Abstract
Intercellular signal transduction pathways regulate the NK-2 family of transcription factors in a conserved gene regulatory network that directs cardiogenesis in both flies and mammals. The Drosophila NK-2 protein Tinman (Tin) was recently shown to regulate Stat92E, the Janus kinase (JAK) and Signal transducer and activator of transcription (Stat) pathway effector, in the developing mesoderm. To understand whether the JAK/Stat pathway also regulates cardiogenesis, we performed a systematic characterization of JAK/Stat signaling during mesoderm development. Drosophila embryos with mutations in the JAK/Stat ligand upd or in Stat92E have non-functional hearts with luminal defects and inappropriate cell aggregations. Using strong Stat92E loss-of-function alleles, we show that the JAK/Stat pathway regulates tin expression prior to heart precursor cell diversification. tin expression can be subdivided into four phases and, in Stat92E mutant embryos, the broad phase 2 expression pattern in the dorsal mesoderm does not restrict to the constrained phase 3 pattern. These embryos also have an expanded pericardial cell domain. We show the E(spl)-C gene HLHm5 is expressed in a pattern complementary to tin during phase 3 and that this expression is JAK/Stat dependent. In addition, E(spl)-C mutant embryos phenocopy the cardiac defects of Stat92E embryos. Mechanistically, JAK/Stat signals activate E(spl)-C genes to restrict Tin expression and the subsequent expression of the T-box transcription factor H15 to direct heart precursor diversification. This study is the first to characterize a role for the JAK/Stat pathway during cardiogenesis and identifies an autoregulatory circuit in which tin limits its own expression domain.
Keywords: E(spl)-C, JAK/Stat, Cardiogenesis, Drosophila
INTRODUCTION
Intercellular signal transduction pathways provide positional information and guidance cues to effect the dynamic cellular changes driving organogenesis. In Drosophila, the Decapentaplegic (Dpp), Wingless (Wg) and Fibroblast growth factor (FGF) signaling pathways activate a highly conserved, cardiac-specific gene regulatory network by inducing the expression of the transcription factor tin in the precardiac, dorsal mesoderm (Olson, 2006). Tin activates the expression of transcription factors in the GATA (Klinedinst and Bodmer, 2003), T-box (Qian et al., 2005a; Reim et al., 2005) and Hand (Han and Olson, 2005) protein families that, in turn, direct cell fate specification and induce muscle structural genes. Although the key components of this cardiac-specific gene regulatory network have been characterized, the precise mechanisms by which Tin regulates cardiac cell diversification are only partially understood.
A genome-wide analysis of Tin binding sites identified novel Tin target genes that probably specify diverse cell fates in the mesoderm (Liu et al., 2009). Of the 400 binding sites reported, two loci, eyes absent and Stat92E, were shown to be direct Tin target genes. Stat92E is the transcriptional effector of the JAK/Stat pathway and, although the JAK/Stat pathway is one of a few conserved signal transduction pathways in Metazoa, a role for JAK/Stat signaling during cardiac morphogenesis has not been described. The identification of Stat92E as a Tin-regulated transcription factor suggests that the JAK/Stat pathway could be a component of the cardiac-specific gene regulatory network.
In Drosophila, the canonical intracellular JAK/Stat response is initiated when an Unpaired (Upd; Os – Flybase) ligand binds the transmembrane receptor Domeless (Dome) (Hou et al., 2002). The Drosophila genome encodes three upd paralogs. Upd and Upd2 have overlapping segmental expression patterns during mesoderm development and, because both ligands signal through Dome in the wing, these proteins are thought to have redundant functions (Hombria et al., 2005). Upon ligand binding, Dome activates a receptor-associated JAK (Hopscotch) that, in turn, phosphorylates preassociated Stat (Stat92E) dimers (Novak et al., 1998; Stancato et al., 1996). Phosphorylated Stat92E homodimers translocate to the nucleus to regulate the expression of specific target genes.
The expression patterns of Upd and Upd2 have probably precluded previous studies of the JAK/Stat pathway during Drosophila heart development. Upd and Upd2 are expressed from the ventral ectoderm, which is relatively distant from the tin expression domain in the dorsal mesoderm, during a brief developmental window (Hombria et al., 2005; Karsten et al., 2002). However, the pulse of Upd and Upd2 expression in the ventral ectoderm is coincident with a dynamic spatial change in tin expression.
tin expression can be divided into four distinct spatial-temporal phases. Phase 1 tin expression initiates after gastrulation during which Twist (Twi) activates pan-mesodermal tin expression via the enhancer tinB (Yin et al., 1997). Phase 2 begins after FGF-mediated mesoderm spreading in which Dpp signals produced by the dorsal ectoderm maintain tin expression throughout the dorsal mesoderm via a second enhancer, tinD (Xu et al., 1998). It is during phase 2 that Tin specifies the major dorsal mesoderm derivatives. Phase 3 initiates after dorsal mesoderm progenitor specification and is characterized by a pronounced restriction of tin to the cardiac and visceral muscle progenitors. Upd and Upd2 are expressed in the ventral ectoderm during the transition from phase 2 to phase 3 expression. Phase 4 initiates after precursor specification and is characterized by further restriction of tin to the cardiac precursors that give rise to the contractile cardiomyocytes and the noncontractile pericardial nephrocytes. Phase 4 expression further directs heart cell diversification and maturation and is dependent on a third enhancer element, tinC (Venkatesh et al., 2000; Zaffran et al., 2006).
To understand whether the JAK/Stat pathway influences cardiac morphology, we performed a systematic characterization of JAK/Stat signaling during mesoderm development. upd and Stat92E embryos have non-functional hearts characterized by a dysfunctional lumen, cell-cell adhesion defects and inappropriate cell aggregations. We show that the JAK/Stat pathway is active in the dorsal mesoderm during the transition from phase 2 to phase 3 tin expression, and that JAK/Stat signals are necessary to restrict tin expression. Stat92E embryos also show an expanded pericardial cell domain owing to inappropriate heart precursor diversification during phase 3. The E(spl)-C gene HLHm5 is expressed in a pattern complementary to tin in the dorsal mesoderm and is JAK/Stat dependent. In addition, E(spl)-C mutant embryos phenocopy the cardiac defects we observe in Stat92E embryos. This study is the first to show that the JAK/Stat pathway regulates heart development and that tin restricts its own expression in the dorsal mesoderm via a novel regulatory circuit.
MATERIALS AND METHODS
Drosophila genetics
All stocks were obtained from the Bloomington Stock Center unless otherwise noted. The stocks used in this study were: upd4, FRT.82B Stat92E85C9, FRT.82B Stat92E397 (Silver and Montell, 2001), UAS.Stat92E-HA (Ekas et al., 2006), hs.Flp, FRT.82B ovoD1, Twi.Gal4, 24B.Gal4, En.Gal4, HLHm5.lacZ, m4.lacZ, E(spl).lacZ, Df(3R)Espl22, Df(3R)EsplΔmd-m6 (Bardin et al., 2010), UAS.Socs36E (Callus and Mathey-Prevot, 2002), Hand.Gal4 (Han and Olson, 2005) and UAS.Twi (Castanon et al., 2001). FM7i, Act.GFP; TM3, Twi.Gal4 2X-UAS.eGFP; and TM6B, AbdA.lacZ were used to identify homozygous embryos.
The crosses to generate GLC experimental embryos include:
Stat92EM–Z–:
Hand.eGFP; FRT.82B Stat92E85C9/397 /TM3, twi.eGFP × hs.Flp/+; FRT.82B Stat92E85C9/397 /FRT.82B ovoD1
Stat92EM–rescue:
twi.Gal4 × hs.Flp/+; UAS-Stat92E-HA/+; FRT.82B Stat92E85C9/FRT.82B ovoD1
twi.Gal4; 24B.Gal4 × hs.Flp/+; UAS-Stat92E-HA/+; FRT.82B Stat92E85C9 /FRT.82B ovoD1
en.Gal4 × hs.Flp/+; UAS-Stat92E-HA/+; FRT.82B Stat92E85C9/FRT.82B ovoD1
en.Gal4; 24B.Gal4 × hs.Flp/+; UAS-Stat92E-HA/+; FRT.82B Stat92E85C9 /FRT.82B ovoD1
HLHm5.lacZ:
HLHm5.lacZ × hs.Flp/+; FRT.82B Stat92E85C9 /FRT.82B ovoD1.
In situ hybridization, immunohistochemistry and imaging
Antibody staining and in situ hybridization were performed as described (Johnson et al., 2007). Antibodies used in this study were rabbit anti-Mef2 (Lilly et al., 1995), rabbit anti-Tin (Venkatesh et al., 2000), rabbit anti-Odd (Ward and Skeath, 2000), rabbit anti-GFP (Torrey Pines Laboratories), mouse anti-HA (Santa Cruz), mouse anti-βgal (Promega) and mouse anti-Fasciclin 3 (FasIII, Developmental Studies Hybridoma Bank). HRP-, Alexa488- or Alexa633-conjugated secondary antibodies were used to detect primary antibodies (Molecular Probes). The TSA system (Molecular Probes) was used to detect HRP-conjugated secondary antibodies. Mef2 was directly conjugated with Zenon633 (Molecular Probes) for co-labeling with rabbit primary antibodies. BDGP clone RE69770 (HLHm5) and IP01538 (H15) were used to generate in situ probes. The m4 transcript was cloned from 0- to 24-hour-old embryo cDNA made with the Superscript III first-strand synthesis system (Invitrogen). Images were generated with LSM 510 (Zeiss) and DMRXE (Leica) microscopes. Wild-type and mutant embryos were prepared and imaged in parallel where possible.
Chromatin immunoprecipitation
Four- to six-hour-old embryos expressing Stat92E-HA under the control of Twi.gal4, 24B.gal4 were collected, dechorionated, cross-linked, devitellinated and homogenized as described (Sandmann et al., 2006). The remainder of the immunoprecipitation was performed with the EZ-ChIP system per manufacturer’s specification (Millipore). Chromatin was precipitated with or without 5 μg of anti-HA for the negative control, PCR amplified, and quantified by standard methods using ImageJ 1.40g software.
Quantitative real-time PCR
Four- to six-hour-old embryos were collected and homogenized in TRizol (Invitrogen) and RNA was extracted per manufacturer’s specification. cDNA was generated using Superscript III and qPCR was performed with SYBR Green (Promega) using an ABI Prism 7000. Primers were designed to span introns for all intron-containing transcripts. qPCR reactions were run in triplicate and normalized to RpL32 expression.
RESULTS
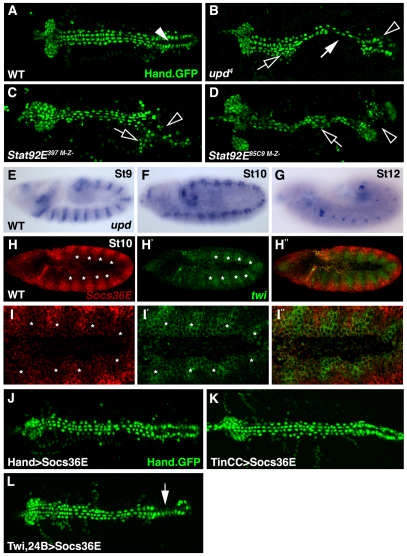
As Tin regulates Stat92E during mesoderm development (Liu et al., 2009), we speculated that the JAK/Stat pathway could be a component of the cardiac-specific gene regulatory network. To test this hypothesis, we characterized cardiac morphology in embryos mutant for the JAK/Stat ligand upd. The reporter gene Hand.GFP expresses a nuclear-localized GFP in the dorsal vessel (Fig. 1A) and serves as a useful tool to identify novel regulators of cardiac development (Yi et al., 2008). Using Hand.GFP as a marker, we found that a subset of upd embryos showed incomplete heart tube formation due to the loss of cell-cell adhesion among cardioblasts (38.1%, n=21; Fig. 1B). The incomplete phenotypic penetrance in upd embryos probably reflects functional redundancy with upd2. In addition, all embryos maternal and zygotic mutant for two distinct Stat92E loss-of-function alleles (hereafter Stat92EM–Z–) showed cardiac defects similar to those of upd embryos (100% n≥24; Fig. 1C,D). Cardiogenesis is, therefore, dependent on JAK/Stat signaling.
Fig. 1.
The Jak/Stat pathway regulates cardiac morphology and is active in the dorsal mesoderm during cardiac precursor specification. (A) WT Hand.GFP embryos express nuclear-localized GFP throughout the dorsal vessel. The contractile ‘heart’ (arrowhead) positioned at the posterior of the dorsal vessel pumps hemolymph through the anterior aorta. (B) Zygotic upd4 embryos lose cardioblast cell-cell adhesion (white arrow) causing inappropriate cell aggregations (open arrow) and incomplete heart lumen formation (open arrowhead). (C,D) Embryos maternal and zygotic mutant for Stat92E397 (C) or Stat92E85C9 (D) phenocopy the dorsal vessel defects of upd embryos. (E-G) upd mRNA expression in WT embryos. upd expression in the ventral ectoderm is segmented during St9 (E). upd expression is restricted to the tracheal system and hindgut in St10 (F) and St12 (G) embryos. (H-I′) St10 WT embryo double labeled for Socs36E (red) and twi (green) mRNA. Socs36E and twi expression is complementary in the dorsal mesoderm. Regions of the mesoderm segment expressing high levels of twi (asterisks) express low levels of Socs36E. (J,K) St17 embryos expressing Hand.GFP. Expressing Socs36E in the heart after cardioblast precursor specification using Hand.gal4 (J) or TinCC.gal4 (K) does not affect heart morphology. (L) Expressing Socs36E in the heart prior to cardioblast precursor specification using Twi.gal4, 24B.gal4 induces mild cardioblast cell-cell adhesion defects. A-G show dorsal views of Hand.GFP expression in live St17 embryos. Embryos are oriented with anterior to the left.
The JAK/Stat pathway does not regulate cardioblast cell polarity, cardioblast cell survival or dorsal closure
The cardiac defects in Stat92EM–Z– embryos suggested that the JAK/Stat pathway might regulate cardiac morphology late in embryogenesis. Cardioblasts originate as unpolarized mesenchymal cells that generate a unique cell polarity after epithelial transition to complete cardiac tube morphogenesis. Embryos that fail to generate proper cardioblast cell polarity, such as slit and robo mutant embryos, show cardiac defects similar to those found in Stat92EM–Z– embryos (Qian et al., 2005b). We used FasIII to assay cell polarity and found that cardioblast cell polarity is normal in Stat92EM–Z– embryos (see Fig. S1 in the supplementary material). We then investigated whether the cardiac defects in Stat92EM–Z– embryos are secondary to dorsal closure defects but found Stat92EM–Z– embryos complete dorsal closure normally (see Fig. S1 in the supplementary material). A third potential explanation for the cardiac defects in Stat92EM–Z– embryos is that the JAK/Stat pathway promotes cardioblast cell survival. However, using TUNEL staining, we saw no apoptotic cardioblasts in wild-type (WT) or Stat92EM–Z– embryos (see Fig. S1 in the supplementary material). Because JAK/Stat signaling does not coordinate cardioblast cell polarity, dorsal closure or cardioblast cell survival, we speculated that JAK/Stat signaling might regulate cardiac development during the early stages of embryogenesis.
The JAK/Stat pathway is active in the dorsal mesoderm
To identify the spatial and temporal requirement of the JAK/Stat pathway during mesoderm development, we assayed upd expression over the course of embryogenesis. Although the upd expression pattern has been described (Karsten et al., 2002), we expected close examination of upd in the context of heart development to reveal expression in the dorsal ectoderm or the dorsal mesoderm. Unexpectedly, we could not detect upd in these tissues during any stage of development. However, brief segmental upd expression was detectable in the ventral ectoderm during St9 (Fig. 1E). In St10-12 embryos, upd was expressed in the developing tracheal system and the hindgut but was no longer detectable in the ventral ectoderm (Fig. 1F,G). These results, in combination with the cardiac defects we observed in upd embryos, suggest that Upd expressed in the ventral ectoderm might signal to cells in the precardiac dorsal mesoderm to regulate heart development.
A standard method to detect JAK/Stat pathway activity is to assay the expression of the Stat92E target gene Socs36E (Baeg et al., 2005). A previous study reported segmental Socs36E expression in the visceral but not in the dorsal mesoderm of St10 embryos (Liu et al., 2009) in a pattern similar to twi. To define accurately the relative dorsal-ventral and anterior-posterior position of the Socs36E expression domain, we performed double fluorescent in situ hybridization with Socs36E and twi. We found that Socs36E is indeed expressed in the dorsal mesoderm of St10 embryos and that Socs36E and twi have complementary expression patterns (Fig. 1H,I). Strikingly, each mesoderm segment comprised one region that expresses high levels of twi and an opposing region that expressed high levels of Socs36E (Fig. 1H,I). We conclude that Upd, and perhaps Upd2, expressed from the ventral ectoderm, translocates to the dorsal mesoderm and activates a segmented JAK/Stat intracellular response.
To support the hypothesis that JAK/Stat signaling during St10 establishes proper cardiac morphology, we expressed Socs36E specifically in the dorsal vessel after St12. Socs36E is not only a Stat92E target gene but is also a negative regulator of JAK/Stat signaling (Callus and Mathey-Prevot, 2002). Expressing Socs36E throughout the heart with Hand.gal4 or in a subset of cardioblasts using TinCC.gal4 did not induce cardiac defects (Fig. 1J,K). Pan-mesodermal expression of Socs36E prior to St10 using Twi.gal4, 24B.gal4 did, however, induce a mild cardioblast cell-cell adhesion phenotype (Fig. 1L). Socs36E expression does not completely block JAK/Stat signaling and the strength of the Twi.gal4, 24B.gal4>Socs36E phenotype is consistent with that reported for other developmental contexts (Bach et al., 2003; Callus and Mathey-Prevot, 2002). Thus, JAK/Stat signaling to the dorsal mesoderm prior to St12 directs proper mesoderm development.
The JAK/Stat pathway regulates phase 3 tin expression
As the JAK/Stat pathway is active in the dorsal mesoderm during St10, we thought that JAK/Stat signaling might modulate the expression of genes that: (1) regulate cardiac precursor specification and (2) show dynamic expression during early mesoderm development. One obvious candidate was tin because the transition from broad phase 2 expression to restricted phase 3 expression is coincident with Socs36E expression in the dorsal mesoderm. By St11, the phase 3 tin expression pattern was evident as Tin-positive (Tin+) cells were juxtaposed with Tin-negative (Tin–) cells in the dorsal mesoderm (Fig. 2A). However, St11 Stat92EM–Z– embryos showed broad, unrestricted Tin expression throughout the dorsal mesoderm (Fig. 2B).
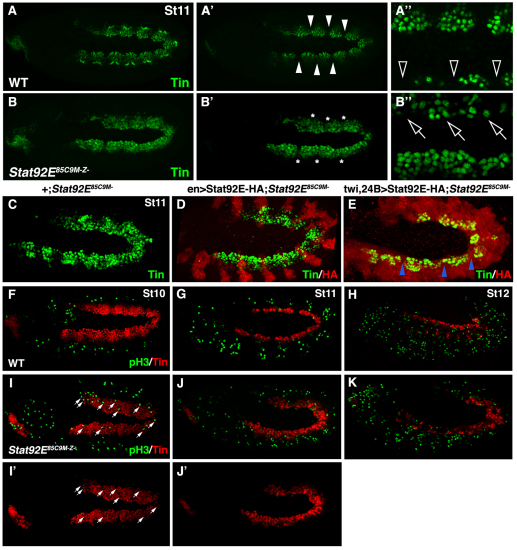
Fig. 2.
JAK/Stat signaling restricts Tin expression in the dorsal mesoderm. (A-B′) Oblique views of St11 embryos labeled for Tin. The confocal micrographs in A and B include the entire Tin expression domain whereas the micrographs in A′, A′, B′ and B′ include only the dorsal mesoderm. (A,A′) Tin expression is restricted to the cardiac and visceral muscle progenitors in WT embryos. Note that Tin is not expressed in a subset of dorsal mesoderm cells (arrowheads). (B,B′) In Stat92EM–Z– St11 embryos, Tin expression does not restrict (asterisks). (A′,B′) High magnification micrographs show that the embryos are slightly rotated (oblique) giving a differential projection of the dorsal mesoderm. (A′) This scan captures more medially positioned Tin+ cells in the top of the image compared with the bottom. Note the large gaps between Tin+ cells in the dorsal-most mesoderm of WT embryos (open arrowheads). (B′) This scan captures more medially positioned Tin+ cells in the bottom of the image compared with the top. Tin-expressing cells can be identified throughout the dorsal-most mesoderm of Stat92EM–Z– embryos (open arrows). (C-E) St11 embryos labeled for HA and Tin. St11 Stat92EM– embryos (C) and Stat92EM– embryos that express Stat92E-HA in the ectoderm (D) show inappropriate Tin expression. Stat92EM– embryos that express Stat92E-HA in the mesoderm show restricted Tin expression (E; blue arrowheads). (F-K) Embryos labeled for pH3 and Tin. pH3 is not detectable in Tin+ cells of WT embryos during St10 (F), St11 (G) or St12 (H) or in Stat92EMZ– embryos at the same stages (I-K). pH3+ cells appear to be present in the Tin expression domain of Stat92EMZ– embryos; however, a majority of these nuclei do not co-express Tin and are likely to be in the ectoderm (arrows). Embryos are oriented with anterior to the left.
To confirm that the altered Tin expression pattern in Stat92EM–Z– embryos is mesoderm cell autonomous, we performed a rescue experiment by crossing UAS.Stat92E-HA/+; FRT82B Stat92E85C9/ FRT82B ovoD1 females to males homozygous for an ectoderm driver (En.Gal4) or a strong mesoderm driver (Twi.Gal4, 24B.Gal4). In this crossing scheme, all embryos lack the maternal contribution of Stat92E (Stat92EM–), but only half of the embryos express the Stat92E-HA transgene. Stat92EM– embryos that do not express Stat92E-HA or that express Stat92E-HA in the ectoderm failed to restrict Tin expression in the dorsal mesoderm (Fig. 2C,D). By contrast, Stat92EM– embryos that express Stat92E-HA in the mesoderm showed a restricted Tin expression domain that was comparable to WT embryos (Fig. 2E). Thus, cells of the dorsal mesoderm transduce JAK/Stat signals to appropriately refine tin expression.
Because the JAK/Stat pathway also regulates cell division (Hou et al., 2002), we assayed phosphorylated Histone3 (pH3; His – FlyBase) as a marker of cell proliferation. Neither WT nor Stat92EM–Z– embryos accumulated pH3 in Tin+ cells prior to or after the restriction of tin in the dorsal mesoderm (Fig. 2F-K). JAK/Stat signaling, therefore, regulates gene expression, and not cell proliferation, to restrict tin in the dorsal mesoderm.
Jak/Stat signals regulate pericardial cell number
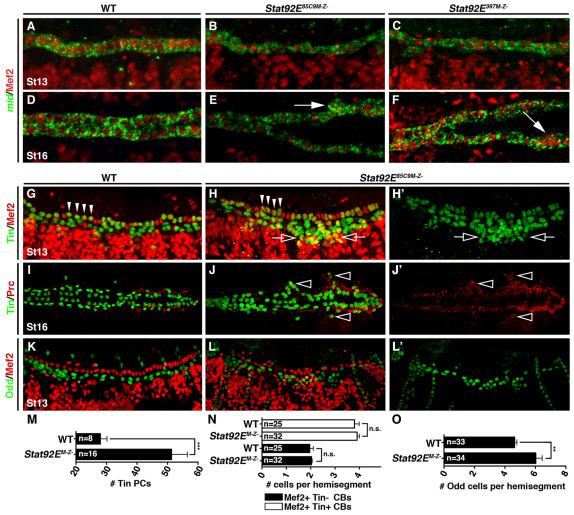
We next wanted to understand whether the JAK/Stat pathway regulates the specification or the diversification of heart precursor cells. The Tbx20 ortholog midline (mid) is regulated by Tin in differentiating cardioblasts and is a useful marker of precursor diversification (Fig. 3A). Stat92EM–Z– embryos showed normal mid expression in the dorsal mesoderm throughout embryogenesis, suggesting that cardioblasts differentiate appropriately in the absence of JAK/Stat signals (Fig. 3B-F). A second indicator of cell fate specification and diversification in the dorsal vessel is the expression of Tin itself. After germ band retraction, embryonic segments A2-A7 contain six cardioblasts, four of which express Tin (Fig. 3G). At this stage, Tin is also expressed in a subset of pericardial cells positioned ventral to the cardioblasts. St13 Stat92EM–Z– embryos expressed Tin in four cardioblasts per hemisegment, but showed a significant increase in the number of Tin+, Mef2–pericardial cells (Fig. 3H,M,N). To confirm that the ectopic Tin+ cells in Stat92EM–Z– embryos are indeed pericardial cells, we looked at the expression of Pericardin (Prc), an extracellular matrix protein expressed in all pericardial cells of St16 embryos. The Prc expression domain was expanded in Stat92EM–Z– embryos and ectopic Tin+ cells co-expressed Prc (Fig. 3I,J). A third pericardial cell marker, Odd-skipped (Odd), was expressed in Tin–pericardial cells and we found that the number of Odd+ pericardial cells was also significantly increased in Stat92EM–Z– embryos at St13 (Fig. 3K,L,O). Interestingly, Stat92EM–Z– embryos also showed a reduced number of somatic muscle founder cells and a smaller visceral muscle domain (see Fig. S2 in the supplementary material). Therefore, proper diversification of heart progenitors requires Stat92E-mediated regulation of tin expression.
Fig. 3.
JAK/Stat signaling regulates pericardial cell number. (A-F) Lateral views of St13 (A-C) and dorsal views of St16 (D-F) embryos labeled for Mef2 protein and mid mRNA. WT and Stat92EM–Z– embryos express mid in all Mef2+ cardioblasts by St13. mid expression persists to St16 even in the aggregated cardioblasts of Stat92EM–Z– embryos (arrows). (G-H′) Lateral views of St13 embryos labeled for Tin and Mef2. (G) WT St13 embryos express Tin in four cardiomyocytes per heart hemisegment (arrowheads) and in a subset of pericardial cells. Mef2 is not expressed in pericardial cells. (H) Stat92EM–Z– embryos express Tin in four cardiomyocytes in a majority of hemisegments but the number of Tin expressing pericardial cells is significantly increased (open arrows). (I-J′) Dorsal views of St16 embryos labeled for Tin and Prc. (I) Prc localizes to the extracellular matrix between cardiomyocytes and pericardial cells in WT embryos. (J) Ectopic Tin cells positioned lateral to the cardioblasts (open arrowheads) co-express Prc in Stat92EM–Z– embryos. (K-L′) Lateral views of St13 embryos labeled for Odd and Mef2. (K) WT embryos express Odd in four pericardial cells per hemisegment. (L) The number of Odd+ pericardial cells is significantly increased in Stat92EM–Z– embryos. (M) Quantification of Tin+ pericardial cells (PCs) in one half of the mesoderm at St13. Tin+ pericardial cells were identified by lack of Mef2 expression. (N,O) Segmental quantification of Tin+ and Tin–cardioblasts (CBs) at St13 (N) and Odd+ pericardial cells at St13 (O). **P<0.005, ***P<0.001, unpaired t-test. n.s., non-significant. Error bars represent s.e.m. Embryos are oriented with anterior to the left.
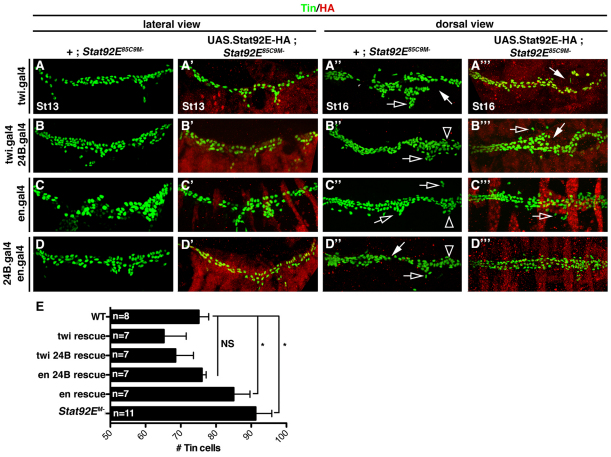
We then extended our rescue experiments to assay Tin+ pericardial cell specification. In Stat92EM– embryos, the number of Tin+ cells was significantly greater than that in WT embryos at St13 (Fig. 4A-E) however, Stat92EM– embryos that express Stat92E-HA in the mesoderm did not show a significant difference in the number of Tin+ cells compared with WT embryos (Fig. 4A′,B′,D′). By contrast, Stat92EM– embryos that express Stat92E-HA solely in the ectoderm showed a significant increase in the number of Tin+ cells (Fig. 4C′). Therefore, expression of Stat92E in the mesoderm is sufficient to rescue the pericardial cell phenotype in Stat92EM– embryos. Surprisingly, expressing Stat92E-HA in either the mesoderm or the ectoderm of Stat92EM– embryos did not fully rescue cardioblast cell-cell adhesion defects (Fig. 4A′′-C′′). Stat92EM– embryos that express Stat92E-HA in both the mesoderm and the ectoderm showed reduced cardiac cell adhesion defects (Fig. 4D′′). The heart tube formation defects in Stat92EM– embryos therefore reflect the pleiotropic functions of the JAK/Stat pathway outside the mesoderm. For example, JAK/Stat signaling regulates segmentation (Harrison et al., 1998), ectoderm morphogenesis (Bertet et al., 2009) and the development of ectoderm derivatives, including the tracheal system (Sotillos et al., 2010). Incomplete ectoderm development probably exacerbates the mesoderm patterning defects in Stat92EM– embryos, causing incomplete cardiac morphogenesis.
Fig. 4.
JAK/Stat signaling in the mesoderm restricts pericardial cell number. Embryos labeled for HA and Tin. The crossing scheme is detailed in the text. (A-D) St13 embryos lacking the maternal contribution of Stat92E (Stat92EM–) have an increased number of Tin+ cells compared with WT embryos. (A′,B′,D′) Stat92EM embryos that express Stat92E-HA in the mesoderm show WT levels of Tin+ cells. (C′) Stat92EM– embryos that express Stat92E-HA in the ectoderm show a significant increase in the number of Tin+ cells compared with WT embryos. (A′-D′) St16 Stat92EM– embryos show a loss of cardioblast cell-cell adhesion (white arrows), collapsed ‘heart’ (open arrowheads) and inappropriate cell aggregations (open arrows). (A′′-C′′) Stat92EM– embryos that express Stat92E-HA in either the mesoderm or the ectoderm also show these three cardiac phenotypes, although they are less severe. (D′′) Stat92EM– embryos that express Stat92E-HA in the ectoderm and the mesoderm show mostly normal heart morphology. (E) Quantification of Tin+ cells in St13 embryos from the genotypes shown in A-D. *P<0.05, unpaired t-test. NS, non-significant. Error bars represent s.e.m. Embryos are oriented with anterior to the left.
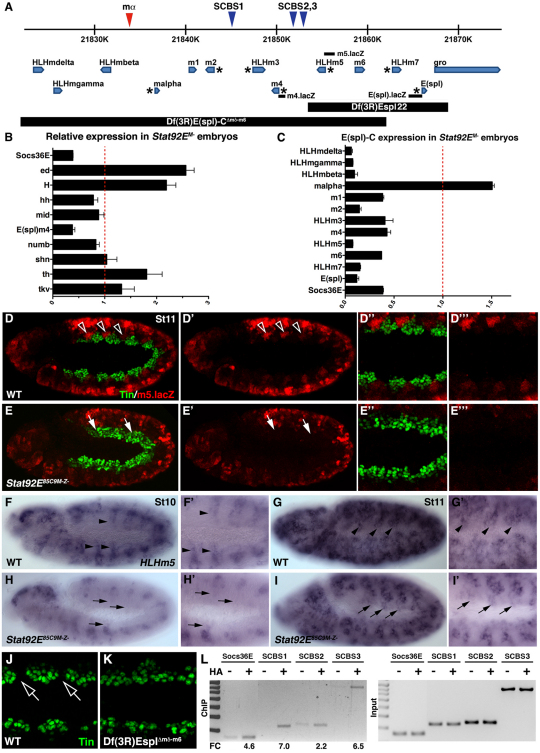
E(spl)-C gene expression is dependent on JAK/Stat signaling
The modENCODE project performed a Stat92E chromatin immunopurification (ChIP)-chip analysis on 0- to 12-hour-old embryos (Celniker et al., 2009) and identified three Stat92E binding regions proximal to the Socs36E locus, including the known Stat92E Response Element (Baeg et al., 2005). The ChIP-chip analysis identified additional Stat92E target genes including ventral veins lacking and trachealess (Sotillos et al., 2010) (see Table S1 in the supplementary material), suggesting that the ChIP-chip data would be a useful tool to identify Stat92E target genes in the mesoderm. Although a total of 107 loci fall within 10 kb of a reported Stat92E binding region (see Table S1 in the supplementary material), the tin transcriptional start site is over 262 kb from the closest Stat92E binding region (3R:17466843). Thus, Stat92E probably regulates tin expression indirectly.
Nine of the 107 loci identified by the ChIP-chip analysis are known regulators of mesoderm development, including components or targets of the Dpp, Hedgehog and Notch signaling pathways (tkv, shn, hh, H, ed, th, numb, mid and m4). We collected RNA from WT and Stat92EM– 4- to 6-hour-old embryos, performed quantitative RT-PCR (qPCR), and found that the E(spl)-C gene m4 was the most downregulated of the nine candidate genes (Fig. 5B). The E(spl)-C genes encode members of the HLH and Bearded family of transcription factors and are well characterized Notch targets that regulate twi expression (Delidakis and Artavanis-Tsakonas, 1992; Lai et al., 2000; Tapanes-Castillo and Baylies, 2004). We checked the expression of the remaining E(spl)-C genes and found that, with the exception of mα, all of the E(spl)-C genes were significantly downregulated in Stat92EM– embryos (Fig. 5C). Thus, E(spl)-C gene expression requires JAK/Stat signaling during the early stages of embryonic development.
Fig. 5.
Stat92E regulates HLHm5 expression in the dorsal mesoderm. (A) The E(spl) gene complex. Blue triangles mark the position of conserved Stat consensus binding sites (SCBSs); the red triangle shows a non-conserved SCBS. E(spl)-C genes with previously reported mesoderm expression are marked with an asterisk. The position of the sequences comprising the m4.lacZ, HLHm5.lacZ and E(spl).lacZ reporters are shown. Sequences deleted by E(spl)-C deficiencies used in this study are indicated. (B,C) qPCR of extracts from 4- to 6-hour-old embryos showing relative mRNA expression in Stat92EM– embryos compared with WT. Socs36E and m4 are downregulated in Stat92EM– embryos (B). With the exception of mα, all E(spl)-C genes are downregulated in Stat92EM– embryos (C). (D-E′′) St11 embryos double-labeled for HLHm5.lacZ and Tin. (D-D′′) WT embryos express HLHm5.lacZ in the neuroectoderm, lateral mesoderm and in the Tin-cells of the dorsal mesoderm (open arrowheads). (E-E′′) HLHm5.lacZ expression is dramatically reduced in St11 Stat92EM– embryos. lacZ is undetectable in the dorsal mesoderm of Stat92EM– embryos. (F-I′) HLHm5 mRNA expression. In WT embryos, HLHm5 expression initiates as stripes in the dorsal mesoderm during St10 (black arrowheads; F,F′) and expands to include large regions of the dorsal mesoderm by St11 (G,G′). HLHm5 expression does not initiate in the dorsal mesoderm of Stat92EM– embryos by St10 (black arrows; H,H′) and is absent from the dorsal mesoderm at St11 (I,I′). (J,K) Oblique view of St11 embryos labeled for Tin similar in orientation to embryos in Fig. 2. Tin expression restricts in the dorsal-most mesoderm of WT embryos (J, open arrows) but not Df(3R)EsplΔmδ-m6 embryos (K). (L) Stat92E ChIP in 4- to 6-hour-old embryos. Stat92E-HA was expressed in the mesoderm with twi,24B.gal4. Cross-linked chromatin was immunoprecipitated with anti-HA or beads alone and PCR amplified. Stat92E binds SCBS1 and SCBS3 but not SCBS2 in the mesoderm. Stat92E does not bind a non-conserved consensus sequence between mβ and mα (see Fig. S4 in the supplementary material). Fold change (FC) was calculated using Image J. Socs36E served as a positive control. Error bars represent s.e.m. Embryos are oriented with anterior to the left.
The JAK/Stat pathway activates HLHm5 expression in the dorsal mesoderm
To identify the E(spl)-C gene(s) that are expressed in the dorsal mesoderm during phase 3 tin expression, we assayed the activity of m4, HLHm5 and E(spl) reporter genes in St11 embryos (Fig. 5A). Strikingly, HLHm5.lacZ directed lacZ expression specifically in Tin–cells of the dorsal mesoderm (Fig. 5D). Expression from HLHm5.lacZ is also detectable in the ventral mesoderm and neuroectoderm. In Stat92EM– embryos, expression from HLHm5.lacZ was reduced throughout the embryo and completely absent from the dorsal mesoderm (Fig. 5E). Moreover, the regions of the dorsal mesoderm that should express HLHm5.lacZ instead expressed Tin in Stat92EM– embryos. By contrast, E(spl).lacZ directs broad lacZ expression in the dorsal mesoderm, whereas m4.lacZ did not drive lacZ expression in the dorsal mesoderm of WT embryos (see Fig. S3 in the supplementary material).
Endogenous HLHm5 expression in the mesoderm initiated at St10 as segmental stripes (Fig. 5F) and expanded to a robust, periodic pattern in the dorsal mesoderm by St11 (Fig. 5G). HLHm5.lacZ therefore recapitulates HLHm5 expression in the dorsal mesoderm. In addition, HLHm5 expression was remarkably similar to Socs36E during St10 (compare Fig. 1I with Fig. 5F). HLHm5 expression did not initiate in the dorsal mesoderm of Stat92EM– embryos during St10 (Fig. 5H) and was absent from the dorsal mesoderm through St11 (Fig. 5I). We then characterized Tin expression in Df(3R)EsplΔmδ-m6 St11 embryos and found that Tin expression did not fully restrict in the dorsal-most mesoderm (Fig. 5J,K). Thus, Stat92E induces HLHm5 expression in the Tin–cells of the dorsal mesoderm and E(spl)-C genes are necessary to restrict Tin expression.
Stat92E binds discrete regions of E(spl)-C in the mesoderm
The 50 kb genomic region encoding E(spl)-C houses a total of three, conserved Stat92E consensus binding sites (SCBS; Fig. 5A; see Fig. S3 in the supplementary material). To determine whether Stat92E binds regions of E(spl)-C, we expressed Stat92E-HA in the mesoderm and performed our own ChIP analysis on 4-6-hour-old embryos. We first assayed Stat92E-HA binding to the Stat92E response element in Socs36E and to the region upstream of m4 identified by the modENCODE ChIP-chip experiment (referred to here as SCBS3; Fig. 5A,L) and confirmed that Stat92E-HA bound both of these regions in the mesoderm. For negative controls, we assayed Stat92E-HA binding to an unconserved SCBS site upstream of mα (see Fig. S4 in the supplementary material) and to a highly conserved SCBS proximal to hedgehog (hh), expression of which is restricted to the ectoderm and the foregut endoderm in 4- to 6-hour-old embryos (see Fig. S4 in the supplementary material). Stat92E-HA did not bind regions near the mα or hh loci, suggesting that our ChIP experiments were identifying bona fide Stat92E chromatin-binding regions in the mesoderm. We then analyzed Stat92E-HA binding at two additional SCBS in E(spl)-C and found that Stat92E-HA binds SCBS1 but not SCBS2 in the mesoderm (Fig. 5L). Thus, E(spl)-C is a direct target of Stat92E in the mesoderm of 4- to 6-hour-old embryos. Although recent work in intestinal stem cells showed that the JAK/Stat pathway activates E(spl)-C genes (Jiang et al., 2009), this is the first evidence that E(spl)-C is a direct target of Stat92E.
Although Stat92E-HA binds regions of E(spl)-C, we did not identify an SCBS in the sequence comprising HLHm5.lacZ. Because E(spl)-C transcription factors regulate gene expression within E(spl)-C itself (Oellers et al., 1994), we conclude that Stat92E indirectly regulates HLHm5 by regulating other E(spl)-C genes. Alternatively, Stat92E might directly regulate HLHm5 via a non-consensus binding site(s).
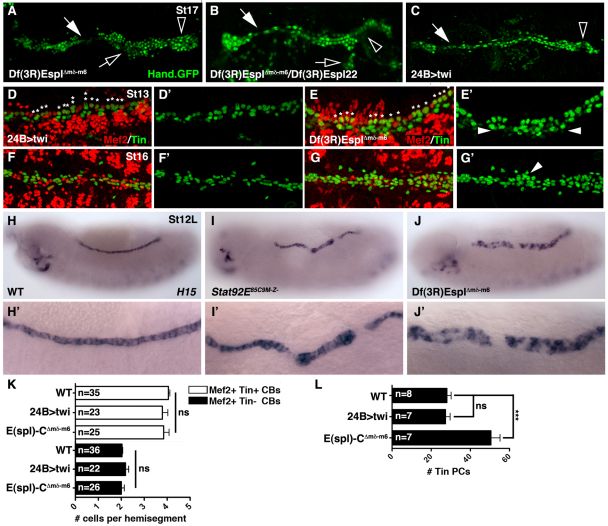
E(spl)-C genes regulate heart morphology
E(spl)-C genes are known regulators of mesoderm development. Previous to our studies, two phenotypes have been reported in E(spl)-C mutant embryos: dysregulated twi expression in the mesoderm (Tapanes-Castillo and Baylies, 2004) and ectopic Ladybird+ cardiac progenitors (Jagla et al., 1997). To understand whether these phenotypes are related through a common mechanism, we characterized heart morphology in E(spl)-C mutant embryos and in embryos misexpressing Twi. Df(3R)EsplΔmδ-m6 deletes all the E(spl)-C genes from HLHmdelta to m6 and presumably disrupts cis-regulatory sequences controlling HLHm7 whereas Df(3R)Espl22 deletes HLHm5, m6, HLHm7, E(Spl) and gro (Fig. 5A). Embryos homozygous for Df(3R)EsplΔmδ-m6 (Fig. 6A) or transheterozygous for Df(3R)EsplΔmδ-m6/Df(3R)Espl22 (Fig. 6B) show cardiac phenotypes similar to Stat92EM–Z– embryos, including loss of cardioblast cell-cell adhesion. Thus, deletion of HLHm5, m6, and perhaps HLHm7, is sufficient to cause heart defects in St17 embryos. If the cardiac defects in E(spl)-C mutant embryos are solely due to dysregulated twi expression, then misexpressing Twi in the mesoderm should induce a cardiac phenotype similar to that of E(spl)-C embryos. Surprisingly, 24B>twi embryos do show a loss of cardioblast cell-cell adhesion (Fig. 6D), but these embryos also lack pericardial cells in the dorsal vessel.
Fig. 6.
E(spl)-C regulates cardiac morphology and pericardial cell number. (A-C) Dorsal views of Hand.GFP expression in live St17 embryos. Df(3R)EsplΔmδ-m6 homozygous embryos (A) and Df(3R)EsplΔmδ-m6/Df(3R)Espl22 heterozygous embryos (B) show cardiac phenotypes similar to Stat92EM–Z– embryos including a collapsed ‘heart’ (open arrowheads), loss of cardioblast cell-cell adhesion (white arrows) and cell aggregations (open arrows). 24B>twi embryos (C) show a collapsed heart and loss of cardioblast cell-cell adhesion but not cell aggregations. Notice that Hand+ pericardial cells appear to be missing in 24B>twi embryos. (D-G′) Embryos double-labeled for Tin and Mef2. (F) 24B>twi St13 embryos have four Tin+ cells in a majority of hemisegments (asterisks). The number of Tin+ pericardial cells is comparable to WT. (E) Df(3R)EsplΔmδ-m6 St13 embryos also show a largely normal pattern of Tin+ cardioblasts; however, the number of Tin+ pericardial cells is increased compared with WT. (F) 24B>twi St16 embryos show a dramatic reduction in Tin+ cardioblasts and pericardial cells. (G) Df(3R)EsplΔmδ-m6 St16 embryos have ectopic Tin+ pericardial cells. (H-J′) H15 mRNA expression in St13 embryos. WT embryos express H15 in a single row of cardioblasts (H). H15 expression expands ventrally in Stat9292EM–Z– (I,I′) and Df(3R)EsplΔmδ-m6 (J,J′) embryos. (K) Segmental quantification of Tin+ and Tin–cardioblasts (CBs) at St13. (L) Quantification of Tin+ pericardial cells (PBs) in one half of the mesoderm at St13. Tin+ pericardial cells were identified by lack of Mef2 expression. ***P<0.001, unpaired t-test. ns, non-significant. Error bars represent s.e.m. Embryos are oriented with anterior to the left.
E(spl)-C genes regulate pericardial cell number independently of twi
We then characterized Tin expression in 24B>twi and Df(3R)EsplΔmδ-m6 St13 embryos. Both 24B>twi and Df(3R)EsplΔmδ-m6 St13 embryos showed four Tin+ cardioblasts in a majority of hemisegments (Fig. 6D,E,K). However, Df(3R)EsplΔmδ-m6 embryos had an increased number of Tin+ pericardial cells compared with WT embryos, whereas 24B>twi embryos showed the appropriate number of Tin+ pericardial cells (Fig. 6E,L). Therefore, the role of E(spl)-C genes in regulating twi expression is independent from their role in regulating pericardial cell number.
Embryos misexpressing Twi in the mesoderm lack pericardial cells at St16 (Castanon et al., 2001); however, the correct number of Tin+ pericardial cells was specified at St13 (Fig. 6D). We assayed Tin expression in St16 24B>twi embryos and found a reduction in the number of Tin+ pericardial cells and Tin+ cardioblasts in the heart (Fig. 6F). By contrast, St16 Df(3R)EsplΔmδ-m6 embryos continued to show ectopic Tin+ pericardial cells (Fig. 6G). Therefore, embryos that misexpress Twi specify the correct number of heart precursors, but over the course of mesoderm migration, precursors are lost from the dorsal mesoderm. We favor a model in which dysregulated twi expression blocks heart precursor cell maturation, resulting in cell-cell adhesion defects during mesoderm migration.
The JAK/Stat pathway and the E(spl)-C genes restrict H15 expression
To gain insights into the mechanism by which JAK/Stat signaling and E(spl)-C regulate pericardial cell number, we assayed H15 expression in Stat92EM–Z– and Df(3R)EsplΔmδ-m6 embryos. The T-box transcription factor H15 induces ectopic Tin+ cells when misexpressed in the mesoderm (Qian et al., 2005a). In St13 WT embryos, H15 was expressed in a single row of cardioblasts in the dorsal mesoderm (Fig. 6H); however, multiple rows of dorsal mesoderm cells expressed H15 in Stat9292EM–Z– and Df(3R)EsplΔmδ-m6 embryos (Fig. 6I,J). As neither Stat92EM–Z– (Fig. 3H) nor Df(3R)EsplΔmδ-m6 (Fig. 6E) embryos specifies supernumerary cardioblasts, we conclude that the JAK/Stat pathway activates E(spl)-C genes to limit the H15 expression domain and, thus, the number of Tin+ pericardial cells.
DISCUSSION
To test the hypothesis that the JAK/Stat pathway functions in the cardiac-specific gene regulatory network, we performed a systematic characterization of JAK/Stat signaling during mesoderm development. We have shown that the JAK/Stat pathway regulates final cardiac morphology as well as heart precursor diversification. Our Stat92E loss-of-function analysis identified a discrete function for the JAK/Stat pathway in restricting tin during the transition from phase 2 to phase 3 expression. In addition, Stat92E embryos have an expanded pericardial cell domain arguing that the JAK/Stat pathway regulates tin to ensure proper heart precursor diversification. Mechanistically, we found that the E(spl)-C gene HLHm5 is expressed in a complementary pattern to tin during phase 3 expression and that the JAK/Stat pathway activates HLHm5 expression in the dorsal mesoderm. The E(spl)-C genes in turn repress twi expression to preserve cardiac morphology and restrict tin and H15 expression to direct heart precursor diversification. These findings provide the first evidence of a role for the JAK/Stat pathway in cardiogenesis and identify a novel tin autoinhibitory circuit involving Stat92E and E(spl)-C.
Stat92E expression in the dorsal mesoderm
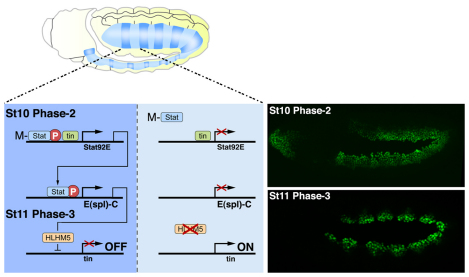
Stat92E is a direct Tin target gene during phase 2 expression; however, Stat92E is expressed in segmented stripes at this stage whereas tin is expressed throughout the dorsal mesoderm (Liu et al., 2009). In addition, embryos lacking only the maternal contribution of Stat92E have mesoderm patterning defects (Fig. 2C). Tin-regulated Stat92E zygotic transcription is therefore insufficient to coordinate mesoderm development. These data suggest that maternally contributed Stat92E is activated in response to segmented Upd and Upd2 activity, binds the Stat92E locus and co-activates Stat92E zygotic transcription with Tin (Fig. 7). In addition, ChIP-chip experiments identified Stat92E binding activity and a conserved SCBS within the Tin-responsive Stat92E mesoderm enhancer (see Table S1 and Fig. S4 in the supplementary material). We conclude that Stat92E and tin co-regulate a brief, spatially restricted JAK/Stat signaling event that patterns the mesoderm.
Fig. 7.
JAK/Stat signaling regulates phase 3 tin expression. During St10, the JAK/Stat pathway is active in one region of each mesoderm segment (dark blue) and largely inactive in the opposing region (light blue). The Tin-responsive Stat92E mesoderm enhancer contains an SCBS (see Fig. S4 and Table S1 in the supplementary material) and directs segmental expression at St10 (Liu et al., 2009). Upd and Upd2 activate the JAK/Stat pathway and induce phosphorylation of maternally contributed Stat92E (M-Stat). Tin and M-Stat co-activate zygotic expression of Stat92E, which induces segmental expression of the E(spl)-C gene HLHm5. HLHm5 then represses tin to establish the phase 3 expression pattern in the dorsal mesoderm. tin expression is sustained in the region of the segment where JAK/Stat signaling is not active. Images on the right show Phase-2 and Phase-3 Tin expression. Embryos are oriented with anterior to the left.
The tin autoinhibitory circuit
Phase 3 tin expression promotes cell-type diversification and differentiation within the dorsal mesoderm and is indirectly activated by Wg via the T-box transcription factors in the Dorsocross complex and the GATA factor Pannier (Jagla et al., 1997; Klinedinst and Bodmer, 2003; Reim and Frasch, 2005). A key finding of this study is that the JAK/Stat pathway activates the transcriptional repressor HLHm5 in the dorsal mesoderm to establish phase 3 tin expression (Fig. 7). Because the HLHm5 cis-regulatory region lacks a conserved SCBS, we predict that Stat92E regulates HLHm5 expression through a non-consensus binding site. Alternatively, Stat92E acts at long distances to regulate gene expression (Sotillos et al., 2010). The SCBSs in E(spl)-C could be a platform from which Stat92E regulates multiple E(spl)-C genes that, in turn, regulate HLHm5 expression. In either event, Stat92E-mediated activation of E(spl)-C genes restricts tin in the dorsal mesoderm to establish phase 3 expression. Tin, therefore, establishes an autoinhibitory circuit by activating its own repressors in the JAK/Stat pathway and in E(spl)-C.
Regulated tin expression directs heart precursor diversification
Both Stat92E and Df(3R)EsplΔmδ-m6 embryos show an increased number of Tin+ pericardial cells and an expanded H15 expression domain (Fig. 2H, Fig. 6G,J-L). Misexpressing mid or H15 in the mesoderm expands the number of Tin+ cells in the dorsal vessel (Qian et al., 2005a; Reim et al., 2005) and embryos misexpressing mid show a phenotype strikingly similar to Stat92E and E(spl) embryos (Reim and Frasch, 2005). As mid, and presumably H15, are positively regulated by Tin during St11/12 (Qian et al., 2005a), unrestricted tin expression in Stat92E or Df(3R)EsplΔmδ-m6 embryos expands the H15 expression domain. Ectopic H15 then specifies supernumerary Tin+ pericardial cells. Because mid expression is not affected in Stat92E embryos, the expression of mid and H15 must be controlled by distinct mechanisms and might have non-redundant functions.
Regulated twi expression directs cardiogenesis
Although the Twi target genes directing mesoderm development and subdivision have been studied in detail (Cripps et al., 1998; Sandmann et al., 2007; Yin et al., 1997), the molecular mechanism that restricts twi expression after gastrulation remains unclear. One regulator of twi is the Notch signaling pathway, which acts through E(spl)-C genes to restrict twi expression (Tapanes-Castillo and Baylies, 2004). However, Notch signaling appears to be active throughout the mesoderm after gastrulation. Our study suggests that segmented JAK/Stat signaling activity (Fig. 1H,I) differentially upregulates E(spl)-C gene expression in concert with Notch to produce periodic twi expression in the mesoderm (Fig. 5H,I). In addition, pan-mesodermal twi expression causes cardiac defects independently of cell fate specification (Fig. 6C,D), suggesting that the cardiac morphology defects in Stat92E embryos are due to dysregulated twi expression. These results highlight a previously unrecognized role for the JAK/Stat pathway and Twi in regulating cardiogenesis.
Sequential JAK/Stat-Dpp signals might regulate heart precursor diversification
Pericardial cell hyperplasia without a concomitant loss of cardioblasts has been reported for dpp hypomorphic embryos (Johnson et al., 2007) and lame duck (lmd) embryos (Sellin et al., 2009). A late Dpp signal, which occurs after the Dpp signal that regulates phase 2 tin expression, instructs the Gli-like transcription factor Lmd to repress Tin expression in fusion competent myoblasts (FCMs; Sellin et al., 2009). Tin expression appears to expand into the FCM domain in Stat92E embryos (see Fig. S2 in the supplementary material); however, the closest Stat92E chromatin binding domain is over 120 kb distal to the lmd transcriptional start site (see Table S1 in the supplementary material). Our study highlights the possibility that sequential JAK/Stat and then Dpp signals regulate Lmd function to direct heart precursor diversification.
E(spl) genes might be conserved Stat targets during mesoderm segmentation
In vertebrates, skeletal myogenesis initiates with the periodic specification of somites in the presomitic mesoderm (Gomez et al., 2008). Cyclical expression of hairy1 in the chick, the hairy- and E(spl)-related family (Her) in zebrafish, and the orthologous Hes family in mice are under the control of Notch-Delta signaling (Giudicelli and Lewis, 2004). Loss of her1 and her7 alters the periodicity with which somite boundaries are specified in fish (Henry et al., 2002), and artificially stabilizing Hes7 causes somites to fuse in the mouse (Hirata et al., 2004). Thus, mesoderm segmentation is governed by Notch-Delta regulation of the E(spl)-C genes in both insects and vertebrates indicating that the two processes share molecular homology. A cell culture model of somitogenesis shows that oscillating Hes1 expression is dependent on Stat activity (Hirata et al., 2002; Yoshiura et al., 2007). Our study supports the exciting possibility that JAK/Stat signaling and E(spl)-C form a conserved developmental cassette directing mesoderm segmentation throughout Metazoa.
Supplementary Material
Acknowledgments
We thank Erika Bach, Mary Baylies, Allison Bardin, Nobert Perrimon, Richard Cripps and the Bloomington Stock Center for fly stocks. We also thank Manfred Frasch, James Skeath and Bruce Paterson for antibodies.
Footnotes
Funding
This work was supported by the National Institutes of Health [2R01HL77439-05 to E.N.O.]; the Robert A. Welch Foundation; and a National Research Service Award fellowship [F32GM083530 to A.N.J.]. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.071464/-/DC1
References
- Bach E. A., Vincent S., Zeidler M. P., Perrimon N. (2003). A sensitized genetic screen to identify novel regulators and components of the Drosophila janus kinase/signal transducer and activator of transcription pathway. Genetics 165, 1149-1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baeg G. H., Zhou R., Perrimon N. (2005). Genome-wide RNAi analysis of JAK/STAT signaling components in Drosophila. Genes Dev. 19, 1861-1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin A. J., Perdigoto C. N., Southall T. D., Brand A. H., Schweisguth F. (2010). Transcriptional control of stem cell maintenance in the Drosophila intestine. Development 137, 705-714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertet C., Rauzi M., Lecuit T. (2009). Repression of Wasp by JAK/STAT signalling inhibits medial actomyosin network assembly and apical cell constriction in intercalating epithelial cells. Development 136, 4199-4212 [DOI] [PubMed] [Google Scholar]
- Callus B. A., Mathey-Prevot B. (2002). SOCS36E, a novel Drosophila SOCS protein, suppresses JAK/STAT and EGF-R signalling in the imaginal wing disc. Oncogene 21, 4812-4821 [DOI] [PubMed] [Google Scholar]
- Castanon I., Von Stetina S., Kass J., Baylies M. K. (2001). Dimerization partners determine the activity of the Twist bHLH protein during Drosophila mesoderm development. Development 128, 3145-3159 [DOI] [PubMed] [Google Scholar]
- Celniker S. E., Dillon L. A., Gerstein M. B., Gunsalus K. C., Henikoff S., Karpen G. H., Kellis M., Lai E. C., Lieb J. D., MacAlpine D. M., et al. (2009). Unlocking the secrets of the genome. Nature 459, 927-930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cripps R. M., Black B. L., Zhao B., Lien C. L., Schulz R. A., Olson E. N. (1998). The myogenic regulatory gene Mef2 is a direct target for transcriptional activation by Twist during Drosophila myogenesis. Genes Dev. 12, 422-434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delidakis C., Artavanis-Tsakonas S. (1992). The Enhancer of split [E(spl)] locus of Drosophila encodes seven independent helix-loop-helix proteins. Proc. Natl. Acad. Sci. USA 89, 8731-8735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekas L. A., Baeg G. H., Flaherty M. S., Ayala-Camargo A., Bach E. A. (2006). JAK/STAT signaling promotes regional specification by negatively regulating wingless expression in Drosophila. Development 133, 4721-4729 [DOI] [PubMed] [Google Scholar]
- Giudicelli F., Lewis J. (2004). The vertebrate segmentation clock. Curr. Opin. Genet. Dev. 14, 407-414 [DOI] [PubMed] [Google Scholar]
- Gomez C., Ozbudak E. M., Wunderlich J., Baumann D., Lewis J., Pourquie O. (2008). Control of segment number in vertebrate embryos. Nature 454, 335-339 [DOI] [PubMed] [Google Scholar]
- Han Z., Olson E. N. (2005). Hand is a direct target of Tinman and GATA factors during Drosophila cardiogenesis and hematopoiesis. Development 132, 3525-3536 [DOI] [PubMed] [Google Scholar]
- Harrison D. A., McCoon P. E., Binari R., Gilman M., Perrimon N. (1998). Drosophila unpaired encodes a secreted protein that activates the JAK signaling pathway. Genes Dev. 12, 3252-3263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry C. A., Urban M. K., Dill K. K., Merlie J. P., Page M. F., Kimmel C. B., Amacher S. L. (2002). Two linked hairy/Enhancer of split-related zebrafish genes, her1 and her7, function together to refine alternating somite boundaries. Development 129, 3693-3704 [DOI] [PubMed] [Google Scholar]
- Hirata H., Yoshiura S., Ohtsuka T., Bessho Y., Harada T., Yoshikawa K., Kageyama R. (2002). Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science 298, 840-843 [DOI] [PubMed] [Google Scholar]
- Hirata H., Bessho Y., Kokubu H., Masamizu Y., Yamada S., Lewis J., Kageyama R. (2004). Instability of Hes7 protein is crucial for the somite segmentation clock. Nat. Genet. 36, 750-754 [DOI] [PubMed] [Google Scholar]
- Hombria J. C., Brown S., Hader S., Zeidler M. P. (2005). Characterisation of Upd2, a Drosophila JAK/STAT pathway ligand. Dev. Biol. 288, 420-433 [DOI] [PubMed] [Google Scholar]
- Hou S. X., Zheng Z., Chen X., Perrimon N. (2002). The Jak/STAT pathway in model organisms: emerging roles in cell movement. Dev. Cell 3, 765-778 [DOI] [PubMed] [Google Scholar]
- Jagla K., Frasch M., Jagla T., Dretzen G., Bellard F., Bellard M. (1997). ladybird, a new component of the cardiogenic pathway in Drosophila required for diversification of heart precursors. Development 124, 3471-3479 [DOI] [PubMed] [Google Scholar]
- Jiang H., Patel P. H., Kohlmaier A., Grenley M. O., McEwen D. G., Edgar B. A. (2009). Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell 137, 1343-1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. N., Burnett L. A., Sellin J., Paululat A., Newfeld S. J. (2007). Defective decapentaplegic signaling results in heart overgrowth and reduced cardiac output in Drosophila. Genetics 176, 1609-1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsten P., Hader S., Zeidler M. P. (2002). Cloning and expression of Drosophila SOCS36E and its potential regulation by the JAK/STAT pathway. Mech. Dev. 117, 343-346 [DOI] [PubMed] [Google Scholar]
- Klinedinst S. L., Bodmer R. (2003). Gata factor Pannier is required to establish competence for heart progenitor formation. Development 130, 3027-3038 [DOI] [PubMed] [Google Scholar]
- Lai E. C., Bodner R., Posakony J. W. (2000). The enhancer of split complex of Drosophila includes four Notch-regulated members of the bearded gene family. Development 127, 3441-3455 [DOI] [PubMed] [Google Scholar]
- Lilly B., Zhao B., Ranganayakulu G., Paterson B. M., Schulz R. A., Olson E. N. (1995). Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science 267, 688-693 [DOI] [PubMed] [Google Scholar]
- Liu Y. H., Jakobsen J. S., Valentin G., Amarantos I., Gilmour D. T., Furlong E. E. (2009). A systematic analysis of Tinman function reveals Eya and JAK-STAT signaling as essential regulators of muscle development. Dev. Cell 16, 280-291 [DOI] [PubMed] [Google Scholar]
- Novak U., Ji H., Kanagasundaram V., Simpson R., Paradiso L. (1998). STAT3 forms stable homodimers in the presence of divalent cations prior to activation. Biochem. Biophys. Res. Commun. 247, 558-563 [DOI] [PubMed] [Google Scholar]
- Oellers N., Dehio M., Knust E. (1994). bHLH proteins encoded by the Enhancer of split complex of Drosophila negatively interfere with transcriptional activation mediated by proneural genes. Mol. Gen. Genet. 244, 465-473 [DOI] [PubMed] [Google Scholar]
- Olson E. N. (2006). Gene regulatory networks in the evolution and development of the heart. Science 313, 1922-1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian L., Liu J., Bodmer R. (2005a). Neuromancer Tbx20-related genes (H15/midline) promote cell fate specification and morphogenesis of the Drosophila heart. Dev. Biol. 279, 509-524 [DOI] [PubMed] [Google Scholar]
- Qian L., Liu J., Bodmer R. (2005b). Slit and Robo control cardiac cell polarity and morphogenesis. Curr. Biol. 15, 2271-2278 [DOI] [PubMed] [Google Scholar]
- Reim I., Frasch M. (2005). The Dorsocross T-box genes are key components of the regulatory network controlling early cardiogenesis in Drosophila. Development 132, 4911-4925 [DOI] [PubMed] [Google Scholar]
- Reim I., Mohler J. P., Frasch M. (2005). Tbx20-related genes, mid and H15, are required for tinman expression, proper patterning, and normal differentiation of cardioblasts in Drosophila. Mech. Dev. 122, 1056-1069 [DOI] [PubMed] [Google Scholar]
- Sandmann T., Jakobsen J. S., Furlong E. E. (2006). ChIP-on-chip protocol for genome-wide analysis of transcription factor binding in Drosophila melanogaster embryos. Nat. Protoc. 1, 2839-2855 [DOI] [PubMed] [Google Scholar]
- Sandmann T., Girardot C., Brehme M., Tongprasit W., Stolc V., Furlong E. E. (2007). A core transcriptional network for early mesoderm development in Drosophila melanogaster. Genes Dev. 21, 436-449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellin J., Drechsler M., Nguyen H. T., Paululat A. (2009). Antagonistic function of Lmd and Zfh1 fine tunes cell fate decisions in the Twi and Tin positive mesoderm of Drosophila melanogaster. Dev. Biol. 326, 444-455 [DOI] [PubMed] [Google Scholar]
- Silver D. L., Montell D. J. (2001). Paracrine signaling through the JAK/STAT pathway activates invasive behavior of ovarian epithelial cells in Drosophila. Cell 107, 831-841 [DOI] [PubMed] [Google Scholar]
- Sotillos S., Espinosa-Vazquez J. M., Foglia F., Hu N., Hombria J. C. (2010). An efficient approach to isolate STAT regulated enhancers uncovers STAT92E fundamental role in Drosophila tracheal development. Dev. Biol. 340, 571-582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stancato L. F., David M., Carter-Su C., Larner A. C., Pratt W. B. (1996). Preassociation of STAT1 with STAT2 and STAT3 in separate signalling complexes prior to cytokine stimulation. J. Biol. Chem. 271, 4134-4137 [DOI] [PubMed] [Google Scholar]
- Tapanes-Castillo A., Baylies M. K. (2004). Notch signaling patterns Drosophila mesodermal segments by regulating the bHLH transcription factor twist. Development 131, 2359-2372 [DOI] [PubMed] [Google Scholar]
- Venkatesh T. V., Park M., Ocorr K., Nemaceck J., Golden K., Wemple M., Bodmer R. (2000). Cardiac enhancer activity of the homeobox gene tinman depends on CREB consensus binding sites in Drosophila. Genesis 26, 55-66 [PubMed] [Google Scholar]
- Ward E. J., Skeath J. B. (2000). Characterization of a novel subset of cardiac cells and their progenitors in the Drosophila embryo. Development 127, 4959-4969 [DOI] [PubMed] [Google Scholar]
- Xu X., Yin Z., Hudson J. B., Ferguson E. L., Frasch M. (1998). Smad proteins act in combination with synergistic and antagonistic regulators to target Dpp responses to the Drosophila mesoderm. Genes Dev. 12, 2354-2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi P., Johnson A. N., Han Z., Wu J., Olson E. N. (2008). Heterotrimeric G proteins regulate a noncanonical function of septate junction proteins to maintain cardiac integrity in Drosophila. Dev. Cell 15, 704-713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Z., Xu X. L., Frasch M. (1997). Regulation of the twist target gene tinman by modular cis-regulatory elements during early mesoderm development. Development 124, 4971-4982 [DOI] [PubMed] [Google Scholar]
- Yoshiura S., Ohtsuka T., Takenaka Y., Nagahara H., Yoshikawa K., Kageyama R. (2007). Ultradian oscillations of Stat, Smad, and Hes1 expression in response to serum. Proc. Natl. Acad. Sci. USA 104, 11292-11297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaffran S., Reim I., Qian L., Lo P. C., Bodmer R., Frasch M. (2006). Cardioblast-intrinsic Tinman activity controls proper diversification and differentiation of myocardial cells in Drosophila. Development 133, 4073-4083 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.