Abstract
With the Sleeping Beauty (SB) DNA transposon, a reconstructed Tc1/mariner element, as the driving force, DNA transposable elements have emerged as new gene delivery vectors with therapeutic potential. The bipartite transposon vector system consists of a transposon vector carrying the transgene and a source of the transposase that catalyzes transposon mobilization. The components of the system are typically residing on separate plasmids that are transfected into cells or tissues of interest. We have recently shown that SB vector technology can be successfully combined with lentiviral delivery. Hence, SB transposons are efficiently mobilized from HIV-based integrase-defective lentiviral vectors by the hyperactive SB100X transposase, leading to the genomic insertion of lentivirally delivered DNA in a reaction controlled by a nonviral integration machinery. This new technology combines the better of two vector worlds and leads to integration profiles that are significantly altered and potentially safer relative to conventional lentiviral vectors. In this short commentary, we discuss our recent findings and the road ahead for hybrid lentivirus-transposon vectors.
Key words: Sleeping Beauty, DNA transposition, lentiviral vector, IDLV, integration profiling
Mobilization of genetic information is the key to any type of genetic engineering including efforts to cure disease by therapeutic gene transfer. Since the concept of gene therapy was introduced in the early 1980s, the unique capacity of viruses to package and carry genetic material has inspired a community of genetic entrepreneurs in the search of virus-based gene vehicles with therapeutic potency. Although the search for perfect, or at least optimized, vector technologies is far from over, the field is starting to see early promises of viral gene therapy fulfilled.1 Successes include hematopoietic stem cell therapies, eradication of blindness and emerging evidence of persistent treatment of hemophilia B.
Concerns related to safety, host immune response and manufacturing costs have fueled interests in nonviral gene transfer technologies as cheaper and biologically less complex alternatives to viral methodologies. With the reconstructed Sleeping Beauty (SB) DNA transposable element in the eye of the storm, nonviral integration systems—mainly based on cut-and-paste DNA transposons and site-specific integrases like PhiC31—have swept into the field of gene therapy. In their simplest form, gene vehicles derived from ancient mobile DNA elements are carried on naked plasmid DNA and offer basically a tool for integrating foreign genes into chromosomal DNA. This capacity has extended the longevity of treatment by nonviral vectors and has solved problems related to the lack of persistency which has in the past been the Achilles' heel of nonviral gene delivery strategies. With the continuous efforts to optimize methods of transfection of plasmid DNA, DNA transposon-based vectors are showing new promises in both hematopoietic stem cells2–4 and primary T-cells,5–7 and clinical trials for treatment of B-cell lymphoma with transposon-engineered T-cells are on the way.8,9
The simplicity of DNA transposon vector systems, consisting of a transposon substrate and a source of transposase protein, is part of the beauty of this technology. However, despite the improved new methods of transfection, delivery of the two-component system remains an important challenge for many applications. Recent work in our laboratory has focused on the development of new hybrid delivery methods that combine the potency of viral gene delivery with the strengths of nonviral integration systems.10–12 Previous work has shown the successful combination of SB transposition with adenoviral gene delivery in mice,13 and this approach was recently found to facilitate persistency of expression of canine factor IX leading to phenotypic correction in a canine model for hemophilia B.14 Based on a human immunodeficiency virus type 1 (HIV-1)-derived lentiviral vector, we have aimed at using this platform for delivery of both SB transposon and transposase. The bipartite rationale for this approach is to engineer lentiviral vectors that show an integration profile that is different from that of conventional lentiviral vectors and to create—at the same time—new methods for delivery of SB vectors. Our most recent contribution, published in the April issue of Molecular Therapy,10 shows successful SB-directed gene insertion from integrase-defective lentiviral vectors (IDLVs), and an unprecedented high-resolution profile of SB integration sites in the human genome displays integration properties that are significantly altered from the properties of conventional lentiviral vectors. Although yet in an early phase and still quite distant from clinical use, the combination of IDLVs and SB marks the first expedition towards uniting retroelements and DNA transposable elements in potent vector technologies with unique biological properties. This plots a new interesting course for creation of hybrid viral vectors in which viral enzymatic proteins are substituted with proteins of nonviral origin. On this course, the creation of engineered virus particles carrying nonviral enzymatic proteins is one of several navigation marks that, if successfully rounded, are bound to make a strong impact on therapeutic gene transfer. In this commentary, we discuss our recent findings and the road ahead for hybrid lentiviral vectors.
Gene Mobilization by Genetic Relics of the Past
Mobile DNA elements, including the predecessors of SB, are genetic relics of the past that have moved around through cut-and-paste transposition. Such primitive DNA elements, normally considered genetic parasites, have colonized all living species and are evolutionarily shaped through a drive for survival and co-existence with their natural hosts. Successful co-existence involves both several mechanisms of self-suppression as well as molecular responses from the host. Although the exact mechanisms may differ between elements and between hosts, many elements have gone through a life span during which invasion of the host is succeeded by spreading within the genome under the continuous control of self-regulatory and cellular mechanisms. Slow accumulation of mutations eventually leads to element defects and cessation of transpositional activity. This is the story also in fish in which inactive members of a family of Tc1/mariner-like transposable elements were identified in several species of teleost fish.15 Phylogenetic analysis and a meticulous work of reconstruction based on numerous rounds of mutagenesis reestablished a functional transposase protein that was found to efficiently catalyze transposition in human cells.16 This basic biological approach obviously led Ivics, Izsvak and co-workers to the ground-breaking discovery of the Sleeping Beauty DNA transposon system which is today not only studied for gene transfer purposes but also for use in animal transgenesis17 and cancer gene discovery.18
From the initial studies of SB, mobilization of a transposon—with outer boundaries defined by two terminal inverted repeats—was easily accomplished in human cells from transfected plasmid DNA in the presence of the original SB10 transposase expressed from a separate plasmid. By using this relatively simple setup, Yant et al. demonstrated in vivo competence of the system in mouse liver by hydrodynamic co-injection of plasmids.19 It was shown later that both components of the system, the transposase expression cassette and the transgene-tagged transposon vector, could be delivered on a single plasmid, leading to higher efficiency of the system.20 Once established by proof-of-principle experiments, the course was charted for improved efficiency and mutagenesis of both transposase gene and inverted repeats quickly established higher efficiency.21,22 An enormous leap of improvement was taken with the development of the SB100X transposase, engineered through a high-throughput, DNA shuffling PCR approach. SB100X was found to be 100-fold more active (hence the name) than the original SB10 transposase under certain experimental conditions.2 This hyperactive transposase is particularly useful—and in some cases crucial, as we will discuss below—when sources of transposon substrate or transposase-encoding nucleic acids are limited, as for example in hard-to-transfect cells like hematopoietic stem cells2 and primary T-cells.7
Mobilization of SB from IDLVs:The Better of Two Worlds
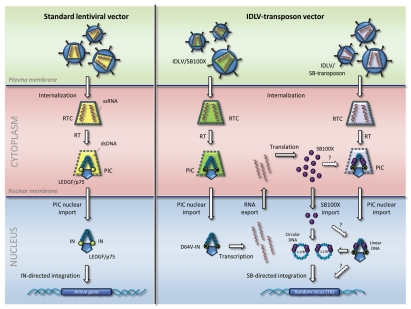
HIV-1 and derived gene vehicles are integrating vectors by the hand of nature. The virus particle carries its own enzymatic proteins which are assembled in the particle as polyproteins and during maturation cleaved to become smaller proteins with specific catalytic functions during viral infection or transduction. One of these proteins is the viral integrase, which recognizes attachment sites in the terminal regions of the reverse-transcribed double-stranded viral DNA and catalyzes its genomic insertion. During the traverse from the cellular to the nuclear membrane of the infected cell, the increasingly porous structure of the preintegration complex (PIC) allows cellular proteins to access the complex and interact with viral components. One of these proteins is the cellular transcriptional coactivator lens epithelium-derived growth factor p75 (LEDGF/p75), which is now widely accepted as playing a crucial role in the viral integration process. Numerous reports have stated that LEDGF/p75 interacts specifically with viral integrase and thereby draws the PIC and its genetic cargo towards active transcriptional units in the genome23 (Fig. 1, left). This interaction is believed to be the main reason that HIV-1 and derived vectors are preferentially inserted into actively transcribed genes.24,25 Typically, between 70 and 80% of the insertions are located within exon or intron sequences, which account for less than 40% of the human genome.
Figure 1.
Schematic representation and comparison of gene delivery mediated by conventional lentiviral vectors (left) and lentivirus-transposon vectors (right). The figure shows the internalization of the nucleocapsid core, shown in yellow, green and purple for the different vectors. Reverse transcription (RT) of vector RNA occurs inside the reverse transcription complex (RTC); after DNA synthesis the increasingly porous complex is referred to as the preintegration complex (PIC). Several cellular proteins become part of the PIC; only LEDGF/p75 is shown. LEDGF/p75 interacts with viral integrase (IN) proteins, shown as yellow circles attached to the terminal regions of the viral DNA. IN proteins harboring the inactivating D64V mutation are indicated by yellow circles marked with a cross. In case of the IDLV-transposon system, cells are transduced with two vectors, one carrying the SB100X expression cassette (green) and one carrying the SB transposon vector (purple). RNA encoding SB100X is produced from episomal viral DNA and exported from the nucleus prior to translation in the cytoplasm. Subunits of SB100X (indicated by purple circles) may be incorporated directly into the PIC (indicated by a stippled arrow and a question mark) or be directly imported into the nucleus where it may interact with both linear DNA and circular DNA intermediates (1- and 2-LTR circles) to facilitate transposition into randomly chosen TA-dinuclotides in the genome. It is not known whether linear viral DNA serves as a substrate for SB transposition. Single-stranded RNA (ssRNA) is shown in red, double-stranded DNA (dsDNA) in blue.
Evidence of gene-directed insertion of HIV-1-derived vectors in a broad range of cell types has prompted attempts of manipulating the integration properties by disengaging vector integration from the actions of the viral integrase. We initially hypothesized that the SB transposase, expressed in vector-treated cells, would be able to gain access to the PIC, or at least to free, episomal, viral DNA intermediates, and facilitate transposition from reverse-transcribed viral substrates. The rationale was that the transposase would be able to fill the role of an inactivated viral integrase and thereby loosen the vector from the biological constraints dictated by the interactions between the integrase and LEDGF/p75 (Fig. 1, right).
We therefore packaged a lentiviral vector harboring an SB transposon in IDLV virus particles carrying the inactive D64V-mutated integrase. Blockage of integration is known to result in the build-up of episomal DNA intermediates in transduced cells. This leads to an increase of vector DNA circles that are generated by recombination between the long terminal repeats (creating 1-LTR circles) or by non-homologous end-joining mediated by cellular repair factors (creating 2-LTR circles).12 Such circularization of potential transposase substrates is convenient since circular DNA forms, relative to linear forms, are superior substrates for SB transposition.13 By providing the SB100X transposase from IDLVs in HEK-293 cells that were also transduced with the transposon-containing IDLV, vector insertion was increased more than 20-fold over background, as measured by colony formation assays.10 Sequence analysis of integration sites verified that insertion into genomic TA-dinucleotides was mediated by the transposase. These findings demonstrated that transposase subunits expressed in IDLV-transduced cells could in fact gain access to viral DNA and mobilize the transposon element carried by the vector. We have previously shown that SB transposition occurs from both 1- and 2-LTR circles,12 but it remains unclear whether also linear viral DNA may directly serve as a transposon donor.
Previous experiments in the laboratory have shown that the original SB10 transposase, expressed in a similar fashion from IDLVs, is not able to facilitate measurable transposition from lentiviral substrates (Staunstrup NH, Moldt B, Mikkelsen JG; unpublished observations). We have also seen that one of the early hyperactive SB transposase variants, HSB3, does not effectively mobilize transposons from IDLV substrates.12 In support, work by Vink and co-workers showed that SB11, another early hyperactive SB transposase, at most resulted in a twofold increase of gene insertion over background when the transposase gene and transposon were delivered to HeLa cells on separate IDLVs.26 These findings together suggest that the high potency of the SB100X transposase is crucial for the functionality of hybrid IDLV-transposon vectors. Lending further support to this notion, dose-response experiments demonstrated that the availability of both IDLV-delivered transposons and transposase are limiting factors for gene insertion.10 Hence, the levels of transposition were increased by increasing the availability of transposon substrate or the amount of IDLV-delivered SB100X. Although SB transposases, including SB100X, are regulated by overproduction inhibition, we did not observe this type of regulation with IDLV-delivered SB100X. Rather, attempts to further increase the potency of the system in vitro were seemingly limited by viral toxicity at increased viral loads. It is not yet clear whether such potential toxicity was caused by insertional mutagenesis or other cellular responses leading to disruption of cell homeostasis.
The formation of circular transposition substrates could potentially be another limiting factor for the activity of IDLV-transposon vectors. The far majority of the viral DNA intermediates in a population of vector-transduced cells are linear and possibly suboptimal substrates for transposition. We therefore created a vector with Flp recombinase recognition target (FRT) sites flanking the SB transposon within the lentiviral vector. By co-expression of the Flp recombinase, we could further enhance circle formation and significantly increase the transposition rate. These findings potentially illustrate the importance of generating circles but may also indicate that substrate exposure, possibly here triggered by Flp-induced release of DNA circles from the PIC, is a key feature for vector function. It is still unclear whether SB transposase subunits are penetrating the PIC to get access to the transposon, or whether transposases preferably interact with free DNA intermediates released from disassembled PICs.
Transposition in HEK-293 could be further improved by replacing the IRs of the original SB vector with the genetically optimized T2 IRs, which have previously been shown to improve transposition from plasmid DNA.21 In colony formation assays, we now measured a 44-fold increase in genomic insertion relative to background. By mobilizing a transposon containing eGFP, we measured levels of eGFP that were almost 80-fold higher than observed in the presence of the inactive mSB transposase. Notably, this transfer effiency was only sixfold lower than the transduction effiency measured with conventional integration-proficient lentiviral vectors.
Efficient mobilization of SB vectors was shown in a variety of cell lines including HeLa, HT-1080, Chang cells and HepG2 as well as in human primary cells. In primary keratinocytes, the transfer efficiency was increased as much as 160-fold in the presence of IDLV-encoded SB100X relative to background, demonstrating a potential of this technology also in primary cell types. Using the vector carrying the eGFP expression cassette, we established robust activity (33-fold over background) in the myelogenous leukemia cell line K562. By taking advantage of the Illumina Genome Analyzer platform, we performed comparative genomic integration profiling in these cells and were able to capture 1,283 unique IDLV-transposon sites and 1,165 conventional lentiviral vector insertions for direct comparison. This dataset showed that 47% of the transposon insertions were located within RefSeq genes, whereas 74% of the sites obtained with the parent vector were identified within RefSeq genes. These findings showed a clear difference in integration profiles and demonstrated a significantly reduced bias toward genes when integration was catalyzed by the transposase. By comparing the frequencies of exonic integrations, insertion by SB transposition was found to reduce the chance of inserting the transgene into a host gene exon by 50% (from 47 out of 1,165 standard vector integrations to 25 out of 1,283 IDLV/SB integrations). Although defining and comparing vector safety features is far from trivial, such findings could indicate that an IDLV-transposon vector technology has a safer integration profile than the conventional lentiviral vectors.
IDLV-Transposon Vectors:The Road Ahead
The idea of combining the properties of lentiviral vectors and the SB integration machinery in a bipartite IDLV-transposon vector system was initially founded on the interest of changing the integration profile of conventional lentiviral vectors. If the SB transposase could get access to lentiviral DNA intermediates, would it then be able to use these intermediates as substrates for transposition and perhaps relieve lentiviral vectors for some of the biological restrictions that challenge the safety of this vector technology? We now know that the answer to both these questions is ‘yes’. When expressed from IDLVs, mRNA encoding the SB100X transposase is transported to the cytoplasm where the transposase is produced. Subunits of the transposase are able to traverse the nuclear membrane and locate the transposon vector delivered by a co-transducing IDLV and facilitate its mobilization from episomal IDLV DNA to TA-dinucleotides in the genome (Fig. 1 and right). Clearly, the integration process is now under guidance of the transposase and potential interactions of the mutated viral integrase with LEDGF/p75 seemingly do not influence the choice of insertion site, as is evident for conventional lentiviral vectors (Fig. 1 and left). Efficiency of the hybrid system is on a rocky road, since activity requires not only co-transduction with two viral vectors but also expression of SB100X from episomal DNA, nuclear import of the transposase, accessibility to IDLV-delivered transposons either within or outside the PIC, possibly circularization of the substrate, and obviously the generation of an active transposition complex. Although the result is a vector technology that is so far not as efficient as standard lentiviral vectors (at best 6-fold less efficient in some cell types), the altered integration properties of this vector system may make it safer than the conventional system and less likely to induce genotoxicity. However, as recently reviewed in reference 27, only a small subset of viral vector integrations near proto-oncogenes has been found to cause transformation and insertional mutagenesis by conventional integrating viral vectors seems to occur only under selective circumstances and only for some cell types and some transgenes. Also, there is not sufficient information so far to facilitate a comparative evaluation of the risks of activating oncogenes associated with viral and nonviral vector integration. Therefore, evidence in support of a safer SB-based integration profile is awaiting further toxicity studies, for example in an appropriate tumor-prone animal model. Furthermore, one may speculate that events of recombination between vectors carrying the SB transposon and the SB transposase gene, respectively, could lead to creation of an autonomous SB transposon. Although this risk cannot formally be ruled out, the conventional pathway of retroviral recombination—by template switching during reverse transcription of vector RNA—is not likely to create an autonomous element in consideration of the fact that the two vectors are produced separately and that the vector RNAs would never be co-packaged in the same particle.
The road ahead for hybrid lentiviral vectors may show evidence of ex vivo functionality in some of the most clinically relevant cell types like human hematopoietic progenitor cells and primary human T lymphocytes. Although we have observed robust activity in human primary cells, including fibroblasts and keratinocytes, it is still too early to tell whether the IDLV transfer efficiency combined with the actions of the SB100X transposase are adequate to facilitate measurable transfer efficiency in hematopoietic stem cells and T lymphocytes. Future studies should also address the potential in vivo applicability of lentivirus-transposon vectors. We have previously demonstrated very efficient lentiviral vector delivery to skin xenotransplanted onto mice.28 Combined with the fact that IDLVs delivered to skin result in only short-term gene expression (Petersen LB, Bak RO, Mikkelsen JG, unpublished observations), it should be possible to measure an effect of transposase-directed gene insertion in skin. However, one of the remaining challenges with IDLV-based gene transfer is that particle production is affected by the mutation incorporated in the integrase protein. Hence, we reproducibly obtain IDLV yields that are reduced, typically in the range of 2- to 5-fold, relative to the yields of conventional integration-competent vectors, and this may reduce gene transfer efficiency. Although this is not directly problematic for ex vivo applications, many in vivo applications require optimal transfer efficiencies and may suffer from reduced IDLV transfer.
Vector systems based on DNA transposable elements rely on the expression of the transposase within treated cells. In many current applications, including those based on plasmid DNA transfer, this implies that the genetic information encoding the transposase may end up being inserted, by mistake, in genomic DNA. Successful delivery and activity of purified SB transposase has not yet been reported, but the problem is potentially solved for some applications by delivery of in vitro-transcribed RNA encoding the transposase.29 One of the remaining challenges also in the case of lentivirus-transposon vectors is to overcome the need for production of the SB transposase from expression cassettes co-delivered to the target cells. To solve this problem, we can perhaps find inspiration in the ability of the virus itself to carry its own integration machinery. Would it be possible to incorporate transposase protein inside the particle and thereby let the virus transport its own nonviral integration machinery? We do not know whether this is possible for an SB-based system but recent findings have shown that the Cre recombinase, fused to the viral Vpr protein, can be assembled into IDLVs and is capable of catalyzing the genomic insertion of viral DNA delivered by the same particles.30 Perhaps genomic insertion in this scenario is supported by the close association of protein and substrate? In any case, it is not unlikely that the mobilization of an IDLV-delivered SB transposon would benefit from the transposase being part of the PIC, although possible negative effects of such inclusion of SB100X in the PIC should not be neglected.
With our recent progress combined with that of others who have examined viral carriers of DNA transposable elements, the first steps have been taken towards a vector technology that fully combines lentiviral gene transfer capacity with the integration properties of the SB system. Efficient mobilization of transposons from episomal viral DNA intermediates induces relatively high transfer efficiencies and completely changes the pattern of integration, resulting in a strongly reduced bias for integration into transcriptional units. This approach could become an alternative to conventional lentiviral vectors and be useful for transposon delivery in cells or tissues that are hard to transfect. We believe that IDLV represents a new seaworthy vessel for SB, an ancient Tc1/mariner element. Lending words from the English poet, Samuel Taylor Coleridge (1,772–1,834), hopes are that our findings may lead to yet another ‘rime of the ancient Tc1/mariner’.
Acknowledgments
The authors would like to thank Brian Moldt and Nicklas Heine Staunstrup for their important contribution to the development of hybrid IDLV-transposon vectors. We also thank Csaba Miskey, Zsuzsanna Izsvák and Zoltán Ivics for their crucial contribution. Work in the laboratory of J.G.M. is made possible through support from The Danish National Advanced Technology Foundation, the Danish Medical Research Council, the Lundbeck Foundation, the Novo Nordisk Foundation, The Danish Heart Foundation, Kgl. Hofbuntmager Aage Bangs Foundation, Helga and Peter Kornings Foundation, and the Augustinus Foundation.
References
- 1.Kay MA. State-of-the-art gene-based therapies: the road ahead. Nat Rev Genet. 2011;12:316–328. doi: 10.1038/nrg2971. [DOI] [PubMed] [Google Scholar]
- 2.Mates L, Chuah MK, Belay E, Jerchow B, Manoj N, Acosta-Sanchez A, et al. Molecular evolution of a novel hyperactive Sleeping Beauty transposase enables robust stable gene transfer in vertebrates. Nat Genet. 2009;41:753–761. doi: 10.1038/ng.343. [DOI] [PubMed] [Google Scholar]
- 3.Sumiyoshi T, Holt NG, Hollis RP, Ge S, Cannon PM, Crooks GM, et al. Stable transgene expression in primitive human CD34+ hematopoietic stem/progenitor cells, using the Sleeping Beauty transposon system. Hum Gene Ther. 2009;20:1607–1626. doi: 10.1089/hum.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xue X, Huang X, Nodland SE, Mates L, Ma L, Izsvak Z, et al. Stable gene transfer and expression in cord blood-derived CD34+ hematopoietic stem and progenitor cells by a hyperactive Sleeping Beauty transposon system. Blood. 2009;114:1319–1330. doi: 10.1182/blood-2009-03-210005. [DOI] [PubMed] [Google Scholar]
- 5.Huang X, Guo H, Tammana S, Jung YC, Mellgren E, Bassi P, et al. Gene transfer efficiency and genome-wide integration profiling of Sleeping Beauty, Tol2 and piggyBac transposons in human primary T cells. Mol Ther. 2010;18:1803–1813. doi: 10.1038/mt.2010.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakazawa Y, Huye LE, Dotti G, Foster AE, Vera JF, Manuri PR, et al. Optimization of the PiggyBac transposon system for the sustained genetic modification of human T lymphocytes. J Immunother. 2009;32:826–836. doi: 10.1097/CJI.0b013e3181ad762b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin Z, Maiti S, Huls H, Singh H, Olivares S, Mates L, et al. The hyperactive Sleeping Beauty transposase SB100X improves the genetic modification of T cells to express a chimeric antigen receptor. Gene Ther. 2011 doi: 10.1038/gt.2011.40. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Izsvak Z, Hackett PB, Cooper LJ, Ivics Z. Translating Sleeping Beauty transposition into cellular therapies: victories and challenges. Bioessays. 2010;32:756–767. doi: 10.1002/bies.201000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh H, Manuri PR, Olivares S, Dara N, Dawson MJ, Huls H, et al. Redirecting specificity of T-cell populations for CD19 using the Sleeping Beauty system. Cancer Res. 2008;68:2961–2971. doi: 10.1158/0008-5472.CAN-07-5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moldt B, Miskey C, Staunstrup NH, Gogol-Döring A, Bak RO, Sharma N, et al. Comparative genomic integration profiling of Sleeping Beauty transposons mobilized with high efficiency from integrase-defective lentiviral vectors in primary human cells. Mol Ther. 2011;19:1499–1510. doi: 10.1038/mt.2011.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moldt B, Staunstrup NH, Jakobsen M, Yanez-Munoz RJ, Mikkelsen JG. Genomic insertion of lentiviral DNA circles directed by the yeast Flp recombinase. BMC biotechnology. 2008;8:60. doi: 10.1186/1472-6750-8-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Staunstrup NH, Moldt B, Mates L, Villesen P, Jakobsen M, Ivics Z, et al. Hybrid lentivirus-transposon vectors with a random integration profile in human cells. Mol Ther. 2009;17:1205–1214. doi: 10.1038/mt.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yant SR, Ehrhardt A, Mikkelsen JG, Meuse L, Pham T, Kay MA. Transposition from a gutless adeno-transposon vector stabilizes transgene expression in vivo. Nat Biotechnol. 2002;20:999–1005. doi: 10.1038/nbt738. [DOI] [PubMed] [Google Scholar]
- 14.Hausl MA, Zhang W, Muther N, Rauschhuber C, Franck HG, Merricks EP, et al. Hyperactive sleeping beauty transposase enables persistent phenotypic correction in mice and a canine model for hemophilia B. Mol Ther. 2010;18:1896–1906. doi: 10.1038/mt.2010.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ivics Z, Izsvak Z, Minter A, Hackett PB. Identification of functional domains and evolution of Tc1-like transposable elements. Proc Natl Acad Sci USA. 1996;93:5008–5013. doi: 10.1073/pnas.93.10.5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- 17.Jakobsen JE, Li J, Kragh PM, Moldt B, Lin L, Liu Y, et al. Pig transgenesis by Sleeping Beauty DNA transposition. Transgenic Res. 2010;20:533–545. doi: 10.1007/s11248-010-9438-x. [DOI] [PubMed] [Google Scholar]
- 18.Carlson CM, Largaespada DA. Insertional mutagenesis in mice: new perspectives and tools. Nat Rev Genet. 2005;6:568–580. doi: 10.1038/nrg1638. [DOI] [PubMed] [Google Scholar]
- 19.Yant SR, Meuse L, Chiu W, Ivics Z, Izsvak Z, Kay MA. Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nat Genet. 2000;25:35–41. doi: 10.1038/75568. [DOI] [PubMed] [Google Scholar]
- 20.Mikkelsen JG, Yant SR, Meuse L, Huang Z, Xu H, Kay MA. Helper-Independent Sleeping Beauty transposon-transposase vectors for efficient nonviral gene delivery and persistent gene expression in vivo. Mol Ther. 2003;8:654–665. doi: 10.1016/s1525-0016(03)00216-8. [DOI] [PubMed] [Google Scholar]
- 21.Cui Z, Geurts AM, Liu G, Kaufman CD, Hackett PB. Structure-function analysis of the inverted terminal repeats of the sleeping beauty transposon. J Mol Biol. 2002;318:1221–1235. doi: 10.1016/s0022-2836(02)00237-1. [DOI] [PubMed] [Google Scholar]
- 22.Yant SR, Park J, Huang Y, Mikkelsen JG, Kay MA. Mutational analysis of the N-terminal DNA-binding domain of sleeping beauty transposase: critical residues for DNA binding and hyperactivity in mammalian cells. Mol Cell Biol. 2004;24:9239–9247. doi: 10.1128/MCB.24.20.9239-9247.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Llano M, Saenz DT, Meehan A, Wongthida P, Peretz M, Walker WH, et al. An essential role for LEDGF/p75 in HIV integration. Science. 2006;314:461–464. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- 24.Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, et al. Retroviral DNA integration: ASLV HIV and MLV show distinct target site preferences. PLoS Biol. 2004;2:234. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 26.Vink CA, Gaspar HB, Gabriel R, Schmidt M, McIvor RS, Thrasher AJ, et al. Sleeping beauty transposition from nonintegrating lentivirus. Mol Ther. 2009;17:1197–1204. doi: 10.1038/mt.2009.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hackett PB, Largaespada DA, Cooper LJ. A transposon and transposase system for human application. Mol Ther. 2010;18:674–683. doi: 10.1038/mt.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bak RO, Stenderup K, Rosada C, petersen LB, Moldt B, Dagnæs-Hansen F, et al. Targeting of human interleukin-12B by small hairpin RNAs in xenografted psoriatic skin. BMC Dermatology. 2011;11:5. doi: 10.1186/1471-5945-11-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilber A, Wangensteen KJ, Chen Y, Zhuo L, Frandsen JL, Bell JB, et al. Messenger RNA as a Source of Transposase for Sleeping Beauty Transposon-mediated Correction of Hereditary Tyrosinemia Type I. Mol Ther. 2007;15:1280–1287. doi: 10.1038/sj.mt.6300160. [DOI] [PubMed] [Google Scholar]
- 30.Michel G, Yu Y, Chang T, Yee JK. Site-specific gene insertion mediated by a Cre-loxP-carrying lentiviral vector. Mol Ther. 2010;18:1814–1821. doi: 10.1038/mt.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]