Abstract
The availability of genomic sequences provided new opportunities to decipher how plant genomes evolve. One recent discovery about plant genomes is the abundance of Pack-MULEs, a special group of transposable elements that duplicate, amplify and recombine gene fragments in many species at a very large scale. Despite the widespread occurrence of Pack-MULEs, their function remains an enigma. Our analysis using maize, rice and Arabidopsis genomic sequences indicates that the acquisition of genic sequences by Pack-MULEs is not random. Pack-MULEs in grasses specifically acquire and amplify GC-rich gene fragments. The resulting GC-rich elements have the ability to form independent transcripts with negative GC gradient, which refers to the decline of GC content along the orientation of transcription of genes. In other cases, Pack-MULEs insert near the 5′ region of “normal” genes, and consequently form additional 5′ exons or replace the original 5′ exon of genes. In this manner, Pack-MULEs raise the GC content of the 5′ termini of genes, modify the gene structure and contribute to the increased number of genes with negative GC gradient in grasses. The possible consequence of such activity is discussed.
Key words: Pack-MULE, GC gradient, acquisition, insertion bias, gene modification
GC Content and GC Gradient of Genes
Living organisms vary dramatically in the GC content of their genomes. The GC content of currently sequenced genomes of bacteria ranges from 17–75%.1 In contrast, eukaryotic genomes vary less in their GC content. Most eukaryotic genomes are relatively GC-poor and have less than 50% GC content.2 Among higher plants, the genomes of monocots are in general more GC-rich than dicots: the average GC content of dicot genomes is 35%, and that of monocots is 44%.3
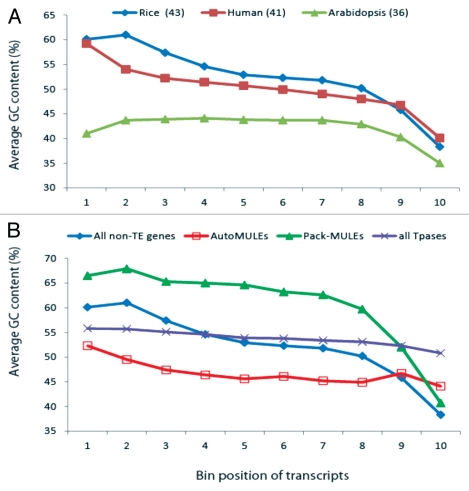
Intriguingly, the increased GC content in monocots is not uniformly distributed in the genome but instead, it largely presents as a negative GC gradient in genes, which refers to the phenomenon whereby the GC content declines along the direction of transcription.4 This is illustrated in Figure 1 where the entire transcripts of genes are divided into 10 equal sized bins from the 5′ end to the 3′ end. As shown in Figure 1A, the average GC content of Arabidopsis (a dicot) genes does not vary markedly along the length of the gene, with the exception of the most 3′ end, where the AT-rich transcription termination signal is located. Therefore, the GC content of the most 3′ end is lower than that of the rest of the gene body. The majority of rice (a monocot) genes, in contrast, are associated with a GC-rich 5′ end (∼60% GC); however, the GC content (∼40%) of their 3′ ends is comparable to that of Arabidopsis genes (Fig. 1A). Despite the prevalence of negative GC gradient in monocot genomes, individual genes demonstrate a dramatic difference in GC gradient. In rice, over 1/3 of the genes are not associated with a significant GC gradient and 10% of the genes have a significant positive GC gradient (the 3′ end is more GC-rich than the 5′ end).5 The difference among individual genes implies that the mechanisms underlying the formation of GC gradient do not act equally on all genes. The divergence in GC gradient among different type of genes is further supported by the fact that transposase genes (including the putative autonomous Mutator-like elements) in the rice genome only display a minor negative GC gradient (Fig. 1B). Thus, a significant negative GC gradient is only limited to a subset of genes.
Figure 1.
Variation in GC gradients of genes. (A) Comparison of genes in rice, human and Arabidopsis. The numbers in parenthesis indicate the genomic average of GC content. (B) Comparison of different types of transcripts in rice. All non-TE genes: all non-transposon genes; AutoMULEs: putative autonomous Mutator-like transposable elements; all Tpases: all annotated transposase genes, including both DNA transposons and retrotransposons. The human gene annotation was downloaded from NCBI on April 1, 2011 (ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/RNA). The Arabidopsis gene annotation information was from TAIR9 (www.arabidopsis.org). Gene annotation of rice is based on MSU release 6.0 (http://rice.plantbiology.msu.edu/).
The presence of GC gradient in genes, however, is not specific to plant genomes. For example, the GC content of the human genome (41%) is slightly lower than that of rice (43%) but higher than that of Arabidopsis (36%), and it is known that there are GC-rich islands in the human genome.6 Like the rice genes, the human genes are associated with a significant negative GC gradient (Fig. 1A). Therefore, a negative GC gradient seems to be prevalent in genomes with local GC content variation.
The Impact of Pack-MULEs on GC Gradient
Mutator elements comprise a superfamily of DNA transposable elements, and most of them are associated with long Terminal Inverted Repeats (TIR). Pack-MULEs refer to the non-autonomous Mutator-like elements (MULEs) carrying genes or gene fragments between the TIR sequences. The genes where the internal regions of Pack-MULEs are derived from are referred to as parental genes. Gene duplication by MULEs was first reported more than 20 years ago when the Mu1/Mu2 element was found to contain part of a gene called MRS-A.7 However, the prevalence of Pack-MULEs was not revealed until fairly recently when a large amount of plant genomic sequences became available.8–10 Unlike other gene duplication processes, most Pack-MULE acquisition events involve gene fragments, not entire genes. Interestingly, the acquired/duplicated fragments are not randomly located in the parental genes; instead, the acquired position is dependent on the GC gradient of the genes—in most cases, the GC-rich sequences are acquired. As a result, the resulting Pack-MULE bears a GC-rich internal sequence, and the GC content of the internal region is much higher than that of the TIR and the genomic average.5
One important feature of Mutator elements is the presence of promoters inside the TIRs.11 These promoters drive the transcription of GC-rich sequences inside the Pack-MULEs, and the relevant transcripts often terminate in the other TIR or in the flanking sequences, which are usually GC-poor.5 For this reason, most Pack-MULE associated transcripts are more GC-rich and are associated with a more dramatic negative GC gradient than other genes or other transposon transcripts in the genome (Fig. 1B). In addition to the formation of independent transcripts, the transcripts initialized within Pack-MULEs often extend to downstream genes to form chimeric transcripts.5 In these transcripts, the internal regions of Pack-MULEs either serve as an additional exon at the 5′ end or replace the original 5′ end exon of the corresponding downstream gene. In either case, the chimeric transcript, which contains both Pack-MULE and adjacent gene sequences, often has a negative GC gradient because of the GC-richness of the internal region of Pack-MULEs. Two important factors permit the formation of such chimeric transcripts: (1) the ability for Pack-MULE TIR to initialize transcription (see above); (2) the insertion preference for Pack-MULEs to land in regions flanking the 5′ termini of genes,5,11–14 which provides spatial convenience for the fusion of Pack-MULEs and their downstream genes.
Through the formation of independent transcripts and chimeric transcripts, it is clear that Pack-MULEs may increase the number of genes with negative GC gradients, or elevate the degree of the existing negative GC gradient in genes of grasses. This raises the question: if Pack-MULEs have been as prevalent in Arabidopsis as they have been in rice, would the influence of Pack-MULEs be powerful enough to turn the Arabidopsis genes into the rice genes in terms of GC gradients? The analysis of Arabidopsis genome did not result in a clear-cut answer because there are few Pack-MULEs in Arabidopsis and most of them appear to represent ancient insertions.5 The acquired regions in Arabidopsis Pack-MULEs are only slightly more GC-rich than the average gene sequences, while those in Pack-MULEs from rice and maize have a much higher GC content than the rest of the gene sequence.5 This may imply that the impact of Pack-MULEs would only be significant when a pre-existing GC gradient or at least a significant variation in GC content in genic regions is present in the genome. In other words, Pack-MULEs represent one of the positive feedback steps in the formation of a negative GC gradient. This is likely because Pack-MULEs preferentially duplicate (and amplify) GC-rich sequences from templates (parental genes) without creating de novo GC-rich sequences. Other genetic forces, such as GC-biased gene conversion,15,16 or codon usage bias,17 might be responsible for creating the GC-rich templates for Pack-MULEs in grasses.
The Consequence of Biased Acquisition and Insertion of Pack-MULEs
In plant genomes, protein-coding sequences are in general more GC-rich than non-coding sequences.18,19 If Pack-MULEs specifically acquire GC-rich sequences, this preference would maximize their opportunities to duplicate and amplify coding sequences, which may provide new raw materials for the evolution of novel coding genes. Given the fact that turnover of TE sequences is rather rapid in the genome, the presence of potentially useful sequences inside these elements may increase the chance for Pack-MULEs to be retained in the genome. Due to the over-representation of GC-rich sequence at the 5′ end of genes in grasses, the acquisition preference of Pack-MULEs results in an excess of 5′ gene region sequences inside Pack-MULEs. In addition, Pack-MULEs can serve as the 5′ termini of genes, so it is likely that the 5′ ends of some “normal” genes represent ancient Pack-MULEs. In this case, the combination of the acquisition bias with the insertion preference of Pack-MULEs could form a recycling mechanism of the 5′ ends of genes and enhance the negative GC gradient.
One apparent consequence of the amplification of GC-rich sequences is the elevation of the global GC content of the genome. Nevertheless, given the fact that Pack-MULE sequences in rice only account for 1.6% of the genome,20 such an increase in GC content should be minor in the short term but could be influential in an evolutionary scale if a considerable amount of Pack-MULE sequences are retained in the genome. On the other hand, the insertion of Pack-MULEs would likely create an immediate shift on the local GC content. This is especially important given the fact that Pack-MULEs preferentially insert in the 5′ region of genes, which may result in a series of genetic and epigenetic alterations on the nearby genes.
The alteration of GC content is accompanied by a variety of physical properties of the relevant DNA sequence. GC-rich sequences tend to have reduced curvature but increased bendability and ability for B to Z transition of the DNA helix.21 All of these features are considered to be related to an open chromatin structure and active transcription, which is consistent with the fact that in general GC-rich genes are more expressed than GC-poor genes.22 Z-DNA forming sequences are also known to induce double strand DNA breaks, translocation and large scale deletion in mammalian cells,23 and therefore are associated with genome instability. In addition, recombination rate is often positively correlated with GC content.24 Finally, GC-rich sequences contain more CG and CHG (H = A, T or C) sites whose methylation status is heritable.25 Thus, Pack-MULEs may stabilize the epigenetic regulation of genes in adjacent regions.
In human and other higher eukaryotes, there are several types of GC-rich structural elements in proximity to the transcription start sites (TSSs) of genes. These include CpG islands, the nuclear factor I (NFI) transcription factor binding sites, and regions with potential to form Z-DNA (ZDRs).26–28 When different genomes are compared, correlation was not found between the organismal complexity and GC or CpG content.2 Nevertheless, there is a significant enrichment of GC and CpG islands adjacent to the TSSs in genomes of higher organisms. Likewise, the NFI binding sites and ZDRs are enriched near the TSS of genes in higher eukaryotes.2 As a result, the emergence of GC-rich transcriptional elements close to the TSS appears to correlate with organismal complexity, coordinate with the evolution of the transcription complex, and provide fine-tuned regulation of gene expression.2 From this point of view, it is tempting to speculate that the abundance of GC-rich Pack-MULEs around the TSSs of genes in monocots may confer regulatory advantages to their host organisms over the dicots.
Acknowledgments
This study was supported by grant DBI-0607123 from the National Science Foundation.
Abbreviations
- MULEs
Mutator-like elements
- NFI
the nuclear factor I
- Pack-MULEs
non-autonomous Mutator-like elements carrying gene fragments
- TIR
terminal inverted repeat
- TSS
transcription start site
- ZDRs
regions with potential to form Z-DNA
References
- 1.McCutcheon JP, McDonald BR, Moran NA. Origin of an alternative genetic code in the extremely small and GC-rich genome of a bacterial symbiont. PLoS Genet. 2009;5:1000565. doi: 10.1371/journal.pgen.1000565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Khuu P, Sandor M, DeYoung J, Ho PS. Phylogenomic analysis of the emergence of GC-rich transcription elements. Proc Natl Acad Sci USA. 2007;104:16528–16533. doi: 10.1073/pnas.0707203104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cavagnaro PF, Senalik DA, Yang L, Simon PW, Harkins TT, Chinna DK, et al. Genome-wide characterization of simple sequence repeats in cucumber (Cucumis sativus L.) BMC Genomics. 2010;11:569. doi: 10.1186/1471-2164-11-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wong GK, Wang J, Tao L, Tan J, Zhang J, Passey DA, et al. Compositional gradients in Gramineae genes. Genome Res. 2002;12:851–856. doi: 10.1101/gr.189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang N, Ferguson AA, Slotkin RK, Lisch D. Pack-Mutator-like transposable elements (Pack-MULEs) induce directional modification of genes through biased insertion and DNA acquisition. Proc Natl Acad Sci USA. 2011;108:1537–1542. doi: 10.1073/pnas.1010814108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 7.Talbert LE, Chandler VL. Characterization of a highly conserved sequence related to Mutator transposable elements in maize. Mol Biol Evol. 1988;5:519–529. doi: 10.1093/oxfordjournals.molbev.a040510. [DOI] [PubMed] [Google Scholar]
- 8.Jiang N, Bao Z, Zhang X, Eddy SR, Wessler SR. Pack-MULE transposable elements mediate gene evolution in plants. Nature. 2004;431:569–573. doi: 10.1038/nature02953. [DOI] [PubMed] [Google Scholar]
- 9.Juretic N, Hoen DR, Huynh ML, Harrison PM, Bureau TE. The evolutionary fate of MULE-mediated duplications of host gene fragments in rice. Genome Res. 2005;15:1292–1297. doi: 10.1101/gr.4064205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holligan D, Zhang X, Jiang N, Pritham EJ, Wessler SR. The transposable element landscape of the model legume Lotus japonicus. Genetics. 2006;174:2215–2228. doi: 10.1534/genetics.106.062752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lisch D. Mutator transposons. Trends Plant Sci. 2002;7:498–504. doi: 10.1016/s1360-1385(02)02347-6. [DOI] [PubMed] [Google Scholar]
- 12.Dietrich CR, Cui F, Packila ML, Li J, Ashlock DA, Nikolau BJ, et al. Maize Mu transposons are targeted to the 5′ untranslated region of the gl8 gene and sequences flanking Mu target-site duplications exhibit nonrandom nucleotide composition throughout the genome. Genetics. 2002;160:697–716. doi: 10.1093/genetics/160.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robbins ML, Sekhon RS, Meeley R, Chopra S. A Mutator transposon insertion is associated with ectopic expression of a tandemly repeated multicopy Myb gene pericarp color1 of maize. Genetics. 2008;178:1859–1874. doi: 10.1534/genetics.107.082503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu S, Yeh CT, Ji T, Ying K, Wu H, Tang HM, et al. Mu transposon insertion sites and meiotic recombination events co-localize with epigenetic marks for open chromatin across the maize genome. PLoS Genet. 2009;5:1000733. doi: 10.1371/journal.pgen.1000733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duret L, Galtier N. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu Rev Genomics Hum Genet. 2009;10:285–311. doi: 10.1146/annurev-genom-082908-150001. [DOI] [PubMed] [Google Scholar]
- 16.Muyle A, Serres-Giardi L, Ressayre A, Escobar J, Glemin S. GC-biased gene conversion and selection affect GC content in the Oryza genus (rice) Mol Biol Evol. 2011 doi: 10.1093/molbev/msr104. [DOI] [PubMed] [Google Scholar]
- 17.Tatarinova TV, Alexandrov NN, Bouck JB, Feldmann KA. GC3 biology in corn, rice, sorghum and other grasses. BMC Genomics. 2010;11:308. doi: 10.1186/1471-2164-11-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Salinas J, Matassi G, Montero LM, Bernardi G. Compositional compartmentalization and compositional patterns in the nuclear genomes of plants. Nucleic Acids Res. 1988;16:4269–4285. doi: 10.1093/nar/16.10.4269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizuno M, Kanehisa M. Distribution profiles of GC content around the translation initiation site in different species. FEBS Lett. 1994;352:7–10. doi: 10.1016/0014-5793(94)00898-1. [DOI] [PubMed] [Google Scholar]
- 20.Hanada K, Vallejo V, Nobuta K, Slotkin RK, Lisch D, Meyers BC, et al. The functional role of Pack-MULEs in rice inferred from purifying selection and expression profile. Plant Cell. 2009;21:25–38. doi: 10.1105/tpc.108.063206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vinogradov AE. DNA helix: the importance of being GC-rich. Nucleic Acids Res. 2003;31:1838–1844. doi: 10.1093/nar/gkg296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arhondakis S, Clay O, Bernardi G. GC level and expression of human coding sequences. Biochem Biophys Res Commun. 2008;367:542–545. doi: 10.1016/j.bbrc.2007.12.155. [DOI] [PubMed] [Google Scholar]
- 23.Wang G, Christensen LA, Vasquez KM. Z-DNA-forming sequences generate large-scale deletions in mammalian cells. Proc Natl Acad Sci USA. 2006;103:2677–2682. doi: 10.1073/pnas.0511084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fullerton SM, Bernardo Carvalho A, Clark AG. Local rates of recombination are positively correlated with GC content in the human genome. Mol Biol Evol. 2001;18:1139–1142. doi: 10.1093/oxfordjournals.molbev.a003886. [DOI] [PubMed] [Google Scholar]
- 25.Law JA, Jacobsen SE. Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet. 2010;11:204–220. doi: 10.1038/nrg2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- 27.Roulet E, Bucher P, Schneider R, Wingender E, Dusserre Y, Werner T, et al. Experimental analysis and computer prediction of CTF/NFI transcription factor DNA binding sites. J Mol Biol. 2000;297:833–84. doi: 10.1006/jmbi.2000.3614. [DOI] [PubMed] [Google Scholar]
- 28.Rich A, Zhang S. Timeline: Z-DNA: the long road to biological function. Nat Rev Genet. 2003;4:566–572. doi: 10.1038/nrg1115. [DOI] [PubMed] [Google Scholar]