Abstract
The ongoing activity of the human retrotransposon Long Interspersed Element 1 (LINE-1 or L1) continues to impact the human genome in various ways. Throughout evolution, mammalian and primate genomes have been under selection to generate strategies to reduce the activity of selfish DNA like L1. Similarly, selfish DNA has evolved to elude these containment systems. This intragenomic conflict has left many inactive versions of LINEs and other Transposable Elements (TEs) littering the human genome, which together account for roughly half of our DNA. Here, we survey the distinct mechanisms operating in the human genome that seem to reduce the mobility of L1s. In addition, we discuss recent findings that strongly suggest epigenetic mechanisms specifically regulate L1 activity in pluripotent human cells.
Key words: LINE-1, epigenetic mechanisms, DNA methylation, histone deacetylation, L1 antisense promoter
The Red Queen's Hypothesis, proposed by Leigh van Valen in 1973,1 is applicable to host-parasite co-evolution. The name of the hypothesis comes from Lewis Carroll's novel “Through the Looking Glass,” where inhabitants of the Red Queen's kingdom have to keep running simply to stay in the same place, as the kingdom is continually moving. This situation is comparable to the state in which hosts and parasites exist, since both are constantly under selection to develop strategies to avoid being eliminated by their opponent, maintaining a state of perpetual confrontation. In such co-evolutionary “arms races” the host evolves new weaponry to eliminate its parasite, or at least minimize the damage caused. However this armament becomes obsolete when it is neutralized by the evolution of the parasite's defenses. Subsequently, selection for new strategies begins again. The conflict is asymmetric because however the parasite evolves to be more efficient in its exploitation of the host, it cannot critically diminish the host's capacity to sustain it, having no independent means of existence. Therefore, both host and parasite evolve, whilst remaining in the same relative positions in their relationship. Resolution of this co-evolutionary conflict can only come if one of the opponents beats its adversary. In the case of intragenomic conflict between parasitic DNA and host genomes we can obviously only observe those cases where the host is victorious.
We can observe host-parasite co-evolution in all levels of life's complexity, from animals, plants and fungi to bacteria, virus and even individual genes. In the same way as a virus can live within an animal, a selfish mobile DNA can live within a species' genome. Transposable Elements (TEs) are mobile DNA elements that are present in all genomes examined to date. TEs are considered a primary example of selfish DNA whose only purpose is to be replicated, passing on increased TE copy number to the next generation, spreading within the population by sexual reproduction and, rarely, to other species (through Horizontal Transfer).2 It is undeniable that TE activity has shaped the human genome and their continuing activity is a source of genomic plasticity that occasionally can be of adaptive value to the host.3 Thus, their impact is not purely parasitic and could be considered beneficial to the host in particular circumstances (see below).3–6
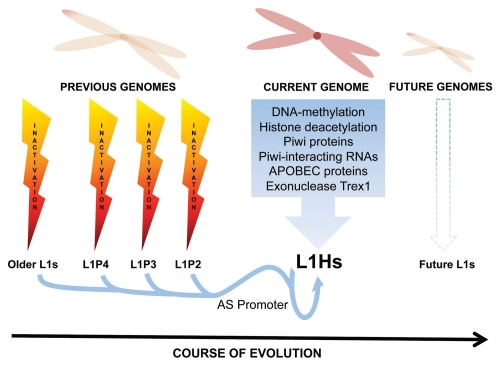
The defining property of a transposable element is to move from one site in the genome to another. This can be achieved by “cut and paste” mechanisms (DNA transposons) or a “copy and paste” mechanisms using an RNA intermediate (RNA transposons or retrotransposons) (reviewed in ref. 6). In general, the activity of TEs has negative impacts on genomes as they can promote several types of alterations at the site of insertion. The effects that new TE insertions produce are quite diverse, from minor sequence alterations to gross structural changes (gene inactivation, alteration of gene expression, genomic instability, etc.; reviewed in refs. 6 and 7). Therefore, it is logical that host genomes are under selection to evolve mechanisms that can eliminate TEs, or at least reduce their negative effects. Conversely TEs are selected to defend themselves against such mechanisms, in order to be perpetuated (Fig. 1).
Figure 1.
Co-evolution of L1s and the human genome. The illustration represents the co-evolution between LINE-1 and the human genome, showing the inactivated L1s neutralized by previous human genomes. Notably, the L1-AS promoter is conserved through LINE-1 evolution. Also shown are currently known L1 control mechanisms, which obligates L1 to evolve to avoid extinction in the human genome.
Besides these negative impacts, it is also clear that TEs have been instrumental in shaping genomes through evolutionary innovation. For example, RAG proteins that participate in V(D)J recombination during antibody class switching, appear to be derived from a DNA transposon transposase enzyme.4,8 Also some species' telomeres are maintained by the mobilization of TEs to chromosome ends.5
One side effect of this ongoing battle is the accumulation of inactive TE copies that can constitute a large proportion of the genome. In the human genome, we find the remains of many different types of DNA transposons (∼3%) and retrotransposons (∼42%) but the only surviving active autonomous transposable element is the non-LTR retrotransposon LINE-1 or L1.9 L1s make up around 17% of the human genome,6,9,10 although on average only 80 to 100 copies per human remain retrotransposition competent (RC-L1s).11 Thus, most L1s have lost the battle to remain active. Currently the L1PA1/L1HS subfamily of L1 includes all known RC-L1s. Moreover, the human genome contains remnants of several inactive subfamilies of L1, that have succeeded each other as a single lineage.12 This picture illustrates the capacity of L1s to survive throughout evolutionary time (Fig. 1). In a similar manner, it stands to reason that mammalian and primate genomes have been selected to generate mechanisms that reduce the mobility of TEs, ultimately resulting in the structure of the contemporary human genome.6,7,9,10
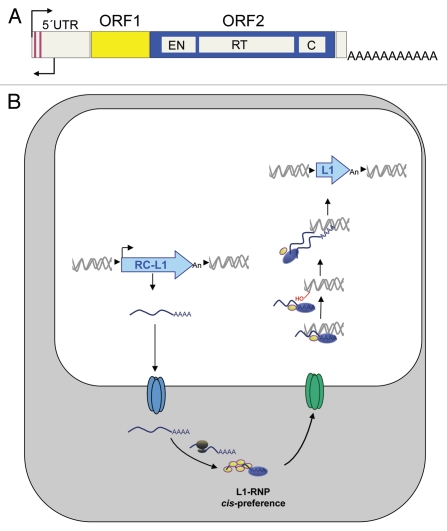
RC-L1s are 6-kb long sequences that contain a 910-bp 5′ untranslated region (UTR) that harbors an internal promoter13 and encodes two Open Reading Frames (ORFs) that are required for L1 retrotransposition10,14 (Fig. 2A). ORF1 encodes an RNA binding protein (ORF1p) while ORF2 encodes a protein (ORF2p) with ENdonuclease (EN) and Reverse Transcriptase (RT) activities (reviewed in refs. 6, 7 and 10). Retrotransposition starts with the generation of a full-length L1 mRNA from a preexisting L1 in the genome that is subsequently translated to generate a RiboNucleoprotein Particle (RNP, Fig. 2B).6,7,10 The L1 RNP is a retrotransposition intermediate that contains both the L1 proteins and their encoding mRNA, associated in cis (Fig. 2B). Indeed this preferential association or cis-preference,6,10,14,15 likely represents a mechanism whereby L1 has ensured its evolutionary success as RC-L1 RNPs will most frequently mobilize RC-L1 mRNAs. Once in the nucleus, retrotransposition occurs by a process known as Target Primed Reverse Transcription (TPRT).16 In TPRT the L1 encoded EN activity recognizes and cleaves a loose consensus sequence, liberating a free 3′OH that is used by the L1 encoded RT activity to prime first strand cDNA synthesis6,10,16 (Fig. 2B). It is thought that the same activities are involved in second strand cDNA synthesis, resulting in a new L1 inserted elsewhere in the genome (Fig. 2B). Notably, most de novo L1 insertions are flanked by variable size Target Site Duplications (TSDs) and are often 5′-truncated.6,10 Thus, it is possible that 5′-truncation could represent a host defense mechanism that reduces the number of potentially active L1s in a genome by aborting reverse transcription before the internal promoter can be copied. Although the host factor/s involved in truncation are currently unknown, L1 insertion and the DNA repair machinery are known to interact at least in cell culture retrotransposition assays.17–20
Figure 2.
LINE-1 retrotransposition. (A) Scheme of an RC-L1. In the scheme, ORF1 and ORF2 are represented as yellow and blue rectangles. Within ORF2, the relative position of the EN, RT and cysteine-rich domains (C) is indicated. Also shown are the S and AS promoters in the L1 5′UTR (black thin arrows). (B) Working model of L1 retrotransposition. In the scheme, for simplicity L1 is drawn as a light blue arrow. Details can be found in the main text.
As a selfish DNA, L1 has just one purpose: to be perpetuated. To achieve this it must retrotranspose during early development before germline partitioning, and/or in germ cells, to ensure transmission to the next generation. Previous studies have shown that full-length L1 mRNA and the L1 encoded ORF1p are expressed in male/female mouse germ cells.21,22 Similarly, L1 mRNA has been detected in human female oocytes23 and in human embryonic stem cells (hESCs,24,25). Interestingly, a mouse model of human L1 retrotransposition indicates that most L1 insertions occur in early embryonic development, and that germ cell insertions are uncommon.26 Thus hESCs and other pluripotent cell lines represent a logical model to study L1 retrotransposition regulation. It is also logical to deduce that L1 restriction mechanisms could act in these cell types, as otherwise new L1 insertions would be transmitted to subsequent generations. Studies have characterized several defense mechanisms that act against L1 mobility, such as APOBEC proteins,27 the exonuclease Trex1,28 Piwi proteins and Piwi-interacting RNAs,29 DNA methylation,30,31 and small RNAs generated by the antisense L1 promoter32,33 (Fig. 1). These host mechanisms clearly act to control currently active L1s and likely could have contributed to reduce the mobility of previously active L1 subfamilies (L1PA2-15 and others), but may require co-evolutionary fine-tuning to control the mobilization of future L1 subfamilies (Fig. 1).
As L1 retrotransposition requires the generation of a full-length L1 mRNA, it is very likely that restricting its expression would strongly repress L1 mobilization. DNA methylation is one way in which cells can silence a gene and is considered to be a general host strategy to defend against TEs.31 L1 is a likely target for DNA methylation as its promoter contains a CpG island. In fact, deletion of the de novo methyltransferase-3 like gene (Dnmt3L), which is implicated in the methylation of LINE-1 and other LTR-retrotransposons, results in overexpression of retrotransposons in germ cells and meiotic failure during gametogenesis in mice.30 Thus, DNA methylation in germ cells clearly contributes to restrict L1 expression and thus retrotransposition, although other post-transcriptional mechanisms, such as piRNAs and APOBEC proteins, may also contribute to L1 control in germ cells. It is tempting to speculate that the low level of L1 mobilization in germ cells is caused by an overlapping combination of these mechanisms, while mobilization events during early embryogenesis cannot be controlled in an analogous manner (see below). Indeed, it is known that the L1 promoter is hypermethylated in human somatic tissues, with the exception of the brain, and certain tumors.34,35
In general, this evidence suggests that the human genome and L1 may have reached an equilibrium involving minimal retrotransposition in somatic cells, so avoiding a reduction of individual fitness that may compromise both host and L1 reproduction. In contrast, recent studies have shown that L1 seems to actively move in certain neuronal cell types.34,36,37 The retrotransposition of engineered human L1s has been observed in Neural Progenitor Cells (NPCs) derived from hESCs or isolated from human fetal brain. These data parallel the increased copy number of endogenous L1s observed in several regions of the adult human brain, compared to other somatic tissues.34 Similar results have also been observed in rats and mice,36,37 leading to the speculation that L1 insertions during neuronal differentiation could cause genetic variability between neurons. Such variability could influence neuronal diversity in the brain, which could be important for learning capacity, memory or behavior.36 Thus L1 activity in the brain may represent a case of somatic domestication, where TE mobility is harnessed to generate genetic diversity. However further investigation will be required to answer the intriguing question: “What does L1 activity in the brain mean (if anything)?” In addition to an increased L1 copy number, there also seems to be a correlation between the level of L1 expression in the human brain and the methylation status of L1 promoters.34 These data thus seem to fit well with the main rule in retrotransposition: if there is L1 mRNA expression, then there is likely L1 retrotransposition, not withstanding the ability of post-transcriptional mechanisms to restrict L1 retrotransposition. In support of this rule, a recent report demonstrated de novo somatic mobilization of endogenous L1 elements in certain human lung tumors. Notably, new somatic L1 insertions seem to accumulate in those tumors that show global hypomethylation.35 Furthermore, it is clear that L1 retrotransposition is dependent upon host factors, and some of these will likely be differentially expressed in different cell types, making it likely that the cellular environment influences the efficiency of L1 mobilization. In sum, as with L1 activity in the brain, it remains to be determined if L1 insertions in tumors could generate somatic variation that can drive cancer cell clonal evolution, or they are simply a consequence of the cancer cell phenotype.
In contrast to the situation in germ cells, early embryonic development appears to be an active battle-ground for L1/host intragenomic conflict, despite the fact that new L1 insertions may be lost due to purifying selection of accompanying deleterious embryonic mutations. The case of a patient affected by a disease-causing L1 insertion inherited from a somatically mosaic parent is proof of principle that endogenous L1s can retrotranspose during human embryonic development,38 in agreement with mouse models.26 Using hESCs as a model of early developmental processes, it has been demonstrated that L1-RNPs are overexpressed in hESCs.24,25 In addition, using a cell-cultured based assay, it was shown that engineered human L1s can retrotranspose at low levels in hESCs.24 Similarly, human embryonic carcinoma cell lines (hECs), which represent a model of hESCs, also show L1 overexpression (mRNA and L1-RNPs,25,39). Consistent with the expression rule, L1 promoters are hypomethylated in both hECs and hESCs.40 Indeed, up to 20% of expressed L1Hs elements in hESCs/hECs are alleles of known RC-L1s that may actively retrotranspose.25 These results suggest that the level of somatic mosaicism that L1 could generate might be significant. As DNA methylation cannot control L1 mobilization in hESCs, it maybe the case that other repression mechanisms come into play in embryogenesis.
Recently a mechanism that epigenetically silences de novo L1 insertions in pluripotent cells has been reported.39 This L1 repression system, which is sensitive to histone deacetylase inhibitors, has been analyzed in hEC cell lines using engineered L1 elements, although it is likely that endogenous L1s are controlled in a similar manner. Briefly, de novo engineered L1 insertions in hECs are silenced during, or shortly after, insertion so preventing further rounds of retrotransposition (Fig. 1). Notably, the process of silencing seems to be specific for TPRT, as non-LTR retroelements from several species are silenced while retroviruses, which insert by a fundamentally different mechanism, are not. Remarkably, this novel epigenetic silencing of L1 insertions seems to operate predominantly in pluripotent cells, as it is strongly attenuated when insertions occur during differentiation. These data may indicate that the L1 silencing response is specifically associated with pluripotent cells, and that the host factors involved are downregulated upon differentiation. We also speculate that the low level of engineered L1 retrotransposition observed in hESCs could be, in part, due to this novel epigenetic silencing response. However, the depth of the silencing response is difficult to estimate in hESCs cells due to technical challenges in their manipulation.24
At the level of an individual, a defense mechanism that can only prevent secondary insertions may not appear to be an effective retroelement control mechanism. However in the context of development, the insertion of a necessarily transposition competent retroelement, into a region of open chromatin brings a risk of further retrotransposition that may evoke strong selection for pluripotent cell specific defense systems. One could envisage that insertions silenced in this way, irrespective of their context, could have two outcomes: insertions into regions that can be silenced without effecting development cannot give rise to further insertions, and insertions in regions whose silence is a lethal epimutation will remove the cell from the embryo. Such a mechanism could restrict runaway retrotransposition of genic L1s until they decay into pseudogenes. That said, likely due to the structure of the human genome, L1 might overcome its complete inhibition by its presence within human genes.25 In addition, and as the host can benefit from the genomic plasticity conferred by the activity of TEs, their epigenetic control could also be seen as a symbiotic mechanism to reduce the load of L1 mobilization during early human embryogenesis. The involvement of chromatin remodeling factors in this process makes it tempting to speculate that some types of epigenetic modifications may represent domestication of a process derived from an anti-TE defense mechanism. By extension, L1 insertions may be exapted to maintain a specific chromatin region in an inactive or active state, as observed with other TEs.41
More recently, we utilized the antisense (AS) promoter located within the L1 5′UTR33 to map individual expressed L1s in hECs/hESCs.25 The antisense L1 promoter is 10 times less active than the sense (S) L1 promoter,25,32,33 but nevertheless drives the transcription of multiple human gene isoforms.33 As the S and AS L1 promoters overlap, it has been speculated that they compete for the binding of transcription factors that modulate the level of sense L1 mRNA production.33 Alternatively, the L1-AS promoter has been associated with the auto-regulation of L1 retrotransposition, by an RNAi-like mechanism,32 as its deletion enhances retrotransposition of L1 constructs. When we analyzed chimeric L1-AS transcripts produced in hECs/hESCs, we found that they belong to several L1 subfamilies, unexpectedly indicating that the L1-AS promoter is conserved through L1 evolution (at least as far back as L1PA10 elements25). This raises the question of why a selfish DNA like L1, under selection to maximize its activity, maintains a self-regulatory sequence? It is possible that inactive L1s, with active AS promoters, may be exapted to regulate the gene expression of neighboring genes, but this is only likely to be true for a proportion of insertions. Alternatively auto-regulation of L1 transcript levels might evolve if the cells supporting retrotransposition have a very sensitive threshold for L1 protein products or their effects (e.g., DNA damage), beyond which cell death occurs. Retrotransposition in early development, where elements that do not auto-regulate kill their host cell, could lead to a selective advantage for the maintenance of an auto-regulating antisense promoter. Of course it remains possible that L1-AS transcripts may have other roles in L1 expression and/or retrotransposition that are yet to be determined.
Perhaps the most notable revelation from examining which L1s are expressed in hECs/hESCs (using L1-AS activity as a reporter) was that most expressed L1s are located within human genes. In addition, we found that despite their abundance, only relatively few L1s are expressed.25 Overall, we found that up to 80% of expressed L1s are located within genes, while only 30% of full-length L1Hs elements reside within human genes. Thus, there is a significant enrichment for expressed L1s located within genes, which suggests epigenetic control of their expression that is distinct from DNA methylation. However we know nothing of the host factor/s that participate in the differential expression of L1s located within genes. Due to the large number of L1 elements dispersed through the genome, we speculate that full-length L1 elements residing within genes actively transcribed in embryonic cells have a greater chance of being transcribed and thus to be perpetuated in the human genome. As a result it is not only important to know the number of active L1s per genome, but also their location within the genome.
In sum, it is evident that mammalian and primate genomes have evolved different strategies to control the activity of L1, and is also possible that L1 has evolved self-control mechanisms. In addition, there are likely other restriction mechanisms directed against TEs that remain to be discovered. With this in mind, how is it possible that the human genome has not been able to eliminate this genetic parasite? Extinction of active L1 elements has been observed in other mammals,42 but this is very rare, so one explanation could be that L1 simply has a higher ability to perpetuate itself in the human genome than other TEs. Alternatively—or additionally—L1s may be maintained for the long term plasticity that they confer to the human genome, although selection for traits that enhance evolvability is a highly controversial subject43,44 and requires positive selection at the level of the individual to get started. The possible short term benefits derived by the host may come from the capacity of L1 to generate useful somatic variability (in the brain, for example). Finally it maybe the case that expression of L1 elements from within genes actively transcribed in pluripotent cells,25 represents a host defense evasion strategy that cannot be effectively countered without disrupting development. Perhaps the epic battle between our genome and its most successful parasite will never end?
Acknowledgments
We apologize to colleagues whose work could not be cited due to space limitations. We thank J.L.G.P. lab members for helpful discussions and critically reading the manuscript. R.M.B. was supported by Wellcome Trust Project Grant (Ref. 075163/Z/04/Z). J.L.G.P.'s laboratory is supported by ISCIII-CSJA-FEDER (EMER07/056), a Marie Curie IRG action (FP7-PEOPLE-2007-4-3-IRG), CICE (P09-CTS-4980), Proyectos en Salud PI-002 from Junta de Andalucia (Spain), and the Spanish Ministry of Health (FIS-FEDER PI08171). M.M.L. is supported by CICE (P09-CTS-4980); A.M. is supported by Marie Curie IRG action (FP7-PEOPLE-2007-4-3-IRG); M.G.C. is supported by Consejeria de Salud (Andalusian Government).
Abbreviations
- TE
transposable element
- LTR
long terminal repeat
- LINE-1 or L1
long interspersed element class 1
- RC-L1
retrotransposition competent L1
- L1Hs
human specific LINE-1
- ORF
open reading frame
- RT
reverse transcriptase
- EN
endonuclease
- TPRT
target primed reverse transcription
- hESCs
human embryonic stem cells
- hECs
human embryonic carcinoma cells
- L1-AS
LINE-1 antisense promoter or transcript
- L1-S
LINE-1 sense promoter or transcript
References
- 1.Van Valen L. A new evolutionary law. Evolutionary Theory. 1973;1:1–30. [Google Scholar]
- 2.Doolittle WF, Sapienza C. Selfish genes, the phenotype paradigm and genome evolution. Nature. 1980;284:601–603. doi: 10.1038/284601a0. [DOI] [PubMed] [Google Scholar]
- 3.Sinzelle L, Izsvak Z, Ivics Z. Molecular domestication of transposable elements: from detrimental parasites to useful host genes. Cell Mol Life Sci. 2009;66:1073–1093. doi: 10.1007/s00018-009-8376-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Melek M, Gellert M, van Gent DC. Rejoining of DNA by the RAG1 and RAG2 proteins. Science. 1998;280:301–303. doi: 10.1126/science.280.5361.301. [DOI] [PubMed] [Google Scholar]
- 5.Curcio MJ, Belfort M. The beginning of the end: links between ancient retroelements and modern telomerases. Proc Natl Acad Sci USA. 2007;104:9107–9108. doi: 10.1073/pnas.0703224104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodier JL, Kazazian HH. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 7.Deininger PL, Moran JV, Batzer MA, Kazazian HH., Jr Mobile elements and mammalian genome evolution. Curr Opin Genet Dev. 2003;13:651–658. doi: 10.1016/j.gde.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 8.Agrawal A, Eastman QM, Schatz DG. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- 9.Mills RE, Bennett EA, Iskow RC, Devine SE. Which transposable elements are active in the human genome? Trends Genet. 2007;23:183–191. doi: 10.1016/j.tig.2007.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Moran JV, Gilbert N. Mammalian LINE-1 retrotransposons and related elements. In: Craig N, Craggie R, Gellert M, Lambowitz A, editors. Mobile DNA II. Washington, DC: ASM Press; 2002. [Google Scholar]
- 11.Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, et al. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141:1159–1170. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boissinot S, Chevret P, Furano AV. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol Biol Evol. 2000;17:915–928. doi: 10.1093/oxfordjournals.molbev.a026372. [DOI] [PubMed] [Google Scholar]
- 13.Swergold GD. Identification, characterization and cell specificity of a human LINE-1 promoter. Mol Cell Biol. 1990;10:6718–6729. doi: 10.1128/mcb.10.12.6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 15.Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, et al. Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 17.Gasior SL, Roy-Engel AM, Deininger P. ERCC1/XPF limits L1 retrotransposition. DNA Repair (Amst) 2008;7:983–989. doi: 10.1016/j.dnarep.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morrish TA, Garcia-Perez JL, Stamato TD, Taccioli GE, Sekiguchi J, Moran JV. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature. 2007;446:208–212. doi: 10.1038/nature05560. [DOI] [PubMed] [Google Scholar]
- 19.Morrish TA, Gilbert N, Myers JS, Vincent BJ, Stamato TD, Taccioli GE, et al. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat Genet. 2002;31:159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- 20.Suzuki J, Yamaguchi K, Kajikawa M, Ichiyanagi K, Adachi N, Koyama H, et al. Genetic Evidence That the Non-Homologous End-Joining Repair Pathway Is Involved in LINE Retrotransposition. PLos Genet. 2009;5:1000461. doi: 10.1371/journal.pgen.1000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Branciforte D, Martin SL. Developmental and cell type specificity of LINE-1 expression in mouse testis: implications for transposition. Mol Cell Biol. 1994;14:2584–2592. doi: 10.1128/mcb.14.4.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trelogan SA, Martin SL. Tightly regulated, developmentally specific expression of the first open reading frame from LINE-1 during mouse embryogenesis. Proc Natl Acad Sci USA. 1995;92:1520–1524. doi: 10.1073/pnas.92.5.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Georgiou I, Noutsopoulos D, Dimitriadou E, Markopoulos G, Apergi A, Lazaros L, et al. Retrotransposon RNA expression and evidence for retrotransposition events in human oocytes. Hum Mol Genet. 2009;18:1221–1228. doi: 10.1093/hmg/ddp022. [DOI] [PubMed] [Google Scholar]
- 24.Garcia-Perez JL, Marchetto MC, Muotri AR, Coufal NG, Gage FH, O'Shea KS, et al. LINE-1 retrotransposition in human embryonic stem cells. Hum Mol Genet. 2007;16:1569–1577. doi: 10.1093/hmg/ddm105. [DOI] [PubMed] [Google Scholar]
- 25.Macia A, Munoz-Lopez M, Cortes JL, Hastings RK, Morell S, Lucena-Aguilar G, et al. Epigenetic control of retrotransposon expression in human embryonic stem cells. Mol Cell Biol. 2011;31:300–316. doi: 10.1128/MCB.00561-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kano H, Godoy I, Courtney C, Vetter MR, Gerton GL, Ostertag EM, et al. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 2009;23:1303–1312. doi: 10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schumann GG. APOBEC3 proteins: major players in intracellular defence against LINE-1-mediated retrotransposition. Biochem Soc Trans. 2007;35:637–642. doi: 10.1042/BST0350637. [DOI] [PubMed] [Google Scholar]
- 28.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134:587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–668. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bourc'his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–99. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 31.Yoder JA, Walsh CP, Bestor TH. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 32.Yang N, Kazazian HH., Jr L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat Struct Mol Biol. 2006;13:763–771. doi: 10.1038/nsmb1141. [DOI] [PubMed] [Google Scholar]
- 33.Speek M. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol Cell Biol. 2001;21:1973–1985. doi: 10.1128/MCB.21.6.1973-1985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coufal NG, Garcia-Perez JL, Peng GE, Yeo GW, Mu Y, Lovci MT, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–1261. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singer T, McConnell MJ, Marchetto MC, Coufal NG, Gage FH. LINE-1 retrotransposons: mediators of somatic variation in neuronal genomes? Trends Neurosci. 2010;33:345–354. doi: 10.1016/j.tins.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 38.van den Hurk JA, Meij IC, Seleme MC, Kano H, Nikopoulos K, Hoefsloot LH, et al. L1 retrotransposition can occur early in human embryonic development. Hum Mol Genet. 2007;16:1587–1592. doi: 10.1093/hmg/ddm108. [DOI] [PubMed] [Google Scholar]
- 39.Garcia-Perez JL, Morell M, Scheys JO, Kulpa DA, Morell S, Carter CC, et al. Epigenetic silencing of engineered L1 retrotransposition events in human embryonic carcinoma cells. Nature. 2010;466:769–773. doi: 10.1038/nature09209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munoz-Lopez M, Garcia-Perez JL. 2011. Unpublished data.
- 41.Sigrist CJ, Pirrotta V. Chromatin insulator elements block the silencing of a target gene by the Drosophila polycomb response element (PRE) but allow trans interactions between PREs on different chromosomes. Genetics. 1997;147:209–221. doi: 10.1093/genetics/147.1.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grahn RA, Rinehart TA, Cantrell MA, Wichman HA. Extinction of LINE-1 activity coincident with a major mammalian radiation in rodents. Cytogenet Genome Res. 2005;110:407–415. doi: 10.1159/000084973. [DOI] [PubMed] [Google Scholar]
- 43.Brookfield JF. Evolution and evolvability: celebrating Darwin 200. Biol Lett. 2009;5:44–46. doi: 10.1098/rsbl.2008.0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Colegrave N, Collins S. Experimental evolution: experimental evolution and evolvability. Heredity. 2008;100:464–470. doi: 10.1038/sj.hdy.6801095. [DOI] [PubMed] [Google Scholar]