Paul R, Gliniak BC, Kabat D. Role of the PU.1 transcription factor in controlling differentiation of Friend erythroleukemia cells. Mol Cell Biol. 1992;12:2967–2975. doi: 10.1128/mcb.12.7.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Galson DL, Hensold JO, Bishop TR, et al. Mouse β-globin DNA-binding protein B1 is identical to a proto-oncogene, the transcription factor Spi-1/PU.1, and is restricted in expression to hematopoietic cells and the testis. Mol Cell Biol. 1993;13:2929–2941. doi: 10.1128/mcb.13.5.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao G, Rekhtman N, Cheng G, Krasikov T, Skoultchi AI. Deregulated expression of the PU.1 transcription factor blocks murine erythroleukemia cell terminal differentiation. Oncogene. 1997;14:123–131. doi: 10.1038/sj.onc.1200807. [DOI] [PubMed] [Google Scholar]
- 24.Delgado MD, Hallier M, Meneceur P, Tavitian A, Moreau-Gachelin F. Inhibition of Friend cells proliferation by spi-1 antisense oligodeoxynucleotides. Oncogene. 1994;9:1723–1727. [PubMed] [Google Scholar]
- 25.Rekhtman N, Radparvar F, Evans T, Skoultchi AI. Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev. 1999;13:1398–1411. doi: 10.1101/gad.13.11.1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang P, Behre G, Pan J, et al. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc Natl Acad Sci U S A. 1999;96:8705–8710. doi: 10.1073/pnas.96.15.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stopka T, Amanatullah DF, Papetti M, Skoultchi AI. PU.1 inhibits the erythroid program by binding to GATA-1 on DNA and creating a repressive chromatin structure. EMBO J. 2005;24:3712–3723. doi: 10.1038/sj.emboj.7600834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rekhtman N, Choe KS, Matushansky I, Murray S, Stopka T, Skoultchi AI. PU.1 and pRB interact and cooperate to repress GATA-1 and block erythroid differentiation. Mol Cell Biol. 2003;23:7460–7474. doi: 10.1128/MCB.23.21.7460-7474.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choe KS, Radparvar F, Matushansky I, Rekhtman N, Han X, Skoultchi AI. Reversal of tumorigenicity and the block to differentiation in erythroleukemia cells by GATA-1. Cancer Res. 2003;63:6363–6369. [PubMed] [Google Scholar]
- 30.Reynolds A, Leake D, Boese Q, Scaringe S, Marshall WS, Khvorova A. Rational siRNA design for RNA interference. Nat Biotechnol. 2004;22:326–330. doi: 10.1038/nbt936. [DOI] [PubMed] [Google Scholar]
- 31.Chen H, Ray-Gallet D, Zhang P, et al. PU.1 (Spi-1) autoregulates its expression in myeloid cells. Oncogene. 1995;11:1549–1560. [PubMed] [Google Scholar]
- 32.Okuno Y, Huang G, Rosenbauer F, et al. Potential autoregulation of transcription factor PU.1 by an upstream regulatory element. Mol Cell Biol. 2005;25:2832–2845. doi: 10.1128/MCB.25.7.2832-2845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gusella J, Geller R, Clarke B, Weeks V, Housman D. Commitment to erythroid differentiation by friend erythroleukemia cells: a stochastic analysis. Cell. 1976;9:221–229. doi: 10.1016/0092-8674(76)90113-6. [DOI] [PubMed] [Google Scholar]
- 34.Matushansky I, Radparvar F, Skoultchi AI. Reprogramming leukemic cells to terminal differentiation by inhibiting specific cyclin-dependent kinases in G1. Proc Natl Acad Sci U S A. 2000;97:14317–14322. doi: 10.1073/pnas.250488697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown EH, Schildkraut CL. Perturbation of growth and differentiation of Friend murine erythroleukemia cells by 5-bromodeoxyuridine incorporation in early S phase. J Cell Physiol. 1979;99:261–278. doi: 10.1002/jcp.1040990213. [DOI] [PubMed] [Google Scholar]
- 36.Nerlov C, Querfurth E, Kulessa H, Graf T. GATA-1 interacts with the myeloid PU.1 transcripti on factor and represses PU.1-dependent transcription. Blood. 2000;95:2543–2551. [PubMed] [Google Scholar]
- 37.Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197. doi: 10.1038/35004599. [DOI] [PubMed] [Google Scholar]
- 38.Graf T. Differentiation plasticity of hematopoietic cells. Blood. 2002;99:3089–3101. doi: 10.1182/blood.v99.9.3089. [DOI] [PubMed] [Google Scholar]
- 39.Tsiftsoglou AS, Pappas IS, Vizirianakis IS. Mechanisms involved in the induced differentiation of leukemia cells. Pharmacol Ther. 2003;100:257–290. doi: 10.1016/j.pharmthera.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 40.Terada M, Fried J, Nudel U, Rifkind RA, Marks PA. Transient inhibition of initiation of S-phase associated with dimethyl sulfoxide induction of murine erythroleukemia cells to erythroid differentiation. Proc Natl Acad Sci U S A. 1977;74:248–252. doi: 10.1073/pnas.74.1.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kiyokawa H, Richon VM, Venta-Perez G, Rifkind RA, Marks PA. Hexamethylenebisacetamide-induced erythroleukemia cell differentiation involves modulation of events required for cell cycle progression through G1. Proc Natl Acad Sci U S A. 1993;90:6746–6750. doi: 10.1073/pnas.90.14.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Atar O, Levi BZ. PU.1 silencin g leads to terminal differentiation of erythroleukemia cells. Biochem Biophys Res Commun. 2005;329:1288–1292. doi: 10.1016/j.bbrc.2005.02.109. [DOI] [PubMed] [Google Scholar]
- 43.McLeod DL, Shreeve MM, Axelrad AA. Improved plasma culture system for production of erythrocytic colonies in vitro: quantitative assay method for CFU-E. Blood. 1974;44:517–534. [PubMed] [Google Scholar]
- 44.Cook WD, McCaw BJ, Herring C, et al. PU.1 is a suppressor of myeloid leukemia, inactivated in mice by gene deletion and mutation of its DNA binding domain. Blood. 2004;104:3437–3444. doi: 10.1182/blood-2004-06-2234. [DOI] [PubMed] [Google Scholar]
- 45.Rosenbauer F, Wagner K, Kutok JL, et al. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat Genet. 2004;36:624–630. doi: 10.1038/ng1361. [DOI] [PubMed] [Google Scholar]
- 46.Mueller BU, Pabst T, Osato M, et al. Heterozygous PU.1 mutations are associated with acute myeloid leukemia. Blood. 2002;100:998–1007. doi: 10.1182/blood.v100.3.998. [DOI] [PubMed] [Google Scholar]
- 47.Vangala RK, Heiss-Neumann MS, Rangatia JS, et al. The myeloid master regulator transcription factor PU.1 is inactivated by AML1-ETO in t(8;21) myeloid leukemia. Blood. 2003;101:270–277. doi: 10.1182/blood-2002-04-1288. [DOI] [PubMed] [Google Scholar]
- 48.Mueller BU, Pabst T, Fos J, et al. ATRA resolves the differentiation block in t(15;17) acute myeloid leukemia by restoring PU.1 expression. Blood. 2006;107:3330–3338. doi: 10.1182/blood-2005-07-3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gopalakrishnan TV, Anderson WF. Mouse erythroleukemia cells. Methods Enzymol. 1979;58:506–511. doi: 10.1016/s0076-6879(79)58165-8. [DOI] [PubMed] [Google Scholar]
- 50.Collins N, Poot RA, Kukimoto I, Garcia-Jimenez C, Dellaire G, Varga-Weisz PD. An ACF1-ISWI chromatin-remodeling complex is required for DNA replication through heterochromatin. Nat Genet. 2002;32:627–632. doi: 10.1038/ng1046. [DOI] [PubMed] [Google Scholar]