Abstract
Moraxella catarrhalis is a Gram-negative diplococcus that is a strict human pathogen, which for a long period of time was regarded as a simple commensal. Research now shows that this organism is a pathogen its own right and is associated with both upper and lower respiratory tract infections. Further, there appears to be a dichotomy in the pathogenic potential of M. catarrhalis with upper respiratory tract infections mainly occurring in children, and lower respiratory tract infections mainly occurring in adults with predisposing pulmonary complications e.g., chronic obstructive pulmonary disease (COPD).
Key words: Moraxella catarrhalis, plasmid, transposon, phag, beta-lactamase
Until recently, very little was known about the number and types of mobile genetic elements present within this species, largely due to the fact that infections by competing bacterial respiratory pathogens e.g., Streptococcus pneumoniae, Haemophilus influenzae, group A streptococci etc tend to generate more serious disease complications. In two recent articles however, whole genome sequencing has been utilized to characterize the type and number of mobile genetic elements present in 12 clinically relevant isolates of M. catarrhalis.1,2
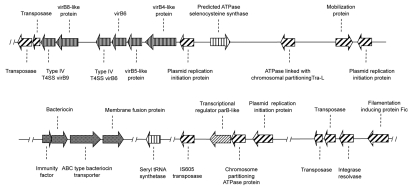
With respect to plasmid mobile elements, only 1 out of 12 isolates sequenced appeared to carry a plasmid, this plasmid comprising 12 ORFs and representing 0.6% of the total number of ORFs predicted for this particular bacterial isolate. Sequence analysis showed that this as yet uncharacterized plasmid possessed homologues of genes found in plasmid pLQ510, but that these homologues were also associated with: (1) an incomplete VirB-family type four secretion system (T4SS), (2) multiple transposases and (3) resolvases; genes not previously reported in plasmid pLQ510. In fact, it is possible that the presence of the extra genes indicate the discovery of a new integrated and conjugative element (ICE) rather than the presence of a plasmid per se (Fig. 1). However, at this moment, no experimental data is available to confirm or refute this hypothesis. In fact, the lack of plasmids found during our sequencing of the 12 isolates is to be expected, as there have only been a few previous reports of plasmids being isolated from M. catarrhalis, and only 2 plasmids (pLQ510 and pEMCJH03) currently having been characterized.3,4 The most intensively studied of the 2 well-characterized M. catarrhalis plasmids is pLQ510, a plasmid first characterized by Beaulieu et al. in 1988, who showed that this was a 12.2 kb plasmid that tended to be species specific.3 Structural analysis of this plasmid was published 10 years later, when Liu and Hansen showed that the plasmid comprises 12,082 bp with a 38% GC content.5 Interestingly, further research in 2009, showed that this plasmid carried putative bacteriocin production and secretion genes via a transcriptionally linked mcbABCI locus.6 It was further shown that the presence of the mcbABCI locus provided a distinct growth advantage upon co-culture of an mcbABCI locus-containing strain with a non-mcbABCI locus-containing strain. Importantly, the mcbABCI locus was also shown to be present in the chromosome of several other M. catarrhalis strains and was therefore not restricted to the pLQ510 mobile genetic element. At the present moment in time, whether other bacteriocin-carrying plasmids exist within the M. catarrhalis species remains to be determined, as does the exact mechanism by which the mcbABCI locus is transferred.
Figure 1.
Schematic diagram of the putative integrated and conjugative element associated with the bacteriocin expressing phenotype in M. catarrhalis isolate Bc7.
In 2005, the author published an article characterising a small 3.5 kb mobilizable plasmid called pEMCJH03, which contained mobilizable genes only (no virulence). Natural transformation of this plasmid was only successful in 25% of strains tested, and the plasmid showed a low transformation efficiency (615 CFU/µg, range 60–1,040 CFU/µg). As with plasmid pLQ510, attempts were subsequently made to convert pEMCJH03 into a suitable cloning an expression vector, though this procedure encountered unexpected obstacles. For example, the author successfully cloned an enhanced green fluorescent protein gene and a kanamycin resistance cassette into pEMCJH03, only to find that within one generation the majority of fluorescent cells decreased by approximately 95% (unpublished data), even in the presence of kanamycin selection pressure. Further, similar attempts using a lux gene have also failed (Personal Communication: R. Harris, Claflin University, USA). Interestingly, Wang and Hansen described plasmid pWW115 (a spontaneous plasmid deletion mutant of an artificial construct of pLQ510 and an unrelated plasmid pACYC184), which was used to clone a uspA2 gene (associated with complement resistance) into a serum sensitive isolate of M. catarrhalis.7 The result was a change in phenotype from serum sensitive to serum resistant. However, similar to the use of antibiotics, serum treatment generates selective pressure on bacteria, such that individual bacteria possessing deleterious mutations are not selected. Taken together, these results most likely indicate that plasmid expression of cloned genes in M. catarrhalis may be hampered by an as yet uncharacterized mechanism, especially for genes that do not provide the host with a phenotypic advantage Further, this observation could provide an explanation as to why there appears to be very few (virulence) plasmids associated with this particular bacterial species, there being natural limits on the acceptance/correct functioning of non-host plasmids and plasmid-borne genes within M. catarrhalis isolates and between co-colonizing species (H. influenzae, S. pneumoniae, S. pyogenes etc.). These mechanisms could operate via restrictions/modification systems,8 or possibly via a high gene mutation frequency. Importantly, this issue could affect the ability to clone and express, for example, putative vaccine candidate genes in M. catarrhalis, suggesting that cloned genes should be regularly re-sequenced in order to establish that no unintended mutations have become established in the putative accine candidate gene being expressed. Alternatively, a different bacterial species may have to be chosen for future vaccine gene expression in order to maintain genetic stability.
Phage ORFs were found in all 12 isolates sequenced ranging from 3–49 ORFs per genome, and representing 0.2–2.5% of the total ORFs per genome. However, precious little phage research has been performed on M. catarrhalis, and indeed there is a lack of understanding of the impact of phages on all species of Moraxella.9,10
Similar to the paucity of plasmids and phages observed during M. catarrhalis whole genome sequencing, the number of transposon open reading frames (ORFs) found in this bacterial species also appears to be very low, with a range of between 3–8 transposon ORFs per genome. In fact, fewer than 10 transposon ORFs were detected in the 12 clinical isolates (isolated from four different countries) that where sequenced, with transposons accounting for only 0.15%–0.44% of all ORFS per genome. This low percentage indicates that transposition may play only a minor role in M. catarrhalis evolution and pathogenicity. In fact, the best-known “mobile genetic element” associated with M. catarrhalis is the bro beta-lactamase gene. This gene was first described in a few clinical M. catarrhalis species in the 1970s, but is now present in approximately 95% of clinical isolates. Further, this spread is not related to clonal expansion as M. catarrhalis isolates show a distinct lack of clonality. Bootsma et al. showed that the acquisition of this gene probably occurred via a homologous recombination event, though the actual origin of the gene itself is still a mystery (BRO beta-lactamase being unrelated to any known beta-lactamase so far described).11–13 However, natural competence has been found to be a feature of M. catarrhalis biology, showing that the bacterium is able to naturally acquire foreign DNA.14 The most likely ‘donor’ candidates for the bro gene are the related Psychrobacter, Acinetobacter and Neisseria genera, and these genera could also possibly act as donors for mobile genetic elements, though the different environmental niches that these genera currently occupy in relation to M. catarrhalis may mean that mobile genetic element transfer between these bacterial genera are rare events.
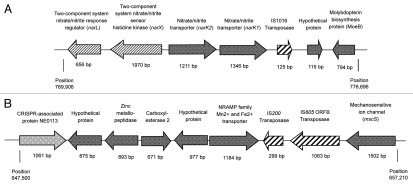
Of the transposon ORFs identified, homologues to IS1016, IS200 and IS4 transposons (sub)families were identified in all 12 genomes sequenced, with five genomes also containing an IS605 family transposon. As mentioned in our sequencing publication, for all 12 isolates, the IS1016 element was found adjacent to a nitrate uptake locus (Fig. 2A), and the IS200 element was found in a locus containing the CRISPR-associated protein (NE0113) and a mechanosensitive ion channel (mscS; Fig. 2B). The exact effect of these insertions (if any) is as yet unknown, though no flanking genes appear to be disrupted. One possible explanation being that deleterious transposition events have been selected-out of the M. catarrhalis population by selection pressure. For example, narK1 and narK2 are involved in the capability of bacteria to grow under anaerobic denitrifying, microaerophilic and occasionally aerobic growth conditions.15 Any disruption of these genes could potentially seriously affect the growth (and hence colonization and virulence) potential of the affected bacterium.
Figure 2.
Schematic representation of the position of transposon IS1016 (A), and IS200 and IS605 (B), in the genome of M. catarrhalis isolate RH4. Isolate RH4 was the first M. catarrhalis isolate to be fully sequenced and annotated as a complete, contiguous, chromosome.
Finally, it should be stressed that most of the scientific research performed to date, and all of the 12 M. catarrhalis isolates sequenced so far, tend to belong to one of the two major genotypic lineages that have been described in M. catarrhalis (rather unimaginatively described as ‘Lineage 1’), which is much more genotypically conserved (as determined using pulsed field gel electrophoresis) than ‘Lineage 2’ isolates.16 Currently, more sequencing work is being performed in order to better characterize Lineage 2 genotype isolates. However, Lineage 1 genotype isolates tend to be more frequently associated with clinical infections, meaning that an increased prevalence of pathogenic gene carriage or increased virulence gene expression in Lineage 2 isolates facilitated by mobile genetic elements is unlikely.
From current research, mobile genetic elements per se appear to play only a minor role in influencing M. catarrhalis evolution, with occasional homologous recombination events having a much more profound, and clinically relevant, effect. However, research is continuing with the current emphasis focussing on whole genome sequencing and comparative genomics of Lineage 2 versus Lineage 1 isolates. Nevertheless, comparative genomics indicates that M. catarrhalis isolates tend to be refractory to the uptake of mobile genetic elements, though the exact mechanism facilitating this phenomenon has yet to be understood.
Acknowledgments
This work was made possible by the contribution of all authors named in references 1 and 2.
References
- 1.Davie JJ, Earl J, de Vries SP, Ahmed A, Hu FZ, Bootsma HJ, et al. Comparative analysis and supragenome modeling of twelve Moraxella catarrhalis clinical isolates. BMC Genomics. 2011;12:70. doi: 10.1186/1471-2164-12-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Vries SP, van Hijum SA, Schueler W, Riesbeck K, Hays JP, Hermans PW, et al. Genome analysis of Moraxella catarrhalis strain RH4, a human respiratory tract pathogen. J Bacteriol. 2010;192:3574–3583. doi: 10.1128/JB.00121-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beaulieu D, Ouellette M, Bergeron MG, Roy PH. Characterization of a plasmid isolated from Branhamella catarrhalis and detection of plasmid sequences within the genome of a B. catarrhalis strain. Plasmid. 1988;20:158–162. doi: 10.1016/0147-619x(88)90020-0. [DOI] [PubMed] [Google Scholar]
- 4.Hays JP, Eadie K, Verduin CM, Verbrugh H, van Belkum A. A novel plasmid (pEMCJH03) isolated from Moraxella catarrhalis possibly useful as a cloning and expression vector within this species. Plasmid. 2005;53:263–268. doi: 10.1016/j.plasmid.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 5.Liu L, Hansen EJ. Structural analysis of plasmid pLQ510 from Moraxella catarrhalis E22. Plasmid. 1999;42:150–153. doi: 10.1006/plas.1999.1411. [DOI] [PubMed] [Google Scholar]
- 6.Attia AS, Sedillo JL, Hoopman TC, Liu W, Liu L, Brautigam CA, et al. Identification of a bacteriocin and its cognate immunity factor expressed by Moraxella catarrhalis. BMC Microbiol. 2009;9:207. doi: 10.1186/1471-2180-9-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang W, Attia AS, Liu L, Rosche T, Wagner NJ, Hansen EJ. Development of a shuttle vector for Moraxella catarrhalis. Plasmid. 2006;55:50–57. doi: 10.1016/j.plasmid.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 8.Seib KL, Peak IR, Jennings MP. Phase variable restriction-modification systems in Moraxella catarrhalis. FEMS Immunol Med Microbiol. 2002;32:159–165. doi: 10.1111/j.1574-695X.2002.tb00548.x. [DOI] [PubMed] [Google Scholar]
- 9.Aalto J, Pelkonen S, Kalimo H, Finne J. Mutant bacteriophage with non-catalytic endosialidase binds to both bacterial and eukaryotic polysialic acid and can be used as probe for its detection. Glycoconj J. 2001;18:751–758. doi: 10.1023/a:1021147316647. [DOI] [PubMed] [Google Scholar]
- 10.Kanawyer WL, Eisenstark A, Splitter EJ. A possible bacteriophage active against Moraxella (Hemophilus) bovis; a preliminary report. J Am Vet Med Assoc. 1953;123:409. [PubMed] [Google Scholar]
- 11.Bootsma HJ, van Dijk H, Vauterin P, Verhoef J, Mooi FR. Genesis of BRO beta-lactamase-producing Moraxella catarrhalis: evidence for transformation-mediated horizontal transfer. Mol Microbiol. 2000;36:93–104. doi: 10.1046/j.1365-2958.2000.01828.x. [DOI] [PubMed] [Google Scholar]
- 12.Bootsma HJ, Aerts PC, Posthuma G, Harmsen T, Verhoef J, van Dijk H, et al. Moraxella (Branhamella) catarrhalis BRO beta-lactamase: a lipoprotein of gram-positive origin? J Bacteriol. 1999;181:5090–5093. doi: 10.1128/jb.181.16.5090-5093.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bootsma HJ, van Dijk H, Verhoef J, Fleer A, Mooi FR. Molecular characterization of the BRO beta-lactamase of Moraxella (Branhamella) catarrhalis. Antimicrob Agents Chemother. 1996;40:966–972. doi: 10.1128/aac.40.4.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meier PS, Troller R, Heiniger N, Hays JP, van Belkum A, Aebi C. Unveiling electrotransformation of Moraxella catarrhalis as a process of natural transformation. FEMS Microbiol Lett. 2006;262:72–76. doi: 10.1111/j.1574-6968.2006.00365.x. [DOI] [PubMed] [Google Scholar]
- 15.Zumft WG. Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verhaegh SJ, Streefland A, Dewnarain JK, Farrell DJ, van Belkum A, Hays JP. Age-related genotypic and phenotypic differences in Moraxella catarrhalis isolates from children and adults presenting with respiratory disease in 2001-2002. Microbiology. 2008;154:1178–1184. doi: 10.1099/mic.0.2007/015057-0. [DOI] [PubMed] [Google Scholar]