Abstract
Forensic dentistry as a science has evolved from simple methods of age estimation and bite-mark analysis, to a new era of genetic and serological investigations. DNA analysis in forensic science requires a sample or source from either an individual (living or dead) or a crime/incident site. The orofacial region is a good source of such material, due to the fact that certain oral tissues are relatively resistant to environmental degradation and destruction by thermal, electrical, and mechanical insult. Dentists may be called upon to provide samples and expert analysis in many such situations. Sources include soft and hard tissues of teeth and jaws, saliva, biopsy material, and mucosal swabs. Tissue samples should be handled with care, and correct protocol in collection and preparation has to be followed. This ensures a high yield of the required DNA. Hard tissues like teeth require specialized procedures to extract the genetic material. Research has shown that there is a wide variation in the quality and quantity of DNA extracted from different individuals from the same site even under similar conditions. This necessitates calibration of the various methods to achieve best results. DNA analysis can provide highly accurate identification if used correctly. Here a description of the various sources in the oral region has been provided from which samples could be forwarded to the forensic laboratory. Most commonly employed techniques of collection and handling for laboratory procedures have been outlined.
Keywords: DNA, forensic dentistry, saliva, teeth
Introduction
Dental professionals are increasingly being employed in expert assessment and testimony in legal proceedings. As the boundary between forensic science and forensic dentistry blurs in part due to technological advances, dentists are impelled to expand their frontiers of knowledge and practice to confront challenging forensic situations.[1,2]
Crime scenes present potential sources of DNA from victims, suspects, and inanimate objects in the vicinity [Figure 1]. The source yielding the highest DNA content is whole blood; however, it may not be available in all situations. The collection and storage of blood requires high-level aseptic protocol and trained personnel. Moreover, in certain criminal jurisprudence systems, blood collection from a suspect may be denied due to privacy laws.[3,4]
Figure 1.
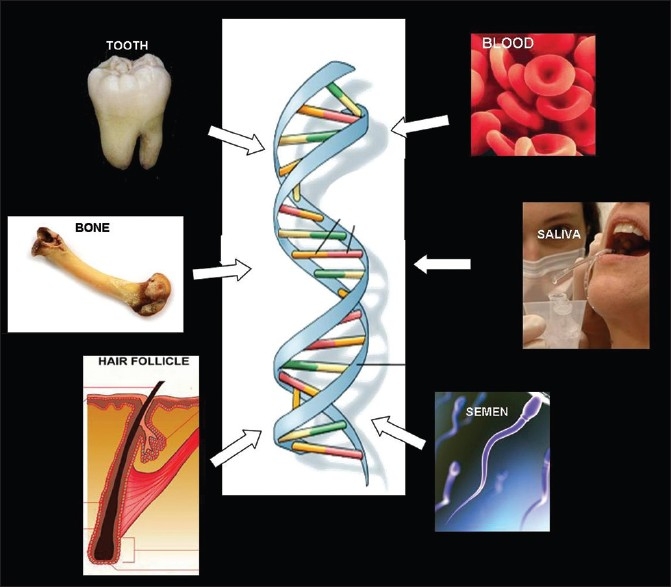
Sources of DNA for forensic analysis
Therefore, noninvasive techniques are more acceptable and popular. The oral cavity is a useful source of evidence. Dentists may be required to provide samples for DNA analysis in many cases. The sources include saliva, mucosal swabs, and teeth [Figure 2].
Figure 2.
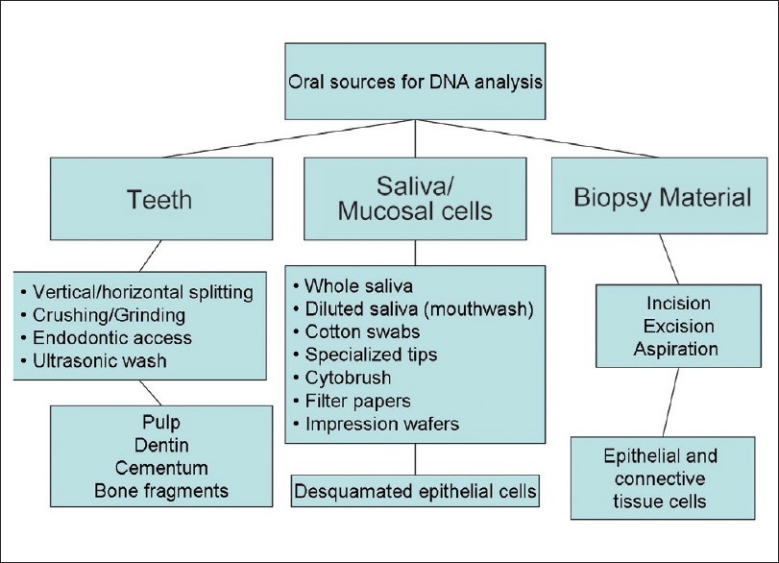
Methods of sample collection in the oral region
DNA techniques involve the detection, quantification, and analysis of DNA from the nucleus and mitochondria. Nuclear DNA is representative of both paternal and maternal inheritance. Mitochondrial DNA (mtDNA) is derived from the ovum and hence purely maternal. In teeth especially, the dentin consists of cellular extensions (Tome's fibers) rich in mitochondria. Dentin powder is therefore presumably a good source of mtDNA.
This article discusses various methods of collection and preparation of such samples for arriving at the result with minimal difficulty and delay. Laboratory procedures and few cases are mentioned. There is a note on the ethical and legal considerations involved in research and court proceedings.
The tooth: Handling, storage, and preparation
The teeth are useful for evidence as the genetic material is protected by the calcified tissues. They are obtained from anthropological surveys and from disaster/crime sites. Teeth are usually cleaned and stored in normal saline and may or may not be refrigerated, depending on time and facilities. In the teeth, the sources of DNA are the pulp, dentin, cementum, periodontal fibers, and attached bone fragments.
Pulp tissue is most commonly used because it is usually abundant and has least chance of contamination by nonhuman DNA. Sampling of the pulp tissue is done in three ways: crushing, vertical or horizontal splitting, and by endodontic access.[5]
Crushing or grinding is not advisable as it gives no scope for further evaluation of the tooth morphology and histology. Moreover, the crushed sample has to undergo multiple cycles of decalcification and purification, and usable quantity of DNA may not be obtained. This is done as a last resort. A complete documentation of the tooth morphology, including photographs from multiple angles, and radiographs, are mandatory before grinding.
Sweet and Hildebrand have pioneered the use of cryogenic grinding of teeth. This technique involved the pulverization of a cleaned tooth in a freezer mill cooled by liquid nitrogen. The workers reported that their method greatly minimizes the risk of contamination and takes as little as 2 min. The powder was solubilized in a proteinase K buffer solution and incubated overnight. Slot blot hybridization was employed to quantify the DNA. The yield of DNA per gram of tooth powder was 18.4 μg.[5]
Vertical or longitudinal splitting of the tooth is done in many cases. This method provides for easy excavation of the entire pulpal tissue with minimal chance of contamination. Crown morphology may be lost. Splitting is done with carborundum discs, with heavy-duty gloves, preferably under a hood, as aerosol may contact the skin or eyes or can be inhaled. The tooth is held securely or may be mounted in dental stone. The carborundum discs are used to split the tooth from the incisal edge, with frequent washing with distilled water. When the pulp cavity is reached, the tooth may be split by a chisel to avoid heat and mechanical damage to the pulp tissue. The pulp is then excavated and transferred to vials. The sliced tooth can be used for histological evaluation [Figure 3].
Figure 3.
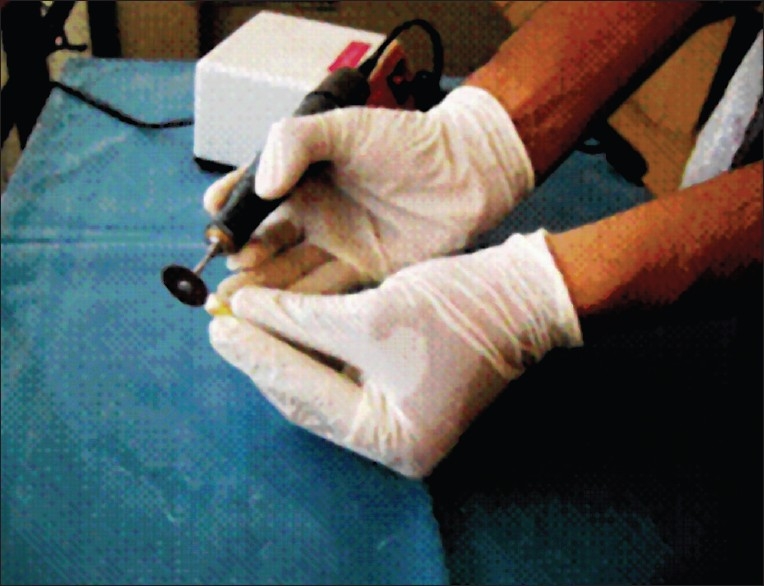
Tooth splitting for pulp extraction
Horizontal splitting is essentially the same procedure and is done when the crown structure is to be preserved. The root is sectioned and sampling of the pulp, dentin, and cementum can be done. The other method is preparing a conventional access cavity on the tooth and excavating the pulp using a suitable hand instrument.[5]
Pulp DNA isolation
After the pulp tissue is obtained, it is placed in Eppendorf vials. One milliliter of sterile deionized water is added and centrifuged three times at 5 min interval. An extraction buffer containing Tris HCl and EDTA is added and incubated with proteinase K for 18 hrs. The contents are subject to multiple stages of centrifugation and purification to obtain the final sediment. This is added to a 1% agarose gel in TBE (Tris HCL, boric acid, and EDTA) buffer and prepared for gel electrophoresis.[6]
Pulp tissue is the easiest to prepare and analyze. However in many cases, the tooth will be devoid of the pulp, or may be endodontically filled. It may also be contaminated by micro-organisms, which introduce nonhuman DNA into the sample. In such cases, dentin or cementum is used to extract DNA. The possibility of encountering nonhuman DNA is lesser in calcified tissues, except in cases of carious lesions. The organic material being calcified, there is more probability of recovering intact DNA. The dentin and cementum samples are obtained by grinding the root, either as a pure dentin powder or as a dentin-cementum powder. The powder is then solubilized and stored in vials. A similar procedure is followed for the extraction of DNA as for pulp tissue.
As previously discussed, dentin powder is a good source of mtDNA. This is more useful than genomic or nuclear DNA. mtDNA has only 13 genes, compared to almost 100,000 in nuclear DNA. Cells contain a high number of mtDNA copies which are survivable for prolonged periods compared to nuclear DNA. It is purely maternal and therefore best for identifying relations and lineage, with case examples dating up to thousands and even million years.[7]
The ultrasonication of teeth without splitting, crushing, or otherwise damaging a tooth, has been found to yield the genetic material for DNA analysis. However, the quantity obtained is usually insufficient and may be used in limited cases when other methods are unjustified.[5]
The techniques for using bone are essentially the same as that for teeth. Crushing is most commonly done. Bone marrow extraction may also be performed.
Sampling and preparation of mucosal cells – Saliva
The objective of using saliva as a source is to analyze DNA from desquamated epithelial cells. Saliva is rich in DNA, being flush with such cells. Whole saliva is collected normally either as resting or stimulated (chewing on paraffin or using a small citric acid crystal). Diluted saliva can also be obtained from the subject by rinsing the mouth with a specified volume of mouthwash and spitting into a test tube. The Oragene test kit (DNA Genotek Inc., Canada) is now increasingly being employed as a simple and safe method for sample collection. It consists of a customized test tube with a flappable lid which has a compartment containing the Oragene solution (for stabilizing DNA at room temperature). Saliva is filled up to the marked level and the lid is then closed, piercing the lid compartment, resulting in the mixing of saliva and the liquid. The test tube is then taken to the lab under normal or refrigerated transport conditions.[8]
The interval between the collection of saliva and the DNA extraction procedure does not significantly affect the result. Studies have shown that the purity of DNA samples and PCR results were unaffected whether the procedure was done in the same day, or after a week at 4°C, or a month at –70°C. The quantification of DNA however was different under various conditions.[9]
The saliva thus collected can be dabbed onto specialized cards. The FTA cards (Flinders Technology Associates, manufactured by Whatman Co, GE Healthcare, UK) are chemically treated filter papers designed for the collection and room temperature storage of biologic samples for molecular analyses; these cards protect DNA and are impregnated with denaturants that guard against oxidation, nuclease and ultraviolet damage, and both bacterial and fungal degradation [Figure 4]. FTA cards provide a convenient and safe method for the collection and storage of DNA from whole blood, tissues, buccal swabs, viral, and bacterial specimens. Beckett et al. observed that DNA from buccal swab samples immobilized on FTA cards remained stable for years at room temperature and therefore, delay in sending the sample to the laboratory did not affect the integrity of the analysis.[10]
Figure 4.

FTA cards
The presence of an ecosystem of microflora in the oral cavity can introduce a huge load of nonhuman DNA (usually bacterial and fungal) into the sample. The newer DNA analysis methods can usually compensate for this and are highly specific. Real-time polymerase chain reaction (RT-PCR) is widely used. Other methods like spectrophotometry and fluorometry have failed to discriminate between human and bacterial DNA.
Sampling of saliva by these methods has been found to yield the highest content of DNA with the lowest rate of fragmentation. Thus, PCR is the most recommended method.
Some studies have identified problems with the PCR procedure especially when the specialized cards have not been used. The problem has been attributed to the presence of PCR inhibitors in saliva. Salivary enzymes or bacterial products can retard amplification and polymerase reactions. The use of FTA cards and repeated washing of the punch specimen appeared to greatly minimize the inhibition.[10]
Sampling and preparation of mucosal cells – Swabs
Swabbing the inside of the mouth with a cotton tip is widely used to obtain samples for DNA. The exfoliated mucosal cells can also be collected using Dacron-coated tips, foam-coated tips, or tongue depressors. The usual technique is to clean the oral cavity with water and a mouthwash and use a firm, repeated scraping motion to collect samples.
Cytobrushes can be used to collect living cells. The cytobrush is pressed gently but firmly on the site, rotated once, and removed. This technique extracts the basal and suprabasal cells with a high nuclear content.
Swabbing and cytobrushes have been found to have greater compliance from young children who may not cooperate with the saliva sampling. There is a presence of nonhuman DNA in samples collected by these methods, but the load is much less compared to salivary samples.[11]
Burger et al. attempted a new method of buccal cell sampling called the buccal DNA collector, which involves the use of a filter paper attached to a plastic handle. A simple swipe and dry technique can provide a sample ready for analysis. These workers found that, even though more number of swipes increased the DNA yield, sufficient DNA was obtained just from swiping twice. The study identified subjects as high shedders and low shedders based on the DNA yield. Repeated sampling from the same site was found to produce decreasing yields.[12]
Salivary sampling from bite-marks and other sources
Salivary deposits left behind as a result of bites can also be a potential source of DNA of a suspect. Depending on the duration, swabbing for sampling may be an additional method after bite-mark replication, especially when impression records are not available. Dried saliva may be difficult to detect, requiring the use of an amylase assay to confirm its presence.
A reliable method for saliva sampling from bite-mark sites in skin is the double swabbing technique. Sweet and co-workers reported that salivary deposits could be extracted by first swabbing with a wet cotton pellet and then using a dry cotton pellet. This technique presumably rehydrates the cells in the salivary deposit. Both the dry and the wet cotton pellets are used for collection and transfer without the risk of contamination or deterioration.[13]
Saliva may also be isolated from various sources in the crime scene, for example, postage stamps and envelopes, glasses, cigarettes, straws, food and chewing gum, toothbrushes and dental floss, and dental impressions.[14,15]
Impression wafers are employed for bite registration as part of prosthodontic and other treatment procedures. They may be of use, however, as a limited source of saliva. Ellis and co-workers determined the yield and quality of DNA obtained from impression wafers that were used for bite registration. Though the DNA quantity was lesser compared to that from buccal swabs and mouthwash rinses, typing for analysis and identification was possible. This is mainly due to the fact that as low as 0.001 μg of sample DNA is sufficient for analysis by RT-PCR.[16]
DNA samples were obtained by using cigarette butts in many criminal cases. Cigarette butts contain many PCR inhibitors like phenolic/tar compounds and color and flavor additives, and therefore gave negative results in the past. Researchers now have developed specific kits for analysis of cigarette butt samples. Hochmeister and co-workers have achieved success in DNA-based identification using cigarettes. In their study, 1 cm of the circumference of the cigarette butt paper was removed using sterile scissors and forceps. This paper was cut into quarters. Each quarter was again cut into four sections and the paper added to a well of a 96-well tray or a 0.2 ml PCR tube. After amplification by PCR, agarose gel electrophoresis was performed for visualization.[17]
Biopsy
Biopsy is the surgical removal of tissue from a living organism for the purpose of microscopic examination and diagnosis. Tissue specimens, especially frozen samples, can be used for genetic analysis. The degradation of the genetic material in the conventional formalin-fixed tissue may limit the usefulness of this source specifically for DNA analysis. This method is rarely used unless it is the only available source.
Laboratory Methods
DNA extraction and quantification - Saliva samples (Oragene kit)
The saliva sample in the vial is incubated at 50°C for 1 hr. About 500 μl of the Oragene/saliva sample is transferred to a 1.5 μl Eppendorf tube. A total of 20 μl of the Oragene purifier (supplied with the kit) is added to each sample, and mixed with inversion. Samples are incubated on ice for 10 min and centrifuged for 3 min at 13,000 rpm. The clear supernatant from each sample is micro-pipetted into a fresh tube and the protein pellet is discarded. A total of 500 μl of 95% ethanol is then added and mixed. The solution is left for 10 min to allow the DNA to precipitate. Centrifugation for 1 min at 13,000 rpm is done again.[8]
The ethanol supernatant is removed, leaving the DNA pellet at the bottom of the Eppendorf tube undisturbed. Samples are centrifuged again for 10 s at 13,000 rpm and more ethanol is removed. A total of 100 μl of the buffer is added to each sample, to dissolve the DNA pellet. Samples are vigorously spun and incubated at 50°C for 10 min to dissolve the DNA. After overnight incubation, samples are taken to the spectrophotometer. Absorbance values at 230, 260, and 280 nm are recorded. These values give the DNA concentration in the given sample.[8]
Polymerase chain reaction procedure
This is a method by which demonstrable quantities of DNA can be generated using the given sample as a template. This is done using an enzyme DNA polymerase and primer sequences of DNA. PCR is useful in cases of very minute and degraded samples to increase the quantity of DNA. Selected sequences required by the forensic scientists can be amplified preferentially. After amplification, the determination of sequences is performed. In forensic science, the laborious DNA typing procedure, the restriction fragment length polymorphism (RFLP) method, has given way to newer techniques like STR (short tandem repeat) analysis.
Also known as simple sequence repeats (SSRs) and microsatellites, STRs are polymorphic sequences of DNA (200–500 base pairs [bp]) that are remarkably conserved among species and are similar especially in first-degree relatives. This technique is especially suited for determining the identity of suspects, victims, and human remains. Developed countries have maintained a national DNA database, and the STR procedure has simplified the work for forensic and other scientists in DNA-based identification.[18,19]
SRT analysis has reduced the susceptibility to contaminant (nonhuman DNA) amplification and helps in analyzing complex sample mixtures. Profiling is performed using gel (agarose, polyacrylamide) electrophoresis, capillary electrophoresis, and chromatography. Electrophoresis is time consuming and requires considerable manual labor. Chromatography therefore is increasingly popular. STR-based systems provide excellent sensitivity down to less than 0.20 ng of DNA.[20]
The automated fluorescent detection of DNA fragments in the sample is done by the “multiplex” method. Several DNA regions can be labeled simultaneously with a different fluorescent colored tag in a single reaction tube during the amplification process. Electrophoresis units use a series of capillaries that direct a single sample at a time through a chemical matrix which contains a polymer “sieve.” Fragments migrate through the capillary according to size. DNA fragments are precisely sized, standardized, and relevant information is entered into a database as they move from point of origin to the detector.[2]
Precision and accuracy are afforded through the use of an internal sizing standard run in the same lane or capillary with each STR sample. The internal lane standard acts as a molecular ruler and is recognized by the computer to generate a fragment size calibration curve. This procedure enables an accurate assessment of not only the size of all the DNA fragments but also the quantity of the DNA fragment based on the amount of a fluorescent signal. The internal lane standard is a precision standard for evaluating any potential aberrant electrophoretic migration pattern. With the aid of the computer and precise digital sizing data, the forensic scientist evaluates each fragment for match or nonmatch [Figure 5]. Future methods include biochip and micro-assay technologies.[2]
Figure 5.
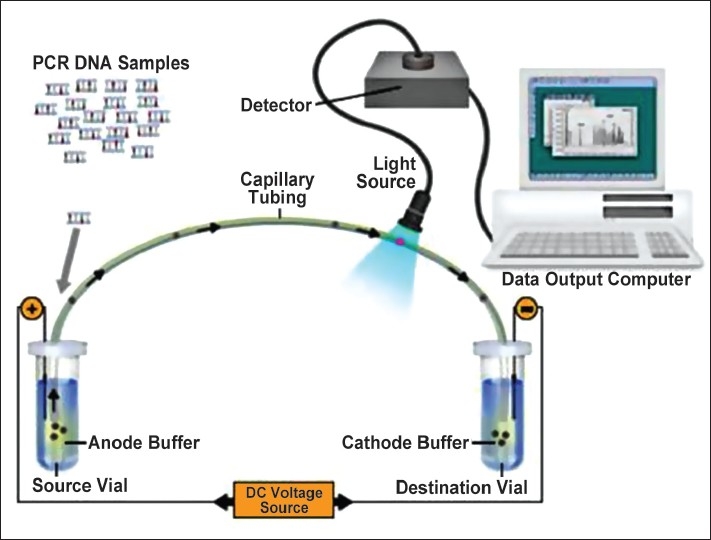
Steps in DNA analysis
There are many sequences in the human genome that can be tagged and analyzed for identification. Scientists use 9–16 sequences in a sample. The larger the number of samples, the higher the accuracy. For example, when using 13 loci, the odds of two different individuals having similar profile are 1 in 250 billion. Markers like D21S11, D7S820, TH01, D13S317, and D19S433 are commonly used for resolving disputed parentage. Y-STR analysis can be used in the case of paternally related males. Mitochondrial markers can be used in the case of maternal lineage.[2]
Amelogenins for gender determination
Amelogenins are proteins essential for the formation of enamel matrix. Genes coding for the proteins are present in the X and Y chromosomes. The AMEL genes are chromosome specific, having 106 bp in X and 112 bp in Y. This provides a basis for the determination of gender in DNA analysis. Moreover, these 106/112 bp AMEL amplicons have been found to be more reliable than other amplicons like telomerases and beta-globins traditionally used for DNA analysis. Tests employing the AMEL genes are rapid, more accurate, and require a small sample quantity.[21,22]
However, some studies have cast doubts on the reliability of the AMEL gene analysis. There are reports of allele dropout of the X amelogenin gene in males due to the polymorphism of primer binding sites. A high prevalence of deletions and other variations in the Y chromosome is known, some of which affect Yp11.2, the locus of the AMEL gene. This results in males being mistakenly identified as females because of a similar AMEL profile. The overall frequency of such errors in gender identification had been as low as 0.006% in males and 0.015% in females. But the errors were observed to be higher in Indian studies (3.2–3.6%) and also in Malaysian (0.6–0.88%) and Austrian groups (0.018%).[23,24]
The gold standard for gender determination is Y-STR and SRY (sex-determining regions of Y) testing and these tests have to be performed in conjunction with the AMEL testing.[24]
A recent study has used the exfoliated epithelial cells recovered from acrylic dentures to extract DNA, and performed SRY analysis for gender determination. The workers reported that such samples were successful in DNA detection and quantification. They reviewed conditions that might give contradictory results, as in chromosomal disorders (Klinefelter syndrome, Turner syndrome, XYY syndrome, and others), chimerism (due to marrow transplantation), and microchimerism, e.g., in pregnancy (male fetus), and organ transplantations. They recommended that techniques employing SRY should be tested for reliability in all the above-mentioned conditions.[25]
Workers have determined the gender of individuals by detecting Barr bodies in the nuclei of the exfoliated/cytobrush sampled cells in buccal smears. Barr bodies are the result of lyonization, in which one of the X chromosomes in female cells gets inactivated and undergoes telomere fusion and condensation. They are usually seen as an extension of the nuclear chromatin material. Positive identification of Barr bodies has been done in 20–78% of cells in females and up to 4% in males. Therefore, it is evident that while the presence of Barr bodies is not confirmatory of the female gender, a high percentage of Barr body-positive cells is characteristic of females. The main disadvantage of this method is the deterioration of the nuclear material with the passage of time. However, this method is simple and inexpensive.[26]
Creditable work of forensic dentists in solving cases
Forensic dentists have helped law enforcement in solving innumerable cases, especially when it seemed impossible to uncover the facts. Paul Revere was probably the first forensic dentist, who helped identify the dead soldiers of the American Revolutionary War. In 1954, a suspect was convicted of a murder after he was made to give a bite-mark sample on cheese to compare with a similar piece at the crime scene. In 1975, a man was convicted of murder based on bite-marks made on the victim's nose.[27]
D Sweet and CHW Sweet were one of the pioneers in DNA-based identification. They reported a case of a completely burnt human body in which the chance of sample collection was next to impossible. However, they discovered an unerupted third molar in the mandible of the victim. They were able to extract the pulp sample from that tooth, which was reasonably preserved, and performed DNA analysis. The amount of DNA recovered was about 1.3 μg, which was more than enough for amplification and typing.[28]
In a recent case of sexual assault in the USA, investigators employed DNA analysis to nab the criminal using the cigarette he smoked during his interrogation for an unrelated case. Forensic scientists use DNA analysis kits specifically designed to receive cigarette butt samples for quicker and standardized results.[29]
There were more than 20,000 specimens of human remains recovered after the 9/11 attacks. Only about half of the victims were identified by 2005 and the rest were considered unidentifiable. But in 2007, the processes were restarted as an advanced STR-based technology became available and even tiny and degraded samples of teeth and bones became adequate for identification.[27]
The Indian Army has initiated DNA profiling of all its soldiers in 2011. This database is to be of invaluable use to identify soldiers for purposes of record, as well as to present the remains to their families. Based on the blood spot tags issued by the US Army, they can be used for identification.[30]
Legal and ethical issues
The forensic dentist is required to follow guidelines set by the regional ethical committee and the law enforcement authority. This is important in medicolegal issues both for the acceptance of forensic evidence and to shield the dentist against claims of malpractice, manipulation, and other liabilities. The American Board of Forensic Odontologists (ABFO) has published manuals to its diplomats detailing the standard guidelines for such procedures. The New Biology and Criminal Investigation unit (NBCI) has given exhaustive legal guidelines for all investigative clinical and laboratory procedures.[31,32]
The code of ethics
As a means to promote the highest quality of personal and professional conduct of ABFO members, the code of ethics was promulgated, which is endorsed and adhered to by all diplomats of the ABFO, and similar rules are followed by a majority of counterpart organizations around the world. Every diplomat
shall refrain from any material misrepresentation of education, training, or area of expertise;
shall refrain from any material misrepresentation of data upon which an expert opinion or conclusion is based;
must provide a signed and notarized ethics statement adopted by the executive committee;
should maintain his/her professional competency through existing programs of continuing education;
may submit formal written allegations of violations concerning a fellow diplomat with proof and correct interpretations;
shall render technically correct statements in all written and oral reports, testimony, public addresses, or publications, and should avoid any misleading or inaccurate claims.[31]
The summary of scientific reasoning is as follows:
Collect all the relevant information needed.
Use the information collected. Do not selectively ignore evidence or place unwarranted credence in unsound or irrelevant information.
Do not allow opinion to be contaminated by unblinding or bias. Maintain impartiality.
Use knowledge effectively when making interpretations, drawing inferences or promulgating an opinion.
In the case of examination of a suspect, the following should be strictly performed:
Obtain a copy of the relevant legal document, e.g., warrant, for the examination of the suspect.
Obtain dental records and take a detailed case history, especially relating to the time of the alleged crime.
Detailed extraoral and intraoral examination with photographs and radiographs.
Make impressions and sample bites.
Sample for DNA analysis.[31]
It should be remembered that voluntary buccal swabs are considered nonintimate samples, while forced buccal swabs, saliva, and dental impressions are considered intimate procedures and may require magistrate warrants. Written consent is also required except under specified conditions.
The contents of a forensic report should be prescribed in a standard format and should cover the following items:
the qualification of the analyst
items examined
result
statistical analysis and
conclusion.[32]
Standard analysis techniques accepted by courts should be followed as scrupulously as possible. Alternative methods have to be verifiable and validated as to their application in the concerned case.
The regional ethics committee monitors all experts and conducts relevant proceedings relating to all issues in this regard. If all guidelines are strictly adhered to, the dental expert will be regarded as a competent authority and will be insulated from any liability. If not followed, the expert might face disciplinary action by the peer board as well as civil/criminal lawsuits.
Conclusion
The oral region is a rich source for simple and noninvasive methods of DNA sampling. The most common method in the case of criminal investigations is buccal swabbing, which is very useful in determining suspects in cases of homicide and physical/sexual assault. In cases of mass disaster and other calamities, skeletal and dental remains are helpful in establishing identity. Whenever dental professionals are required or requested to provide such samples, knowledge of sampling and storage for the optimization of the DNA analysis procedure can greatly enhance the efficiency of the investigation. The dental expert needs to be aware of relevant legal and ethical issues and operate within the ambit of the law.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared
References
- 1.Smith BC, Holland MM, Sweet D, DiZinno JA. DNA and the forensic odontologist. In: Bowers CM, Bell GL, editors. Manual of forensic odontology. 3rd ed. Colorado Springs: American society of forensic odontology; 1997. pp. 283–98. [Google Scholar]
- 2.Fourney RM. Forensic reality and the practical experience of DNA typing update. Article provided to the international society for the reform of criminal law 2002. National DNA data bank of Canada, Forensic Laboratory Services. [cited in 2002]. Available from: http://www.isrcl.org/Papers/Fourney.pdf .
- 3.Leung KK. Forensic Odontology. Dent Bull Hong Kong Med Diary. 2008;13:16–20. [Google Scholar]
- 4.Silva RH, Peres AS, Oliveira RN, Oliveira FT, Sales-Peres SH. Use of DNA technology in forensic dentistry. J App Oral Sci. 2007;15:156–61. doi: 10.1590/S1678-77572007000300002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sweet DJ, Hildebrand DP. Recovery of DNA from human teeth by cryogenic grinding. J Forensic Sci. 1998;43:1199–202. [PubMed] [Google Scholar]
- 6.Preseki A, Brkic H, Primorac D, Drmick I. Methods of preparing the tooth for DNA isolation. Acta Stomatol Croat. 2000;34:21–4. [Google Scholar]
- 7.Silva LAF, Passos NS. DNA forense: Coleta de amostras biolicas em locais de crime para estudo do DNA. Macei Ed UFAL. 2002 [Google Scholar]
- 8.Mitsouras K, Faulhaber EA. Saliva as an alternative source of high yield canine genomic DNA for genotyping studies. BMC Res Notes. 2009;2:219. doi: 10.1186/1756-0500-2-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daniel PK, David K, Serena GL, Vivian N, Qiuyun F. Effect of storage conditions on the extraction of PCR-quality genomic DNA from saliva. Clin Chem Acta. 2004;343:191–9. doi: 10.1016/j.cccn.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 10.Beckett SM, Laughton SJ, Pozza LD, McCowage GB, Marshall G, Cohn RJ, et al. Buccal swabs and treated cards: Methodological considerations for molecular epidemiologic studies examining pediatric populations. Am J Epidemiol. 2008;167:1260–7. doi: 10.1093/aje/kwn012. [DOI] [PubMed] [Google Scholar]
- 11.Closas MG, Egan KM, Abruzzo J, Newcomb PA, Ernstoff LT, Franklin T, et al. Collection of genomic DNA from adults in epidemiological studies by buccal cytobrush and mouthwash. Cancer Epidemiol Biomarkers Prev. 2001;10:687–96. [PubMed] [Google Scholar]
- 12.Burger MF, Song EY, Schumm JW. Buccal DNA samples for DNA typing: New collection and processing methods. BioTech. 2005;39:257–66. doi: 10.2144/05392PF01. [DOI] [PubMed] [Google Scholar]
- 13.Sweet D, Lorente M, Lorente JA, Valenzuela A, Villanueva E. An improved method to recover saliva from human skin: the double swab technique. J Forensic Sci. 1997;42:320–2. [PubMed] [Google Scholar]
- 14.Kanto A, Hirata MH, Hirata RD, Nunes FD, Melani RFH, Oliveira RN. DNA extraction from human saliva deposited on skin and its use in forensic identification procedures. Braz Oral Res. 2005;19:216–22. doi: 10.1590/s1806-83242005000300011. [DOI] [PubMed] [Google Scholar]
- 15.Silva RHA, Musse JO, Francisco RH, Melani RF, Oliveira RN. Human bite mark identification and DNA technology in forensic dentistry. Braz J Oral Sci. 2006;5:1193–7. [Google Scholar]
- 16.Ellis MA, Song F, Parks ET, Eckert GJ, Dean JA, Windsor LJ. An evaluation of DNA yield, DNA quality and bite registration from a dental impression wafer. J Am Dent Assoc. 2007;138:1234–40. doi: 10.14219/jada.archive.2007.0349. [DOI] [PubMed] [Google Scholar]
- 17.Hochmeister MN, Rudin O, Ambach E. PCR analysis from cigarette butts, postage stamps, envelope sealing flaps, and other salivastained material. In: Lincoln PJ, Thomson J, editors. Forensic DNA profiling protocols. Totowa: Humana Press; 1998. pp. 27–32. [Google Scholar]
- 18.Polymerase Chain Reaction. [cited in 2010]. Available from: http://www.ncbi.nlm.nih.gov/projects/ genome/probe/doc/TechPCR.shtml .
- 19.Fregeau CJ, Fourney RM. Update to: DNA typing with fluorescently tagged short tandem repeats: A sensitive and accurate approach to human identification. In: Burczak JD, Mardis E, editors. Polymophism Detection Analysis, Biotechniques Books, Update Series. Natick, MA: Eaton Publishing; 2000. pp. 149–92. [PubMed] [Google Scholar]
- 20.Moretti TR, Baumstark AL, Defenbaugh DA, Keys KM, Smerick JB, Budowle BJ. Validation of STRs for forensic usage: Performance testing of fluorescent multiplex STR systems and analysis of authentic and simulated forensic samples. Forensic Sci. 2001;46:647–60. [PubMed] [Google Scholar]
- 21.Kobayashi K, Hecker KH. Gender determination by amelogenin specific PCR and product analysis by DNA chromatography. Application note 113; Transgenomic Inc (Internet) [cited in 2010]. Available from: http://www.transgenomic.com/pdf/AN113.pdf .
- 22.Anwar N, Goodman M, Hulme P, Elsmor P, Greenhalgh M, McKeown B. Amelogenin as a target for real time PCR quantitation of forensic templates. Int Cong Ser. 2006;1288:768–70. [Google Scholar]
- 23.Kao LG, Tsai LC, Lee JC, Hsieh HM. Controversial cases of human gender identification by amelogenin test. Forensic Sci J. 2007;6:69–71. [Google Scholar]
- 24.Steinlechner M, Berger B, Niederstatter H, Parson W. Rare failures in the amelogenin sex test. Int J Legal Med. 2002;116:117–20. doi: 10.1007/s00414-001-0264-9. [DOI] [PubMed] [Google Scholar]
- 25.George R, Sriram G, Saraswathi TR, Sivapathasundharam B. Isolation of epithelial cells from acrylic removable dentures and gender identification by amplification of SRY gene using real time PCR. J Forensic Dent Sci. 2010;2:326. doi: 10.4103/0974-2948.71055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das N, Gorea RK, Gargi J, Singh JR. Sex determination from pulpal tissue. J Indian Forensic Med. 2004;26:50. [Google Scholar]
- 27.Mishra A. Recent trends used in medical forensic science and Indian law. [cited in 2010]. Available from: http://www.lawyersclubindia.com/articles/recent-trends-used-in-medical-forensic-science-indianlaw-2701 .
- 28.Sweet D, Sweet CHW. DNA analysis of dental pulp to link incinerated remains of homicide victim to crime scene. J Forensic Sci. 1995;40:310–4. [PubMed] [Google Scholar]
- 29.DNA from cigarette butt man discarded while being questioned in another matter leads to rape charge. [cited in 2010]. Available from: http://www.sfgate.com/cgi-bin/article.cgi?f=/n/a/2010/12/17/state/n064941S87 .
- 30.Press Trust of India. Indian Army to begin DNA profiling of troops. [cited in 2010]. Available from: ibnlive.in.com/news/indian-army-to-begin-dnaprofiling-of-troops .
- 31.Hampl JA. American Board of Forensic Odontology. Diplomates Reference Manual. Article II: Code of Ethics and Conduct. 2010. [cited in 2010]. pp. 20–4. Available from: www.abfo.org .
- 32.Abichandani RK. New biology and criminal investigation (with a schedule of model DNA legislation proposed). No:18: Forensic Report. 2010. [cited in 2010]. p. 50. Available from: http://gujarathighcourt.nic.in/Articles/NBCI.pdf .


