Abstract
A major virulence mechanism used by pathogenic Gram-negative bacteria is the delivery of effector proteins from the bacterial cytoplasm into host cells by type III secretion. Typically, genes encoding type III secretion systems (T3SS) and effectors have been horizontally acquired by the bacteria that employ them. In proteobacteria, and especially Salmonella, and attaching and effacing (A/E) pathogens, the genetic structure of these systems presents as a large locus encoding a T3SS with a small number of effectors, plus numerous small unlinked loci encoding additional individual effectors. We discuss the generation of novel effectors, and the evolution of G+C content following acquisition. We also consider the currently held view that each locus has been acquired individually, as well as propose an alternative where recombination may have redistributed and broken up clusters of effectors. It is clear that the evolution of this virulence strategy is highly complex and challenging to analyze.
Key words: type III secretion system, type III effector, G+C content, codon usage, horizontal gene transfer, ORFan
Type III secretion systems and the effector proteins that they deliver into host cells are generally considered to have been acquired by horizontal gene transfer. The approximately 20 genes that encode a functional T3SS are linked to one locus that commonly constitutes a chromosomal pathogenicity island or less frequently is located on a virulence associated plasmid.1,2 Pathogenicity islands are defined as loci encoding virulence functions that bare hallmarks of horizontal acquisition, including a G+C content different from the genome mean,2 although it is noteworthy that this is strongly biased towards a lower G+C content. The clearest exception to this is the Chlamydia T3SS, which is encoded in a handful of loci distributed around the genome, and does not differ significantly in G+C content from the genome mean, which itself is quite low.1,3 Genes encoding type III effectors also show the signs of horizontal acquisition, but are only occasionally linked with T3SS loci or even other effectors. Effectors are often associated with phage,4 which perhaps most commonly act as the vehicles transferring effectors between different bacteria. The Salmonella effector SopE has also been found to be encoded by distinct phage families,5 indicating that horizontal transfer events of effector genes involve more than phage lysogeny.
Owing to the apparent horizontal acquisition of these genes, the origin of these sequences has been challenging to study and there is little commentary on the subject. It is well accepted that the T3SS is related to the bacterial flagellum,1,6 but the focus of this article is the origin of the contemporary proteobacterial systems. The simplest notion about the origin of these genes is that they originate from an organism whose genome has a low G+C content, with the best candidate being Chlamydia.3,6 However, while Chlamydia has a low G+C genome, it does not appear to have acquired its T3SS or effectors horizontally, perhaps because its obligate intracellular lifestyle poses a significant barrier to horizontal gene transfer. It is a phylogenetically ancient system, and it is more likely a distant descendant of the source of the original horizontally transferred T3SS, rather than being a recent origin of the proteobacterial systems.6 Primary reasons for this are the obvious extent of evolutionary distance between the Chlamydia systems and those present in proteobacteria,6 and that the Chlamydia system is not confined to one locus. Hence, likely candidate organisms that have acted as a source of proteobacterial effectors and T3SSs remain elusive.
A challenge to the concept that these genes, in particular effector genes, originate from a low G+C organism comes from a hypothesis on how new genes evolve in E. coli. These new genes, termed ORFans, have no homologs in the sequence databases and tend to be linked to phage attachment sites,7 a common theme for type III effectors. ORFans tend towards a shorter length than effector genes, and are rapidly evolving sequences that are considered to occasionally become integrated into cellular functions.7 A means by which these novel functions could become integrated into T3SS dependent virulence mechanisms is by terminal reassortment, a process where functional domains are fused together to provide novel functional combinations.8,9 Numerous current examples exist where two or more effectors may be related to each other over only a discrete domain, such as the WxxxE or WEK(I/M)XXFF motif containing domains (Fig. 1).8,10–12 Based on this information, it is possible for an ORFan to be fused to a domain that is targeted to a T3SS for delivery into host cells, followed by rapid evolution into an effector functioning inside host cells. Of course this evolution could only proceed in bacteria that use T3SSs to interact with a host organism, and it can be viewed as being a means by which novel effector functions are forged.
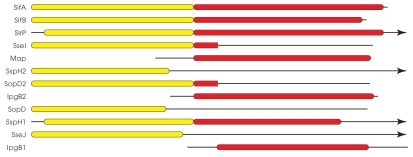
Figure 1.
Scale diagram of WEK(I/M)xxFF (yellow) and WxxxE (red) domain containing effector proteins. Data for the alignment was generated from a Position Specific Iterated-BLAST search using SifA as the query, iterated five times. While SlrP and SspH1 are shown as possessing full WxxxE domains, it is noteworthy that they lack the actual WxxxE motif that is required to mimmick the function of Ras superfamily members. Similarly, SseI and SopD2 show similarity to WxxxE domains over only a short length of sequence as indicated, and this region does not include the WxxxE motif itself. Black lines represent sequence which is not similar to either WEK(I/M)xxFF or WxxxE domains, and arrowheads shown at the right end of a line indicate that the respective protein sequence extends beyond the space of the diagram.
An interesting comparison to make when considering the origin of T3SS and effectors is between the system of Salmonella pathogenicity island 1 (SPI1) and its xenolog from Burkholderia pseudomallei, termed T3SS3.13 What makes this comparison interesting is that clearly these systems are related (good synteny, ∼30–58% protein identity),14,15 but the loci encoding the two systems differ dramatically in G+C content, with SPI1 being 44.97% and the T3SS3 locus being 69.77%. The significance of this difference is reinforced when the genomic G+C content are also considered; with SPI1 differing significantly from the Salmonella genome mean of ∼52%, whereas the T3SS3 locus does not differ significantly from the B. pseudomallei genome mean of ∼69%. The T3SS3 locus does appear to have been acquired horizontally since it lies adjacent to numerous transposase genes. Has T3SS3 rapidly evolved a high G+C content or has SPI1 evolved a lower G+C content? These two scenarios need not be mutually exclusive, yet they have evolved drastically different G+C contents. It is unlikely that this can be explained by a large difference in time frame for the acquisition of each system: SPI1 was acquired as Salmonella split from Escherichia16 and T3SS3 is present in the very closely related Burkholderia species pseudomallei, mallei and thailandensis but absent from the remainder of the genus.13 This leads to the interesting question of how two closely related systems have come to differ in G+C content to such an extent, and how do these differences affect the function of these two systems?
The evolution of differences in G+C content can be driven by numerous factors. In general, the principle factor is the optimisation of translation by evolution towards the use of preferred codons based on tRNA abundance.17–19 It has also been found that horizontal gene transfer is more likely to occur if the codon usage of the transferred gene resembles the tRNA pool of the host organism.20
B. pseudomallei T3SS3 appears to have evolved effectively in this direction, yet SPI1 has not. Hence, it is worth considering that the disparity in G+C content between the SPI1 and B. pseudomallei T3SS3 systems might provide benefits specific to each species. One notable possibility in the case of Salmonella is the repression of low G+C gene expression by H-NS.21,22 Loss of H-NS leads to growth rate reduction in Salmonella, and this phenotype can be partially relieved by knocking out T3SS genes,21 providing a possible selectable advantage for maintaining a low G+C content. In the case of B. pseudomallei, little data is available for gene regulation by the many H-NS homologs it possesses. In any case, clearly any barrier preventing evolution towards the chromosomal G+C content has been overcome in the case of T3SS3. With an effective barrier in place for Salmonella, it relies on pathogenicity island encoded transcription factors to overcome H-NS repression, though this leaves T3SS and effector genes enriched for suboptimal codons. The significance of, and extent to which translation of these genes is slowed as a result, or an alternative possibility that the tRNA pool maybe regulated by environmental signals to overcome suboptimal codon usage, remains to be explored.
Another interesting aspect of effector evolution are the forces that shape the effector repertoire translocated by a given system. In particular, there are some striking examples of what, at least superficially, appear to be convergent evolution by horizontal gene transfer. An outstanding example of this are the attaching and effacing pathogens that use a T3SS and an extensive repertoire of effectors for virulence.23,24 Some A/E pathogens, such as Citrobacter rodentium and E. coli, diverged as species well before the horizontal acquisition of what are now over 20 distinct virulence loci that the two organisms have in common.23,24 Thus far, the explanation put forward for this is that the T3SS and separate effector encoding loci have all been acquired separately.25,26 Whilst there is good evidence indicating the strength for selection of these effectors,26 we also acknowledge the remote odds that such distinct organisms would be exposed to such a similar combination of horizontally transferred DNA by proposing an alternative scenario: The number of separate horizontal gene transfer events where effectors were acquired by these organisms may have been less, perhaps considerably so, than the number of separate loci encoding effectors. Recently the extent of genome rearrangement in C. rodentium has come to light, including events likely involving phage.27 Perhaps such genome rearrangements, together with a more modest number of horizontal acquisition events could result in the genetic structure of virulence factors we observe in these pathogens today.
In conclusion, we understand enough about the evolution of effectors and T3SSs to comprehend the complexity involved, but this is based on relatively little data. Recent evidence that T3SSs mediate not only bacterial interactions with animals and plants, but also fungi,28 further extends the environmental niches where selective pressures may come to bare. It seems most likely that Chlamydia has the only vertically inherited T3SS thus far identified, and therefore is the best estimate of the origin of the T3SS. Effectors, on the other hand seem harder to track. They either have no database homologs, or their only homologs are easily identified xenologous effectors or effector domains from other bacteria who have also acquired the genes horizontally. The balance of whether effectors are continually being synthesized from fusion of ORFans with T3SS targeting domains, or being handed horizontally from strain to strain from a long ago extinct ancestor (or uncultivated bacterium) is certainly a fascinating question.
Acknowledgments
Work in N.F.B.'s lab is supported by the National Health and Medical Research Council of Australia, and work in B.B.F.'s lab is supported by the Canadian Institutes of Health Research and the Howard Hughes Medical Institute.
Abbreviations
- T3SS
type III secretion system
- SPI1
salmonella pathogenicity island 1
- A/E
attaching and effacing
References
- 1.Hueck CJ. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hacker J, Blum-Oehler G, Mühldorfer I, Tschäpe H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 3.Kim JF. Revisiting the chlamydial type III protein secretion system: clues to the origin of type III protein secretion. Trends Genet. 2001;17:65–69. doi: 10.1016/s0168-9525(00)02175-2. [DOI] [PubMed] [Google Scholar]
- 4.Ehrbar K, Hardt WD. Bacteriophage-encoded type III effectors in Salmonella enterica subspecies 1 serovar Typhimurium. Infect Genet Evol. 2005;5:1–9. doi: 10.1016/j.meegid.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Mirold S, Rabsch W, Tschäpe H, Hardt WD. Transfer of the Salmonella type III effector sopE between unrelated phage families. J Mol Biol. 2001;312:7–16. doi: 10.1006/jmbi.2001.4950. [DOI] [PubMed] [Google Scholar]
- 6.Pallen MJ, Beatson SA, Bailey CM. Bioinformatics, genomics and evolution of non-flagellar type-III secretion systems: a Darwinian perspective. FEMS Microbiol Rev. 2005;29:201–229. doi: 10.1016/j.femsre.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Daubin V, Ochman H. Bacterial genomes as new gene homes: the genealogy of ORFans in E. coli. Genome Res. 2004;14:1036–1042. doi: 10.1101/gr.2231904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stavrinides J, Ma W, Guttman DS. Terminal reassortment drives the quantum evolution of type III effectors in bacterial pathogens. PLoS Pathog. 2006;2:104. doi: 10.1371/journal.ppat.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCann HC, Guttman DS. Evolution of the type III secretion system and its effectors in plant-microbe interactions. New Phytol. 2008;177:33–47. doi: 10.1111/j.1469-8137.2007.02293.x. [DOI] [PubMed] [Google Scholar]
- 10.Ham JH, Majerczak DR, Nomura K, Mecey C, Uribe F, He SY, et al. Multiple activities of the plant pathogen type III effector proteins WtsE and AvrE require WxxxE motifs. Mol Plant Microbe Interact. 2009;22:703–712. doi: 10.1094/MPMI-22-6-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miao EA, Miller SI. A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc Natl Acad Sci USA. 2000;97:7539–7544. doi: 10.1073/pnas.97.13.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alto NM, Shao F, Lazar CS, Brost RL, Chua G, Mattoo S, et al. Identification of a bacterial type III effector family with G protein mimicry functions. Cell. 2006;124:133–145. doi: 10.1016/j.cell.2005.10.031. [DOI] [PubMed] [Google Scholar]
- 13.Sun GW, Gan YH. Unraveling type III secretion systems in the highly versatile Burkholderia pseudomallei. Trends Microbiol. 2010;18:561–568. doi: 10.1016/j.tim.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 14.Stevens MP, Wood MW, Taylor LA, Monaghan P, Hawes P, Jones PW, et al. An Inv/Mxi-Spa-like type III protein secretion system in Burkholderia pseudomallei modulates intracellular behaviour of the pathogen. Mol Microbiol. 2002;46:649–659. doi: 10.1046/j.1365-2958.2002.03190.x. [DOI] [PubMed] [Google Scholar]
- 15.Attree O, Attree I. A second type III secretion system in Burkholderia pseudomallei: who is the real culprit? Microbiology. 2001;147:3197. doi: 10.1099/00221287-147-12-3197. [DOI] [PubMed] [Google Scholar]
- 16.Mirold S, Ehrbar K, Weissmüller A, Prager R, Tschäpe H, Rüssmann H, et al. Salmonella host cell invasion emerged by acquisition of a mosaic of separate genetic elements, including Salmonella pathogenicity island 1 (SPI1), SPI5 and sopE2. J Bacteriol. 2001;183:2348–2358. doi: 10.1128/JB.183.7.2348-2358.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akashi H. Gene expression and molecular evolution. Curr Opin Genet Dev. 2001;11:660–666. doi: 10.1016/s0959-437x(00)00250-1. [DOI] [PubMed] [Google Scholar]
- 18.Kreitman M, Comeron JM. Coding sequence evolution. Curr Opin Genet Dev. 1999;9:637–641. doi: 10.1016/s0959-437x(99)00034-9. [DOI] [PubMed] [Google Scholar]
- 19.Hershberg R, Petrov DA. Selection on codon bias. Annu Rev Genet. 2008;42:287–299. doi: 10.1146/annurev.genet.42.110807.091442. [DOI] [PubMed] [Google Scholar]
- 20.Tuller T, Girshovich Y, Sella Y, Kreimer A, Freilich S, Kupiec M, et al. Association between translation efficiency and horizontal gene transfer within microbial communities. Nucleic Acids Res. 2011;39:4743–4755. doi: 10.1093/nar/gkr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lucchini S, Rowley G, Goldberg MD, Hurd D, Harrison M, Hinton JC. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2006;2:81. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Navarre WW, Porwollik S, Wang Y, McClelland M, Rosen H, Libby SJ, Fang FC. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–238. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 23.Deng W, Puente JL, Gruenheid S, Li Y, Vallance BA, Vázquez A, et al. Dissecting virulence: systematic and functional analyzes of a pathogenicity island. Proc Natl Acad Sci USA. 2004;101:3597–3602. doi: 10.1073/pnas.0400326101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tobe T, Beatson SA, Taniguchi H, Abe H, Bailey CM, Fivian A, et al. An extensive repertoire of type III secretion effectors in Escherichia coli O157 and the role of lambdoid phages in their dissemination. Proc Natl Acad Sci USA. 2006;103:14941–14946. doi: 10.1073/pnas.0604891103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reid SD, Herbelin CJ, Bumbaugh AC, Selander RK, Whittam TS. Parallel evolution of virulence in pathogenic Escherichia coli. Nature. 2000;406:64–67. doi: 10.1038/35017546. [DOI] [PubMed] [Google Scholar]
- 26.Wickham ME, Brown NF, Boyle EC, Coombes BK, Finlay BB. Virulence is positively selected by transmission success between mammalian hosts. Curr Biol. 2007;17:783–788. doi: 10.1016/j.cub.2007.03.067. [DOI] [PubMed] [Google Scholar]
- 27.Petty NK, Feltwell T, Pickard D, Clare S, Toribio AL, Fookes M, et al. Citrobacter rodentium is an Unstable Pathogen Showing Evidence of Significant Genomic Flux. PLoS Pathog. 2011;7:1002018. doi: 10.1371/journal.ppat.1002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim Y, Mylonakis E. Killing of Candida albicans Filaments by Salmonella enterica serovar Typhimurium Is Mediated by the sopB Effectors, Parts of a Type III Secretion System. Eukaryot Cell. 2011;10:782–790. doi: 10.1128/EC.00014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]