Abstract
Nutrient stress is generally considered from the standpoint of how cells detect and respond to an insufficient supply of nutrients to meet their bioenergetic needs. However, cells also experience stress as a result of nutrient excess, during which reactive oxygen species (ROS) production exceeds that required for normal physiological responses. This may occur due to oncogene activation or chronic exposure to growth factors combined with high levels of nutrients. As a result, multiple mechanisms have evolved to allow cells to detect and adapt to elevated levels of intracellular metabolites, including: promotion of signaling and proliferation by ROS, amino acid-dependent mTOR activation, and regulation of signaling and transcription through metabolite-sensitive protein modifications. We discuss how each of these responses can contribute to the development and/or progression of cancer under conditions of cellular nutrient excess and their potential roles in linking chronic organismal over-nutrition (obesity) with cancer.
Introduction
Core metabolic pathways have been generally well conserved among eukaryotes; organisms from yeast to mammals make use of both glycolytic and mitochondrial metabolism depending on extracellular conditions and cues, cellular needs, and stage of metabolic or circadian cycle (DeBerardinis et al., 2008; Sahar and Sassone-Corsi, 2009; Tu et al., 2005) Despite these similarities, fundamental differences exist between unicellular and multicellular organisms in the acquisition of nutrients and control of metabolism. While unicellular organisms must deal with the potentially large fluctuations in nutrient availability in the extracellular environment, cells within larger multicellular organisms have access to a relatively stable supply of nutrients from the bloodstream. While some cell types such as liver, muscle, and fat cells have the capacity to store excess carbon in the form of glycogen or lipid, most cells are unable to assimilate excess nutrients in the absence of engaging in cell growth and/or proliferation. Nutrient uptake in metazoan cells is controlled primarily by growth factor signaling. Thus too much or too little growth factor signaling-induced nutrient uptake can profoundly affect cellular bioenergetic fitness. A major readout of growth factor-regulated nutrient uptake is the level of reactive oxygen species (ROS) produced by mitochondria. In this review, we discuss how high levels of nutrient metabolism can stress the cell, the mechanisms used by the cell to detect and respond to elevated intracellular metabolite levels, and the contribution of cellular and organismal nutrient excess to cancer.
Nutrient excess and cellular stress
ROS are produced at a low level by the electron transport chain as a normal part of cellular metabolism and play a physiologically important role in the regulation of cell signaling, proliferation, and differentiation. ROS production can rise, however, with changes in oxidative mitochondrial metabolism, potentially causing damage to cellular components and cell death (Hamanaka and Chandel, 2010; Trachootham et al., 2009; Veal et al., 2007). Cells experience stress as a result of “nutrient excess” when ROS production exceeds that needed for normal physiological responses (Figure 1). It is not coincidental that diseases characterized by altered cellular metabolism, such as cancer and diabetes, are also characterized by elevated ROS levels that may contribute to disease pathogenesis (Brandon et al., 2006; Halliwell, 2007; Nathan, 2008; Roberts and Sindhu, 2009; Trachootham et al., 2009; Wallace, 2005).
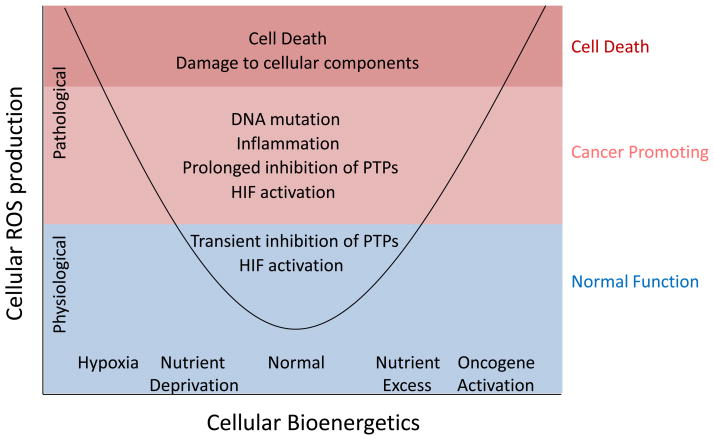
Figure 1. Both nutrient deficiency and nutrient excess can cause cellular stress.
Mitochondrial ROS production from the electron transport chain increases in response to either hypoxia or oncogene activation and nutrient excess. Other mitochondrial sources of ROS such as proline oxidase are also involved in stress responses. At low levels, ROS production is critical for normal physiological processes, such as proliferation and differentiation, through regulation of signaling. At higher levels, ROS can induce changes that promote the development of cancer, such as mutation of DNA, prolonged signaling, and activation of inflammatory pathways. High levels of ROS can also lead to irreversible damage to cellular components and cell death.
How do changes in metabolism impact mitochondrial ROS production? The tricarboxylic acid (TCA) cycle oxidizes nutrients, and the resulting electrons are transferred to nicotinamide adenine dinucleotide (NAD+) and flavin adenine dinucleotide (FAD) to produce NADH and FADH2. These electrons are donated to the electron transport chain (ETC) at complexes I and II, respectively. Electrons are shuttled from complexes I and II to complex III via ubiquinone and from complex III to complex IV via cytochrome c. Two electrons are finally donated to molecular oxygen (½ O2), generating H2O. Incomplete, one-electron reduction of oxygen can also occur at complexes I, II, and III, producing superoxide (O2−•). Superoxide production increases with the concentration of oxygen and electron donors (Turrens, 2003). Hence, rising levels of NADH/NAD+ is a major factor leading to increased mitochondrial ROS production (Murphy, 2009). When breakdown of metabolites in the TCA cycle exceeds the capacity of the ETC to assimilate the resulting electrons, ROS production increases. In addition, stalling of the ETC, due to mutated oxidative phosphorylation (OXPHOS) genes can cause build-up of electrons and increased propensity to produce superoxide (Brandon et al., 2006; Wallace, 2005). Since the mitochondrial genome, which encodes several OXPHOS genes, is exposed to elevated levels of ROS due to its proximity to the ETC, conditions that favor increased ROS production such as excessive metabolite-dependent conversion of NAD+ to NADH could potentially lead to a feed-forward loop of increased ROS production and mitochondrial DNA (mtDNA) mutation (Trachootham et al., 2009).
While superoxide mediates its effects within a short range of its production, it can also be converted by superoxide dismutases (SODs) into hydrogen peroxide (H2O2), which is more stable and can diffuse throughout the cell. H2O2 is a known regulator of signaling through oxidation of redox sensitive proteins, such as protein tyrosine phosphatases (PTPs) (Meng et al., 2002); hence, electrons generated by excess mitochondrial metabolism can be used to regulate intracellular signaling through production of ROS. H2O2 can also be converted into the highly reactive hydroxyl radical (OH•) in the presence of reduced transition metals, via the Fenton reaction; in this manner, ROS production in the mitochondria can lead to damage to macromolecules throughout the cell, including nuclear DNA, membrane lipids, and proteins.
Mitochondrial ROS production also rises under hypoxic conditions, and these ROS are involved in activation of hypoxia inducible factors (HIFs), which mediate a transcriptional response that promotes adaptation to hypoxia (Brunelle et al., 2005; Guzy et al., 2005; Guzy and Schumacker, 2006; Mansfield et al., 2005). ROS production under hypoxic conditions occurs primarily at complex III, although the precise mechanisms involved are not yet clear (Bell et al., 2007; Guzy and Schumacker, 2006). Other mitochondrial sources of ROS may also be important in addition to that produced from the ETC. For example, during nutrient deprivation, the mitochondrial enzyme proline oxidase (POX) is induced and can augment TCA cycle function by converting proline into pyrroline-5-carboxylate, which can then be converted into the TCA cycle metabolite α-ketoglutarate (Phang et al., 2010). The POX reaction is also a source of superoxide, and POX-dependent ROS production has been shown to regulate signaling and promote apoptosis under stress conditions (Phang et al., 2010). Tumor cells may encounter both types of ROS-inducing nutrient stress: 1) oncogene-driven increases in nutrient metabolism and 2) nutrient-deprivation and hypoxia as tumor growth outpaces angiogenesis.
In addition to rate of production, the cumulative effects of ROS are contingent on antioxidant activity. As discussed, superoxide accumulation is limited by the presence of superoxide dismutases (SODs), which rapidly convert superoxide into hydrogen peroxide (H2O2) and oxygen. Hydrogen peroxide can be reduced to water by enzymes such as catalase, glutathione peroxidases, and peroxiredoxins distributed throughout the cell. Under normal conditions, ROS production and scavenging are balanced by the cell to maintain ROS levels within a low physiological range. In contrast, many cancer cells have altered antioxidant defenses. The tumor suppressor p53, for example, plays a critical role in cellular antioxidant defenses, and its downregulation is associated with reduced levels of antioxidant production and increased DNA oxidation and mutation (Sablina et al., 2005). Akt activation, which occurs commonly in cancer as a result of mutations in upstream signaling components, is also associated with higher mitochondrial oxygen consumption and ROS generation, coupled with reduced ROS scavenging, due to Akt-dependent inhibition of FoxO transcription factors (Noguiera et al., 2008). FoxO transcription factors, which are themselves tumor suppressors, regulate the expression of multiple ROS detoxifying enzymes, among other effects (Dansen and Burgering, 2008). While increased ROS production contributes to tumorigenesis, ROS levels must be restrained even in cancer cells if they are to avoid the damaging effects of ROS on the integrity of vital intracellular macromolecules. Therapeutic augmentation of ROS levels has the potential to be selectively toxic to cancer cells; both Ras transformation and Akt activation sensitize cells to phenethyl isothiocyanate (PEITC), a substance which inactivates the glutathione antioxidant system (Nogueira et al., 2008; Trachootham et al., 2006).
The point at which ROS production crosses the line from physiological to pathological is often not clear. Sensitivity to ROS varies among cell types, and location of production and species of ROS can lead to different biological outcomes. This is exemplified by the regulation of insulin sensitivity by ROS. Activation of NADPH oxidases at the plasma membrane upon insulin binding to its receptor promotes insulin action, via H2O2-dependent inactivation of PTPs that inhibit insulin signaling (Goldstein et al., 2005). This temporal and highly regulated ROS generation promotes sensitivity to insulin. In contrast, chronic ROS production has been demonstrated to cause insulin resistance, and increased ROS production has been implicated in linking obesity with diabetes (Houstis et al., 2006; Roberts and Sindhu, 2009). Indeed, eating a high fat diet (HFD) has been shown to increase the ROS-emitting potential of mitochondria in both rats and humans (Anderson et al., 2009). Reducing this ROS production through antioxidant treatment or through over-expression of mitochondria-targeted catalase in muscle improves insulin sensitivity in obese rodents (Anderson et al., 2009; Houstis et al., 2006). Interestingly, even in the context of HFD, ROS production can also promote insulin sensitivity; mice lacking the H2O2 scavenger glutathione peroxidase 1 are protected from HFD-induced insulin resistance due to increased oxidation of PTEN, a redox-sensitive PTP, thus enabling increased signaling downstream of the insulin receptor (Loh et al., 2009). These findings highlight the complexity of ROS biology and point to the importance of location and duration of ROS production, as well as species of ROS in determining its cellular impact.
Sensing nutrient excess: Cellular metabolism can promote cancer development
Nutrient uptake in mammalian cells is controlled by growth factor signaling, allowing nutrient consumption to increase transiently when growth factors bind to their cell surface receptors and activate downstream signaling cascades (Figure 2). Prolonged high levels of nutrient metabolism can occur due either to 1) a chronic combination of excess nutrients and insulin/growth factors or 2) to oncogenic activation of signaling pathways.
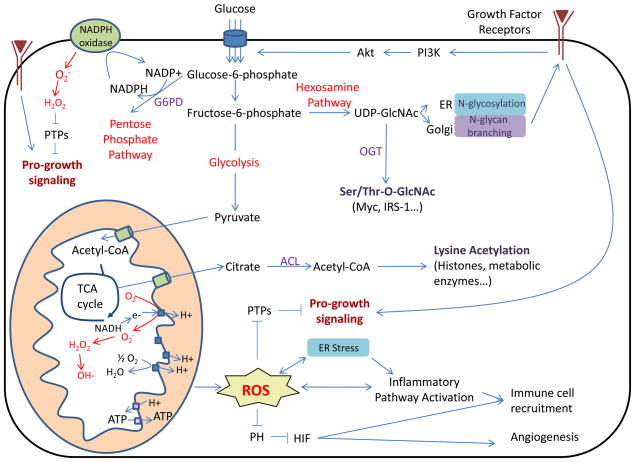
Figure 2. High levels of nutrient uptake induced by chronic growth factor signaling or oncogenic mutations cause cellular ROS stress and can contribute to cancer development through multiple mechanisms.
Elevated mitochondrial or NADPH oxidase-dependent ROS production can contribute to cancer through multiple mechanisms, including DNA mutation, activation of inflammatory pathways, HIF activation, and promotion of pro-growth signaling. ER stress can also activate inflammatory pathways and regulate oxidative stress response. Increased glucose metabolism leading to changes in protein acetylation and O-GlcNAcylation can regulate signaling, transcription, and metabolism. Metabolic regulation of N-glycan branching modulates growth factor receptor surface expression and downstream signaling. Increased pro-growth signaling, HIF stabilization, or mutation resulting in oncogene activation can all promote increased glucose uptake and metabolism, leading to a vicious cycle of increased nutrient uptake, stress responses, and cancer promotion.
Nutrient availability is sensed at multiple levels by the cell. AMP-activated protein kinase (AMPK) inhibits growth and proliferation in response to ATP depletion and/or AMP accumulation. Mammalian target of rapamycin (mTOR) is activated by growth factor signaling in order to promote protein synthesis and cell growth, in a manner dependent on intracellular amino acid levels. These kinases are sensitive to nutrient deficiency and instruct cells as to whether available nutrients are sufficient to permit growth. In addition, both mitochondrial and endoplasmic reticulum (ER) stress responses can regulate or induce adaptation to the ROS production initiated by nutrient excess. Finally, mounting evidence suggests that metabolically sensitive protein modifications, such as acetylation, O-GlcNAcylation, and N-glycosylation, may each play key roles in cellular response to nutrient excess and are likely to be relevant in metabolic diseases and cancer (Haigis and Sinclair, 2010; Lau and Dennis, 2008; Schwer and Verdin, 2008; Slawson et al., 2010) (Figure 3).
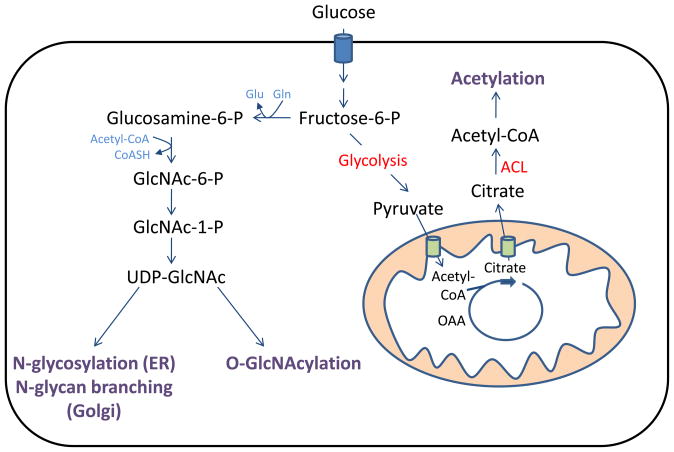
Figure 3. Metabolically-sensitive protein modifications can participate in cellular nutrient sensing.
Production of acetyl-CoA for acetylation is regulated by ATP-citrate lyase (ACL), in a manner dependent on the availability of mitochondria-derived citrate. Both N-linked glycosylation and O-GlcNAcylation of proteins relies on production of UDP-GlcNAc by the hexosamine biosynthetic pathway.
Excess Mitochondria-Produced ROS and Disease
Mitochondrial ROS production increases with oncogene-induced metabolic stress, and substantial evidence indicates that elevated ROS levels can promote tumorigenesis (Halliwell, 2007; Wallace, 2005). While cancer cells are often thought of as relying exclusively on glycolytic metabolism, it is now clear that mitochondrial metabolism is also essential, particularly in supporting cellular biosynthesis (DeBerardinis et al., 2008). In proliferating cells utilizing aerobic glycolysis, glutamine plays a key role in supplying substrate to the TCA cycle (DeBerardinis et al., 2007). Glutamine uptake and mitochondrial metabolism are both regulated by pro-growth signaling pathways and oncoproteins. For example, Myc stimulates glutamine uptake, glutaminolysis, and mitochondrial biogenesis (Gao et al., 2009; Li et al., 2005; Wise et al., 2008). mTOR also plays a key role in promoting increased mitochondrial metabolism and oxygen consumption and can stimulate net uptake of amino acids (Edinger and Thompson, 2002; Schieke et al., 2006). As we will discuss, increased production of ROS, largely from the mitochondria, may promote cancer by increasing DNA mutation, by regulating signaling and transcription, and by promoting inflammation.
Direct damage to and mutation of DNA due to elevated ROS production can contribute to cancer development. A number of mouse models containing reduced levels of antioxidants exhibit increased cancer development or progression, associated with increased oxidative damage to DNA and other macromolecules (Chu et al., 2004; Elchuri et al., 2005; Frohlich et al., 2008; Neumann et al., 2003; Sablina et al., 2005). ROS-induced damage to DNA and DNA repair enzymes can induce genomic instability, and mutation of nuclear encoded genes such as TP53 promotes carcinogenesis (Colotta et al., 2009; Hussain et al., 2003). Mutation of mtDNA can also increase ROS production and promote cancer progression, as demonstrated in experiments using cybrid (cytoplasmic hybrid) technology, which allows the generation of cells that retain their own nuclear genome but contain mtDNA from another source (Brandon et al., 2006). Cybrid PC-3 prostate cancer cells were generated containing either wild type cytochrome oxidase subunit I (COI) or COI containing a mutation found in multiple independent prostate tumors. Remarkably, when these cells were grown in nude mice, the mutant COI-containing tumors grew to roughly 7 times the size of the tumors expressing wild type COI (Petros et al., 2005). Furthermore, similar experiments in which cybrids were generated containing mutations in NADH dehydrogenase subunit 6 (ND6), causing deficient complex I activity and elevated ROS production, led to increased metastasis in poorly metastatic recipient cells (Ishikawa et al., 2008). Thus, mutation of either mtDNA or nuclear DNA in the presence of ROS can contribute to cancer development.
Signaling and transcriptional changes in response to elevated ROS are also relevant to cancer development. ROS-dependent inactivation of PTPs has been reported to be increased in cancer cells and may also enhance pro-growth signaling and contribute to cancer in this way (Lou et al., 2008; Wu, 2006). Regulation of signaling by ROS has consequences for cancer cell proliferation; for example, mitochondrial ROS production was shown to be critical for anchorage-independent growth driven by oncogenic Kras, likely through the regulation of ERK1/2 signaling (Weinberg et al., 2010). Additionally, hypoxia-induced ROS is important for activation of hypoxia inducible factors (HIFs), which can regulate tumorigenesis through transcriptional induction of genes involved in angiogenesis, invasion and metastasis, dedifferentiation, and glycolytic metabolism, as well as through interactions with proteins such as Myc and p53 (reviewed in (Majmundar, 2010)). ROS-induced stabilization of HIF-1α was also shown to be critical for Myc-dependent tumorigenesis and an important target of antioxidants in inhibiting tumor growth (Gao et al., 2007).
Activation of inflammatory pathways is another key link between ROS and cancer. Much evidence indicates that cancer is frequently associated with a state of chronic inflammation, and that inflammation plays a key role in every stage of cancer development and progression (Coussens and Werb, 2002; Grivennikov et al., 2010; Karin and Greten, 2005; Mantovani et al., 2008). ROS are implicated in activating the transcription factors NF-κB and AP-1, mainly through activation of the upstream kinases that control their activation, inhibitor of kappa B kinase(IKK) and c-Jun N-terminal kinase (JNK) (Gloire et al., 2006; Kamata et al., 2005; Sen and Packer, 1996). Inflammation in cancer involves a close interplay between tumor-associated immune cells and the tumor cells themselves. Activation of NF-κB and AP-1 in immune cells induces production of inflammatory cytokines such as TNFα and IL-6. Inflammatory cytokine signaling in tumor cells promotes further increases in ROS production, stimulation of proliferation through Stat3, and promotion of cell survival and further recruitment of immune cells through transcriptional targets of NF-κB. The importance of the IKK-NF-κB pathway, TNFα, IL-6, and Stat3 in tumorigenesis has been demonstrated in multiple murine cancer models (Grivennikov et al., 2010).
Nutrient adaptation and the ER stress response
Cellular redox homeostasis and inflammatory status are also regulated by the unfolded protein response (UPR) in response to ER stress caused by factors such as nutrient deprivation or nutrient excess. The UPR is mediated primarily by three ER-membrane associated proteins PERK, IRE1, and ATF6, which together act to relieve ER stress by reducing protein translation and assisting protein folding and degradation of misfolded proteins (Buchberger, 2010; Kaufman et al., 2002). Inflammatory pathways crosstalk with the UPR through multiple mechanisms; each of the three arms of the UPR has been implicated in activation of NF-κB, and JNK can be activated through IRE1α (reviewed in (Hotamisligil, 2010)). Adaptation to oxidative stress can in part be regulated by the UPR through PERK-dependent activation of Nrf2, a transcription factor that induces antioxidant gene expression (Cullinan and Diehl, 2006). Markers of ER stress are altered in cancer and substantial evidence implicates the involvement of ER stress in cancer (Healy et al., 2009; Lee, 2007; Moenner et al., 2007). Evidence that regulation of redox homeostasis by the PERK-Nrf2 arm of the UPR may impact cancer development has recently been provided. Suppression of PERK in cancer cells was associated with increased ROS levels and activation of a DNA damage checkpoint, and in MMTV-Neu-driven mammary tumors, deletion of PERK inhibited tumor growth (Bobrovnikova-Marjon et al., 2010). Interestingly, mammary gland-specific PERK deletion also led to an increase in spontaneous tumors, likely due to increased genomic instability (Bobrovnikova-Marjon et al., 2010).
Acetylation as a Nutrient-Regulated Protein Modification
Protein modifications such as acetylation and glycosylation can be regulated in a metabolically responsive manner and might also contribute to adaptation to metabolic stress (Figure 3). In mammalian cells, nucleo-cytosolic acetyl-CoA is generated by two enzymes, acetyl-CoA synthetase 1 (AceCS1), which produces acetyl-CoA from acetate, and ACL, which generates acetyl-CoA through the cleavage of citrate. Citrate is produced in the TCA cycle and exported by the tricarboxylate transporter from the mitochondria into the cytosol, where it is accessible to ACL. The regulation of histone acetylation is sensitive to nutrient availability, in a manner dependent on ACL (Wellen et al., 2009). Moreover, ACL-dependent acetylation may be sensitive to differences in nutrient metabolism at the high end of the spectrum; higher levels of histone acetylation were observed when adipocytes were differentiated in 25 mM as compared to 4 mM glucose, correlating with specific changes in gene expression (Wellen et al., 2009). Recent studies have demonstrated that nutrient-dependent acetylation extends beyond histones; lysine acetylation of various metabolic enzymes is also metabolically responsive (Wang et al., 2010a; Zhao et al., 2010). Proteomic analyses have revealed that thousands of proteins in mammalian cells are acetylated (Choudhary et al., 2009; Zhao et al., 2010), and the list of proteins acetylated in a nutrient-responsive manner will undoubtedly continue to grow. Interestingly, removal of acetyl groups from proteins is also metabolically sensitive, through the action of NAD+-dependent sirtuin protein deacetylatases. NAD+ is consumed during glycolytic and oxidative metabolism, and calorie-restricted conditions promote activation of sirtuins (Haigis and Sinclair, 2010; Schwer and Verdin, 2008). The combined action of deacetylation by sirtuins under low nutrient conditions and ACL-dependent acetylation under nutrient-replete conditions offers the possibility for protein acetylation levels to participate in coupling metabolism to protein function over a wide range of nutritional conditions.
Is metabolic regulation of acetylation relevant to cancer? Certainly more work is needed. RNAi-mediated suppression of ACL has been demonstrated to inhibit tumor growth (Hatzivassiliou et al., 2005). While this is due in large part to the critical role of ACL in de novo lipogenesis, which is important for membrane synthesis in proliferating cells, it is possible that effects on acetylation could also play a more direct role in carcinogenesis. Suppression of ACL promotes differentiation in cancer cells and myoblasts (Bracha et al., 2010; Hatzivassiliou et al., 2005), and it is an intriguing possibility that changes in acetylation of histones or other proteins might be part of the mechanism through which ACL can regulate differentiation. Evidence for this possibility was suggested in the study in myoblasts; treatment with the histone deacetylase inhibitor trichostatin A could reverse ACL suppression-dependent differentiation (Bracha et al., 2010).
Glycosylation as a Nutrient-Sensitive Modification
Protein N-linked glycosylation and O-linked N-acetylglucosamine (O-GlcNAc) modification can also each be regulated in a nutrient-responsive manner, at least in part through the flux of glucose into the hexosamine biosynthetic pathway. The hexosamine pathway is a relatively minor branch of glucose metabolism, representing the fate of roughly 3–5% of glucose entering the cell (Marshall et al., 1991). In addition to glucose, glutamine and acetyl-CoA are necessary for pathway activity, suggesting that the hexosamine pathway is highly attuned to cellular metabolism. The hexosamine pathway diverges from glycolysis at fructose-6-phosphate, where glutamine-fructose-6-phosphate amidotransferase (GFAT) transaminates fructose-6-phosphate to produce glucosamine-6-phosphate. The end product of the pathway is UDP-GlcNAc, which is critical for both nucleocytoplasmic O-GlcNAc protein modification and N-linked glycosylation in the ER and Golgi (Dennis et al., 2009; Love and Hanover, 2005).
N-linked glycosylation of proteins occurs cotranslationally in the ER, and further remodeling of N-glycans takes place in the Golgi apparatus (Kornfeld and Kornfeld, 1985). UDP-GlcNAc is used for production of the lipid linked oligosaccharide (LLO) that is transferred onto asparagines residues of nascent polypeptides in order to initiate N-glycosylation in the ER (Kornfeld and Kornfeld, 1985). UDP-GlcNAc is also transported into the Golgi, where it is used by 4 N-acetylglucosaminyltransferases (MGAT1, 2, 4, and 5) to sequentially modify N-glycan branching (Dennis et al., 2009). These enzymes possess differential affinities for UDP-GlcNAc and the glycoprotein acceptors, such that the activities of MGAT4 and MGAT5 depend on the concentration of UDP-GlcNAc in the Golgi (reviewed in (Dennis et al., 2009).
Importantly, MGAT5 activity and UDP-GlcNAc availability have been shown to modulate the surface expression of various growth factor receptors such as the EGFR, directly regulating growth factor-dependent signaling (Lau et al., 2007; Partridge et al., 2004). In addition, MGAT4-dependent regulation of glycosylation of the glucose transporter Glut2 has been shown to promote its retention at the cell surface of pancreatic β cells. In the absence of MGAT4, Glut2 is found primarily in endosomes and lysosomes rather than at the cell surface, and glucose-stimulated insulin secretion is markedly inhibited (Ohtsubo et al., 2005).
O-linked GlcNAc modification occurs in a reversible manner on serine and threonine residues of a wide range of intracellular proteins, from signaling proteins and transcription factors to metabolic enzymes, and accumulating evidence indicates that O-GlcNAc is key to many cellular functions (Love and Hanover, 2005; Zachara and Hart, 2004). Proteins are modified by O-GlcNAc through the action of a single enzyme, O-GlcNAc transferase (OGT). Efforts to generate OGT null mice revealed that this protein is necessary for viability of embryonic stem cells, and conditional mutagenesis of OGT in mice demonstrated that O-GlcNAc is critical for the viability and/or function all cell types examined (O’Donnell et al., 2004; Shafi et al., 2000). The ability of increased hexosamine pathway activation to mediate insulin resistance has been known for nearly two decades, and subsequent characterization has identified a key role for the O-GlcNAc modification in mediating these effects (Marshall, 2006; Teo et al., 2010). OGT is recruited to the plasma membrane in response to insulin, where it can bind phosphatidylinositide-(3,4,5)-triphosphate (PIP3), presumably in order to place OGT in proximity to proteins in the insulin signaling cascade (Yang et al., 2008). OGT has also been reported to associate with PDK-1 and the insulin receptor (IR), which phosphorylates and activates OGT (Whelan et al., 2010; Whelan et al., 2008). O-GlcNAc modification has been demonstrated on a number of proteins in the insulin signaling cascade, and at least one mechanism that appears to be involved in O-GlcNAc-mediated inhibition of insulin action is the impairment of IRS-1 tyrosine phosphorylation at a PI3K p85 binding site, resulting in reduced interaction between IRS-1 and PI3K (Whelan et al., 2010).
It is anticipated that hexosamine pathway flux would be elevated in cancer cells exhibiting increased glucose uptake, and indeed, evidence has been emerging that hexosamine pathway activity increases under conditions of oncogene activation and likely contributes to tumorigenesis. Recent studies have shown that O-GlcNAcylation increases upon Myc activation and is elevated in breast cancer and leukemia cells (Caldwell et al., 2010; Gu et al., 2010; Morrish et al., 2009; Shi et al., 2010). In two xenograft breast cancer models, RNAi-mediated suppression of OGT led to either an almost complete suppression of tumor growth or reduced metastasis, despite equal growth of the primary tumor (Caldwell et al., 2010; Gu et al., 2010). These different phenotypes may reflect differences between the breast cancer cell lines or perhaps degree of OGT suppression. Given that OGT is known to modify a number of tumor suppressors and oncoproteins, including p53 and c-Myc, and that O-GlcNAcylation is critical for proper cell cycle control and cytokinesis, it appears likely that O-GlcNAcylation will prove to be an important player during tumorigenesis (Caldwell et al., 2010; Chou et al., 1995; Slawson et al., 2005; Wang et al., 2010b; Yang et al., 2006).
Additionally, UDP-GlcNAc may become available at increased levels in the Golgi in cancer cells, in order to regulate N-glycan modification. Such changes would have the potential to be functionally significant to the cancer cell. For example, tumor growth in mouse models of cancer driven either by PTEN heterozygosity or by polyomavirus middle T (PyMT) transgenic expression was significantly delayed in the absence of MGAT5 (Cheung and Dennis, 2007; Granovsky et al., 2000). Studies in mammary epithelial tumor cells from PyMT transgenic Mgat5−/− and Mgat5+/+ mice showed that the absence of MGAT5 resulted in impaired response to multiple growth factors, which could be rescued by re-expression of Mgat5 (Partridge et al., 2004). The β1,6GlcNAc –branched N-glycans produced by MGAT5 interact with galectins at the cell surface and can oppose glycoprotein endocytosis; both EGFR and TGFβR were found at lower levels at the cell surface of PyMT Mgat5−/− than PyMT Mgat5+/+ cells (Partridge et al., 2004). Since whole body metabolic homeostasis and cell surface expression of glucose transporters such as Glut2 can be regulated at the level of N-glycan branching (Ohtsubo et al., 2005, Cheung et al., 2007), it is possible that metabolic fluxes in cancer cells may also regulate nutrient transporter surface expression and cellular acquisition of nutrients. Clearly, more research is needed to understand how metabolic regulation of glycosylation may contribute to cancer.
Thus, nutrient excess affects multiple mechanisms that couple metabolism to pathways that modulate cell growth and proliferation (Figure 2). The mitochondrial and ER responses to high levels of nutrients can contribute to cancer through ROS-mediated mutagenesis of DNA, regulation of signaling, and activation of inflammatory pathways. Metabolically sensitive protein modifications such as acetylation and glycosylation are also emerging as key players in regulating cell function in accordance with intracellular nutrient availability. Significant research effort will be required to understand the complex roles that these modifications are playing.
Obesity contributes to cancer development
As we have discussed in this article, changes in cellular metabolism contribute to the development and progression of cancer in a number of different ways. Are all of these metabolic changes mediated exclusively through cell autonomous mechanisms such as oncogenic mutations, or can over-nutrition at an organismal level also contribute to cancer development? Epidemiological evidence clearly indicates that it can (Calle and Kaaks, 2004). Obesity is associated with increased risk for several types of cancer, with percentage of cases attributable to overweight and obesity in the United States and Europe estimated at over 20% for several types of cancer and between 40–60% for both endometrial and oesophageal cancers (Calle and Kaaks, 2004). Several mechanisms have been proposed to explain the link between metabolic disease and cancer, particularly hyperinsulinemia and chronic low-grade inflammation. As insulin resistance develops in obese individuals, insulin secretion from pancreatic beta cells increases in compensation, causing hyperinsulinemia. High levels of insulin itself might promote tumorigenesis, though hyperinsulinemia can also lead to increased production of insulin-like growth factor I (IGF-I), as well as suppression of IGF binding proteins 1 and 2, which bind to IGF, inhibiting its bioavailability (Gallagher and Leroith, 2010; Renehan et al., 2006). High circulating levels of free IGF-I can stimulate PI3K-Akt signaling, thus promoting increased glucose metabolism, increased mitochondrial ROS production, and cell growth and survival. Much data indicates that IGF-I can promote tumorigenesis (Gallagher and Leroith, 2010), and a recent study using a mouse model of non-obese insulin resistance has demonstrated that hyperinsulinemia can enhance IR/IGF-IR and Akt phosphorylation in mammary tissue and promote mammary tumor growth (Novosyadlyy et al., 2010). Notably, calorie restriction, which reduces circulating levels of insulin and IGF-1, was shown to impair tumor growth only from cells that were responsive to insulin and IGF-1 and lacked mutations that resulted in activation of the PI3K/Akt pathway (Kalaany and Sabatini, 2009). The relationship between insulin, IGF, and obesity is complex, however. While insulin levels show a positive linear correlation with both body mass index (BMI) and free IGF-I, non-linear relationships exist between BMI and either total or free IGF-I, so the degree to which this mechanism contributes to obesity-linked cancer remains to be completely defined (Renehan et al., 2006).
Obesity is also associated with a state of chronic low-grade inflammation, likely due to changes wrought by nutrient excess, such as mitochondrial ROS production and ER stress (Wellen and Hotamisligil, 2005). It is now well-established that obesity-linked inflammation is a key mediator of insulin resistance (Hotamisligil, 2006; Shoelson et al., 2006). Given the clear role that chronic inflammation plays in cancer development, it stands to reason that obesity-linked inflammation may also promote cancer.
Recent studies have provided genetic evidence for the involvement of obesity-induced inflammation in liver and pancreatic cancer. Both high fat diet (HFD)-induced obesity and genetic obesity were shown to promote hepatocellular carcinoma development following injection of the pro-carcinogen diethylnitrosamine (DEN) into mice (Park et al., 2010). Obese mice exhibited increased levels of inflammatory cytokines in liver and serum, and mice lacking either interleukin-6 (IL-6−/−) or tumor necrosis factor signaling (TNFR1−/−) were protected from the tumor-promoting effects of HFD (Park et al., 2010). Similarly, HFD was also shown to promote K-ras-induced pancreatic cancer in mice, in a manner dependent on TNF receptor signaling (Khasawneh et al., 2009). Interestingly, these mice do not exhibit hyperinsulinemia on HFD, suggesting that obesity-linked inflammation can promote tumorigenesis independent of effects on insulin sensitivity (Khasawneh et al., 2009). Thus, the chronic, low-grade inflammation associated with obesity may contribute not only to the development of metabolic diseases such as diabetes, but also to cancer.
Conclusion
High levels of cellular nutrient metabolism result in ROS production and oxidative stress that can contribute to the development and progression of cancer. While this frequently occurs as a result of oncogene activation, it is becoming evident that organismal over-nutrition also leads to cellular stress responses that promote carcinogenesis. The obesity epidemic does not seem to be abating and contributes significantly to the cancer burden. At present, it appears likely that obesity can promote cancer development through a combination of hyperinsulinemia and nutrient-stress induced chronic low-grade inflammation, with each of these factors playing lesser or greater roles in different cancer types. It is likely that additional mechanisms also participate in linking chronic over-nutrition with cancer development and progression. For example, it will be interesting to examine how nutritionally-induced changes in cell surface proteins and their glycosylation regulate tumor cell response to immune cells and the microenvironment. Additionally, assessing which specific proteins are differentially modified by acetylation and/or O-GlcNAcylation during conditions of nutrient excess might provide insight into the potential roles of these modifications in the development of cancer in obesity. Over the past several years, our knowledge of the interaction between metabolic and signaling pathways and the metabolic changes associated with cancer has increased dramatically, suggesting that the time is right to address how altered metabolism promotes cancer in the context of metabolic disease.
Acknowledgments
Work in the Thompson lab is supported by grants from the NCI and NIH. K.E.W. was supported by a postdoctoral fellowship from the Damon Runyon Cancer Research Foundation. We thank Chao Lu and Scott Olejniczak for critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anderson EJ, Lustig ME, Boyle KE, Woodlief TL, Kane DA, Lin CT, Price JW, 3rd, Kang L, Rabinovitch PS, Szeto HH, et al. Mitochondrial H2O2 emission and cellular redox state link excess fat intake to insulin resistance in both rodents and humans. J Clin Invest. 2009 doi: 10.1172/JCI37048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell EL, Klimova TA, Eisenbart J, Moraes CT, Murphy MP, Budinger GR, Chandel NS. The Qo site of the mitochondrial complex III is required for the transduction of hypoxic signaling via reactive oxygen species production. J Cell Biol. 2007;177:1029–1036. doi: 10.1083/jcb.200609074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrovnikova-Marjon E, Grigoriadou C, Pytel D, Zhang F, Ye J, Koumenis C, Cavener D, Diehl JA. PERK promotes cancer cell proliferation and tumor growth by limiting oxidative DNA damage. Oncogene. 2010;29:3881–3895. doi: 10.1038/onc.2010.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracha AL, Ramanathan A, Huang S, Ingber DE, Schreiber SL. Carbon metabolism-mediated myogenic differentiation. Nat Chem Biol. 2010;6:202–204. doi: 10.1038/nchembio.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandon M, Baldi P, Wallace DC. Mitochondrial mutations in cancer. Oncogene. 2006;25:4647–4662. doi: 10.1038/sj.onc.1209607. [DOI] [PubMed] [Google Scholar]
- Brunelle JK, Bell EL, Quesada NM, Vercauteren K, Tiranti V, Zeviani M, Scarpulla RC, Chandel NS. Oxygen sensing requires mitochondrial ROS but not oxidative phosphorylation. Cell Metab. 2005;1:409–414. doi: 10.1016/j.cmet.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Buchberger A, Bukau B, Sommer T. Protein quality control in the cytosol and the Endoplasmic Reticulum: Brothers in Arms. Mol Cell. 2010;40:xxx. doi: 10.1016/j.molcel.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Caldwell SA, Jackson SR, Shahriari KS, Lynch TP, Sethi G, Walker S, Vosseller K, Reginato MJ. Nutrient sensor O-GlcNAc transferase regulates breast cancer tumorigenesis through targeting of the oncogenic transcription factor FoxM1. Oncogene. 2010;29:2831–2842. doi: 10.1038/onc.2010.41. [DOI] [PubMed] [Google Scholar]
- Calle EE, Kaaks R. Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms. Nat Rev Cancer. 2004;4:579–591. doi: 10.1038/nrc1408. [DOI] [PubMed] [Google Scholar]
- Cheung P, Dennis JW. Mgat5 and Pten interact to regulate cell growth and polarity. Glycobiology. 2007;17:767–773. doi: 10.1093/glycob/cwm037. [DOI] [PubMed] [Google Scholar]
- Chou TY, Hart GW, Dang CV. c-Myc is glycosylated at threonine 58, a known phosphorylation site and a mutational hot spot in lymphomas. J Biol Chem. 1995;270:18961–18965. doi: 10.1074/jbc.270.32.18961. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Chu FF, Esworthy RS, Chu PG, Longmate JA, Huycke MM, Wilczynski S, Doroshow JH. Bacteria-induced intestinal cancer in mice with disrupted Gpx1 and Gpx2 genes. Cancer Res. 2004;64:962–968. doi: 10.1158/0008-5472.can-03-2272. [DOI] [PubMed] [Google Scholar]
- Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis. 2009;30:1073–1081. doi: 10.1093/carcin/bgp127. [DOI] [PubMed] [Google Scholar]
- Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullinan SB, Diehl JA. Coordination of ER and oxidative stress signaling: the PERK/Nrf2 signaling pathway. Int J Biochem Cell Biol. 2006;38:317–332. doi: 10.1016/j.biocel.2005.09.018. [DOI] [PubMed] [Google Scholar]
- Dansen TB, Burgering BM. Unravelling the tumor-suppressive functions of FOXO proteins. Trends Cell Biol. 2008;18:421–429. doi: 10.1016/j.tcb.2008.07.004. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis JW, Nabi IR, Demetriou M. Metabolism, cell surface organization, and disease. Cell. 2009;139:1229–1241. doi: 10.1016/j.cell.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edinger AL, Thompson CB. Akt maintains cell size and survival by increasing mTOR-dependent nutrient uptake. Mol Biol Cell. 2002;13:2276–2288. doi: 10.1091/mbc.01-12-0584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elchuri S, Oberley TD, Qi W, Eisenstein RS, Jackson Roberts L, Van Remmen H, Epstein CJ, Huang TT. CuZnSOD deficiency leads to persistent and widespread oxidative damage and hepatocarcinogenesis later in life. Oncogene. 2005;24:367–380. doi: 10.1038/sj.onc.1208207. [DOI] [PubMed] [Google Scholar]
- Frohlich DA, McCabe MT, Arnold RS, Day ML. The role of Nrf2 in increased reactive oxygen species and DNA damage in prostate tumorigenesis. Oncogene. 2008;27:4353–4362. doi: 10.1038/onc.2008.79. [DOI] [PubMed] [Google Scholar]
- Gallagher EJ, Leroith D. The proliferating role of insulin and insulin-like growth factors in cancer. Trends Endocrinol Metab. 2010 doi: 10.1016/j.tem.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Tchernyshyov I, Chang TC, Lee YS, Kita K, Ochi T, Zeller KI, De Marzo AM, Van Eyk JE, Mendell JT, Dang CV. c-Myc suppression of miR-23a/b enhances mitochondrial glutaminase expression and glutamine metabolism. Nature. 2009;458:762–765. doi: 10.1038/nature07823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao P, Zhang H, Dinavahi R, Li F, Xiang Y, Raman V, Bhujwalla ZM, Felsher DW, Cheng L, Pevsner J, et al. HIF-dependent antitumorigenic effect of antioxidants in vivo. Cancer Cell. 2007;12:230–238. doi: 10.1016/j.ccr.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloire G, Legrand-Poels S, Piette J. NF-kappaB activation by reactive oxygen species: fifteen years later. Biochem Pharmacol. 2006;72:1493–1505. doi: 10.1016/j.bcp.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Goldstein BJ, Mahadev K, Wu X. Redox paradox: insulin action is facilitated by insulin-stimulated reactive oxygen species with multiple potential signaling targets. Diabetes. 2005;54:311–321. doi: 10.2337/diabetes.54.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granovsky M, Fata J, Pawling J, Muller WJ, Khokha R, Dennis JW. Suppression of tumor growth and metastasis in Mgat5-deficient mice. Nat Med. 2000;6:306–312. doi: 10.1038/73163. [DOI] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Mi W, Ge Y, Liu H, Fan Q, Han C, Yang J, Han F, Lu X, Yu W. GlcNAcylation plays an essential role in breast cancer metastasis. Cancer Res. 2010;70:6344–6351. doi: 10.1158/0008-5472.CAN-09-1887. [DOI] [PubMed] [Google Scholar]
- Guzy RD, Hoyos B, Robin E, Chen H, Liu L, Mansfield KD, Simon MC, Hammerling U, Schumacker PT. Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 2005;1:401–408. doi: 10.1016/j.cmet.2005.05.001. [DOI] [PubMed] [Google Scholar]
- Guzy RD, Schumacker PT. Oxygen sensing by mitochondria at complex III: the paradox of increased reactive oxygen species during hypoxia. Exp Physiol. 2006;91:807–819. doi: 10.1113/expphysiol.2006.033506. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B. Oxidative stress and cancer: have we moved forward? Biochem J. 2007;401:1–11. doi: 10.1042/BJ20061131. [DOI] [PubMed] [Google Scholar]
- Hamanaka RB, Chandel NS. Mitochondrial reactive oxygen species regulate cellular signaling and dictate biological outcomes. Trends Biochem Sci. 2010 doi: 10.1016/j.tibs.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatzivassiliou G, Zhao F, Bauer DE, Andreadis C, Shaw AN, Dhanak D, Hingorani SR, Tuveson DA, Thompson CB. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8:311–321. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Healy SJ, Gorman AM, Mousavi-Shafaei P, Gupta S, Samali A. Targeting the endoplasmic reticulum-stress response as an anticancer strategy. Eur J Pharmacol. 2009;625:234–246. doi: 10.1016/j.ejphar.2009.06.064. [DOI] [PubMed] [Google Scholar]
- Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444:860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010;140:900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houstis N, Rosen ED, Lander ES. Reactive oxygen species have a causal role in multiple forms of insulin resistance. Nature. 2006;440:944–948. doi: 10.1038/nature04634. [DOI] [PubMed] [Google Scholar]
- Hussain SP, Hofseth LJ, Harris CC. Radical causes of cancer. Nat Rev Cancer. 2003;3:276–285. doi: 10.1038/nrc1046. [DOI] [PubMed] [Google Scholar]
- Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, Yamaguchi A, Imanishi H, Nakada K, Honma Y, Hayashi J. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661–664. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- Kalaany NY, Sabatini DM. Tumours with PI3K activation are resistant to dietary restriction. Nature. 2009;458:725–731. doi: 10.1038/nature07782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamata H, Honda S, Maeda S, Chang L, Hirata H, Karin M. Reactive oxygen species promote TNFalpha-induced death and sustained JNK activation by inhibiting MAP kinase phosphatases. Cell. 2005;120:649–661. doi: 10.1016/j.cell.2004.12.041. [DOI] [PubMed] [Google Scholar]
- Karin M, Greten FR. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat Rev Immunol. 2005;5:749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- Kaufman RJ, Scheuner D, Schroder M, Shen X, Lee K, Liu CY, Arnold SM. The unfolded protein response in nutrient sensing and differentiation. Nat Rev Mol Cell Biol. 2002;3:411–421. doi: 10.1038/nrm829. [DOI] [PubMed] [Google Scholar]
- Khasawneh J, Schulz MD, Walch A, Rozman J, Hrabe de Angelis M, Klingenspor M, Buck A, Schwaiger M, Saur D, Schmid RM, et al. Inflammation and mitochondrial fatty acid beta-oxidation link obesity to early tumor promotion. Proc Natl Acad Sci U S A. 2009;106:3354–3359. doi: 10.1073/pnas.0802864106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornfeld R, Kornfeld S. Assembly of asparagine-linked oligosaccharides. Annu Rev Biochem. 1985;54:631–664. doi: 10.1146/annurev.bi.54.070185.003215. [DOI] [PubMed] [Google Scholar]
- Lau KS, Dennis JW. N-Glycans in cancer progression. Glycobiology. 2008;18:750–760. doi: 10.1093/glycob/cwn071. [DOI] [PubMed] [Google Scholar]
- Lau KS, Partridge EA, Grigorian A, Silvescu CI, Reinhold VN, Demetriou M, Dennis JW. Complex N-glycan number and degree of branching cooperate to regulate cell proliferation and differentiation. Cell. 2007;129:123–134. doi: 10.1016/j.cell.2007.01.049. [DOI] [PubMed] [Google Scholar]
- Lee AS. GRP78 induction in cancer: therapeutic and prognostic implications. Cancer Res. 2007;67:3496–3499. doi: 10.1158/0008-5472.CAN-07-0325. [DOI] [PubMed] [Google Scholar]
- Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O’Donnell KA, Kim JW, Yustein JT, Lee LA, Dang CV. Myc stimulates nuclearly encoded mitochondrial genes and mitochondrial biogenesis. Mol Cell Biol. 2005;25:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh K, Deng H, Fukushima A, Cai X, Boivin B, Galic S, Bruce C, Shields BJ, Skiba B, Ooms LM, et al. Reactive oxygen species enhance insulin sensitivity. Cell Metab. 2009;10:260–272. doi: 10.1016/j.cmet.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou YW, Chen YY, Hsu SF, Chen RK, Lee CL, Khoo KH, Tonks NK, Meng TC. Redox regulation of the protein tyrosine phosphatase PTP1B in cancer cells. Febs J. 2008;275:69–88. doi: 10.1111/j.1742-4658.2007.06173.x. [DOI] [PubMed] [Google Scholar]
- Love DC, Hanover JA. The hexosamine signaling pathway: deciphering the “O-GlcNAc code”. Sci STKE. 2005;2005:re13. doi: 10.1126/stke.3122005re13. [DOI] [PubMed] [Google Scholar]
- Majmundar A, Wong WJ, Simon MC. Hypoxia inducible factors and the response to hypoxic stress. Mol Cell. 2010:40. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield KD, Guzy RD, Pan Y, Young RM, Cash TP, Schumacker PT, Simon MC. Mitochondrial dysfunction resulting from loss of cytochrome c impairs cellular oxygen sensing and hypoxic HIF-alpha activation. Cell Metab. 2005;1:393–399. doi: 10.1016/j.cmet.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- Marshall S. Role of insulin, adipocyte hormones, and nutrient-sensing pathways in regulating fuel metabolism and energy homeostasis: a nutritional perspective of diabetes, obesity, and cancer. Sci STKE. 2006;2006:re7. doi: 10.1126/stke.3462006re7. [DOI] [PubMed] [Google Scholar]
- Marshall S, Bacote V, Traxinger RR. Discovery of a metabolic pathway mediating glucose-induced desensitization of the glucose transport system. Role of hexosamine biosynthesis in the induction of insulin resistance. J Biol Chem. 1991;266:4706–4712. [PubMed] [Google Scholar]
- Meng TC, Fukada T, Tonks NK. Reversible oxidation and inactivation of protein tyrosine phosphatases in vivo. Mol Cell. 2002;9:387–399. doi: 10.1016/s1097-2765(02)00445-8. [DOI] [PubMed] [Google Scholar]
- Moenner M, Pluquet O, Bouchecareilh M, Chevet E. Integrated endoplasmic reticulum stress responses in cancer. Cancer Res. 2007;67:10631–10634. doi: 10.1158/0008-5472.CAN-07-1705. [DOI] [PubMed] [Google Scholar]
- Morrish F, Isern N, Sadilek M, Jeffrey M, Hockenbery DM. c-Myc activates multiple metabolic networks to generate substrates for cell-cycle entry. Oncogene. 2009;28:2485–2491. doi: 10.1038/onc.2009.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan C. Epidemic inflammation: pondering obesity. Mol Med. 2008;14:485–492. doi: 10.2119/2008-00038.Nathan. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann CA, Krause DS, Carman CV, Das S, Dubey DP, Abraham JL, Bronson RT, Fujiwara Y, Orkin SH, Van Etten RA. Essential role for the peroxiredoxin Prdx1 in erythrocyte antioxidant defence and tumour suppression. Nature. 2003;424:561–565. doi: 10.1038/nature01819. [DOI] [PubMed] [Google Scholar]
- Nogueira V, Park Y, Chen CC, Xu PZ, Chen ML, Tonic I, Unterman T, Hay N. Akt determines replicative senescence and oxidative or oncogenic premature senescence and sensitizes cells to oxidative apoptosis. Cancer Cell. 2008;14:458–470. doi: 10.1016/j.ccr.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novosyadlyy R, Lann DE, Vijayakumar A, Rowzee A, Lazzarino DA, Fierz Y, Carboni JM, Gottardis MM, Pennisi PA, Molinolo AA, et al. Insulin-mediated acceleration of breast cancer development and progression in a nonobese model of type 2 diabetes. Cancer Res. 2010;70:741–751. doi: 10.1158/0008-5472.CAN-09-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell N, Zachara NE, Hart GW, Marth JD. Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol Cell Biol. 2004;24:1680–1690. doi: 10.1128/MCB.24.4.1680-1690.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo K, Takamatsu S, Minowa MT, Yoshida A, Takeuchi M, Marth JD. Dietary and genetic control of glucose transporter 2 glycosylation promotes insulin secretion in suppressing diabetes. Cell. 2005;123:1307–1321. doi: 10.1016/j.cell.2005.09.041. [DOI] [PubMed] [Google Scholar]
- Park EJ, Lee JH, Yu GY, He G, Ali SR, Holzer RG, Osterreicher CH, Takahashi H, Karin M. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell. 2010;140:197–208. doi: 10.1016/j.cell.2009.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge EA, Le Roy C, Di Guglielmo GM, Pawling J, Cheung P, Granovsky M, Nabi IR, Wrana JL, Dennis JW. Regulation of cytokine receptors by Golgi N-glycan processing and endocytosis. Science. 2004;306:120–124. doi: 10.1126/science.1102109. [DOI] [PubMed] [Google Scholar]
- Petros JA, Baumann AK, Ruiz-Pesini E, Amin MB, Sun CQ, Hall J, Lim S, Issa MM, Flanders WD, Hosseini SH, et al. mtDNA mutations increase tumorigenicity in prostate cancer. Proc Natl Acad Sci U S A. 2005;102:719–724. doi: 10.1073/pnas.0408894102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phang JM, Liu W, Zabirnyk O. Proline metabolism and microenvironmental stress. Annu Rev Nutr. 2010;30:441–463. doi: 10.1146/annurev.nutr.012809.104638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renehan AG, Frystyk J, Flyvbjerg A. Obesity and cancer risk: the role of the insulin-IGF axis. Trends Endocrinol Metab. 2006;17:328–336. doi: 10.1016/j.tem.2006.08.006. [DOI] [PubMed] [Google Scholar]
- Roberts CK, Sindhu KK. Oxidative stress and metabolic syndrome. Life Sci. 2009;84:705–712. doi: 10.1016/j.lfs.2009.02.026. [DOI] [PubMed] [Google Scholar]
- Sablina AA, Budanov AV, Ilyinskaya GV, Agapova LS, Kravchenko JE, Chumakov PM. The antioxidant function of the p53 tumor suppressor. Nat Med. 2005;11:1306–1313. doi: 10.1038/nm1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahar S, Sassone-Corsi P. Metabolism and cancer: the circadian clock connection. Nat Rev Cancer. 2009;9:886–896. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- Schieke SM, Phillips D, McCoy JP, Jr, Aponte AM, Shen RF, Balaban RS, Finkel T. The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J Biol Chem. 2006;281:27643–27652. doi: 10.1074/jbc.M603536200. [DOI] [PubMed] [Google Scholar]
- Schwer B, Verdin E. Conserved metabolic regulatory functions of sirtuins. Cell Metab. 2008;7:104–112. doi: 10.1016/j.cmet.2007.11.006. [DOI] [PubMed] [Google Scholar]
- Sen CK, Packer L. Antioxidant and redox regulation of gene transcription. Faseb J. 1996;10:709–720. doi: 10.1096/fasebj.10.7.8635688. [DOI] [PubMed] [Google Scholar]
- Shafi R, Iyer SP, Ellies LG, O’Donnell N, Marek KW, Chui D, Hart GW, Marth JD. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci U S A. 2000;97:5735–5739. doi: 10.1073/pnas.100471497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Tomic J, Wen F, Shaha S, Bahlo A, Harrison R, Dennis JW, Williams R, Gross BJ, Walker S, et al. Aberrant O-GlcNAcylation characterizes chronic lymphocytic leukemia. Leukemia. 2010 doi: 10.1038/leu.2010.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoelson SE, Lee J, Goldfine AB. Inflammation and insulin resistance. J Clin Invest. 2006;116:1793–1801. doi: 10.1172/JCI29069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slawson C, Copeland RJ, Hart GW. O-GlcNAc signaling: a metabolic link between diabetes and cancer? Trends Biochem Sci. 2010 doi: 10.1016/j.tibs.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slawson C, Zachara NE, Vosseller K, Cheung WD, Lane MD, Hart GW. Perturbations in O-linked beta-N-acetylglucosamine protein modification cause severe defects in mitotic progression and cytokinesis. J Biol Chem. 2005;280:32944–32956. doi: 10.1074/jbc.M503396200. [DOI] [PubMed] [Google Scholar]
- Teo CF, Wollaston-Hayden EE, Wells L. Hexosamine flux, the O-GlcNAc modification, and the development of insulin resistance in adipocytes. Mol Cell Endocrinol. 2010;318:44–53. doi: 10.1016/j.mce.2009.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat Rev Drug Discov. 2009;8:579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- Trachootham D, Zhou Y, Zhang H, Demizu Y, Chen Z, Pelicano H, Chiao PJ, Achanta G, Arlinghaus RB, Liu J, Huang P. Selective killing of oncogenically transformed cells through a ROS-mediated mechanism by beta-phenylethyl isothiocyanate. Cancer Cell. 2006;10:241–252. doi: 10.1016/j.ccr.2006.08.009. [DOI] [PubMed] [Google Scholar]
- Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- Turrens JF. Mitochondrial formation of reactive oxygen species. J Physiol. 2003;552:335–344. doi: 10.1113/jphysiol.2003.049478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veal EA, Day AM, Morgan BA. Hydrogen peroxide sensing and signaling. Mol Cell. 2007;26:1–14. doi: 10.1016/j.molcel.2007.03.016. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010a;327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Udeshi ND, Slawson C, Compton PD, Sakabe K, Cheung WD, Shabanowitz J, Hunt DF, Hart GW. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Sci Signal. 2010b;3:ra2. doi: 10.1126/scisignal.2000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M, Kalyanaraman B, Mutlu GM, Budinger GR, Chandel NS. Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity. Proc Natl Acad Sci U S A. 2010;107:8788–8793. doi: 10.1073/pnas.1003428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Hotamisligil GS. Inflammation, stress, and diabetes. J Clin Invest. 2005;115:1111–1119. doi: 10.1172/JCI25102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan SA, Dias WB, Thiruneelakantapillai L, Lane MD, Hart GW. Regulation of insulin receptor substrate 1 (IRS-1)/AKT kinase-mediated insulin signaling by O-Linked beta-N-acetylglucosamine in 3T3-L1 adipocytes. J Biol Chem. 2010;285:5204–5211. doi: 10.1074/jbc.M109.077818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan SA, Lane MD, Hart GW. Regulation of the O-linked beta-N-acetylglucosamine transferase by insulin signaling. J Biol Chem. 2008;283:21411–21417. doi: 10.1074/jbc.M800677200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, Thompson CB. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WS. The signaling mechanism of ROS in tumor progression. Cancer Metastasis Rev. 2006;25:695–705. doi: 10.1007/s10555-006-9037-8. [DOI] [PubMed] [Google Scholar]
- Yang WH, Kim JE, Nam HW, Ju JW, Kim HS, Kim YS, Cho JW. Modification of p53 with O-linked N-acetylglucosamine regulates p53 activity and stability. Nat Cell Biol. 2006;8:1074–1083. doi: 10.1038/ncb1470. [DOI] [PubMed] [Google Scholar]
- Yang X, Ongusaha PP, Miles PD, Havstad JC, Zhang F, So WV, Kudlow JE, Michell RH, Olefsky JM, Field SJ, Evans RM. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature. 2008;451:964–969. doi: 10.1038/nature06668. [DOI] [PubMed] [Google Scholar]
- Zachara NE, Hart GW. O-GlcNAc a sensor of cellular state: the role of nucleocytoplasmic glycosylation in modulating cellular function in response to nutrition and stress. Biochim Biophys Acta. 2004;1673:13–28. doi: 10.1016/j.bbagen.2004.03.016. [DOI] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]