Abstract
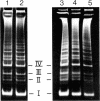
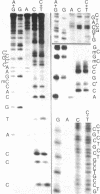
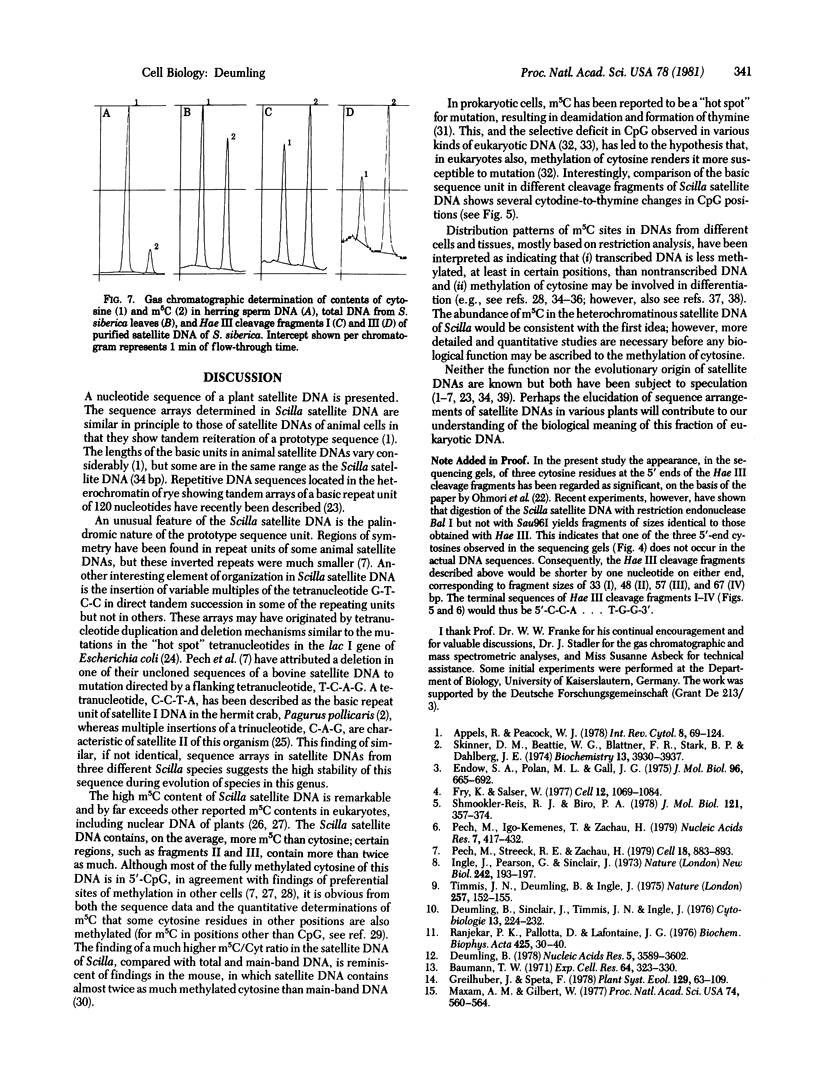
G+C-rich satellite DNA, representing about 19% of total nuclear DNA, was isolated from various tissues of the monocotyledonous plant, Scilla siberica, by using Ag+-Cs2SO4 gradient techniques. This satellite DNA had an unusually high melting point and a high methylcytosine (m5C) content (≈25% of total bases; m5C/cytosine ratio ≈1.5) and was localized, by in situ hybridization, in the heterochromatin regions of the chromosomes. Digestion with restriction endonuclease Hae III yielded a series of fragments ranging from 35 to several hundred nucleotide pairs. The major fragments, I-IV (35, 50, 59, and 69, nucleotide pairs, respectively), were isolated, and their nucleotide sequences were determined. The dominant fragment I was a highly symmetrical molecule, with a basically palindromic arrangement. This sequence represented the basic unit of Scilla satellite DNA and was tandemly repeated many times, with some base substitutions and multiple successive insertions of the tetranucleotide G-T-C-C. The dinucleotide CpG was the commonest nearest-neighbor sequence. Thin layer chromatography, DNA sequence analysis, and gas chromatography combined with mass spectrometry showed the high m5C content (m5C/Cyt = 2.2 and 2.8, respectively, for fragments II and III). Identical cleavage fragments were found in satellite DNAs from two other species of this genus (S. amoena and S. ingridae), which suggests that this constitutively methylated sequence is evolutionarily stable. The sequence arrangement of this plant satellite DNA is compared with those reported for several animal satellite DNAs.
Keywords: plant chromosomes, DNA sequences, gene arrangement, methylcytosine, mutation
Full text
PDF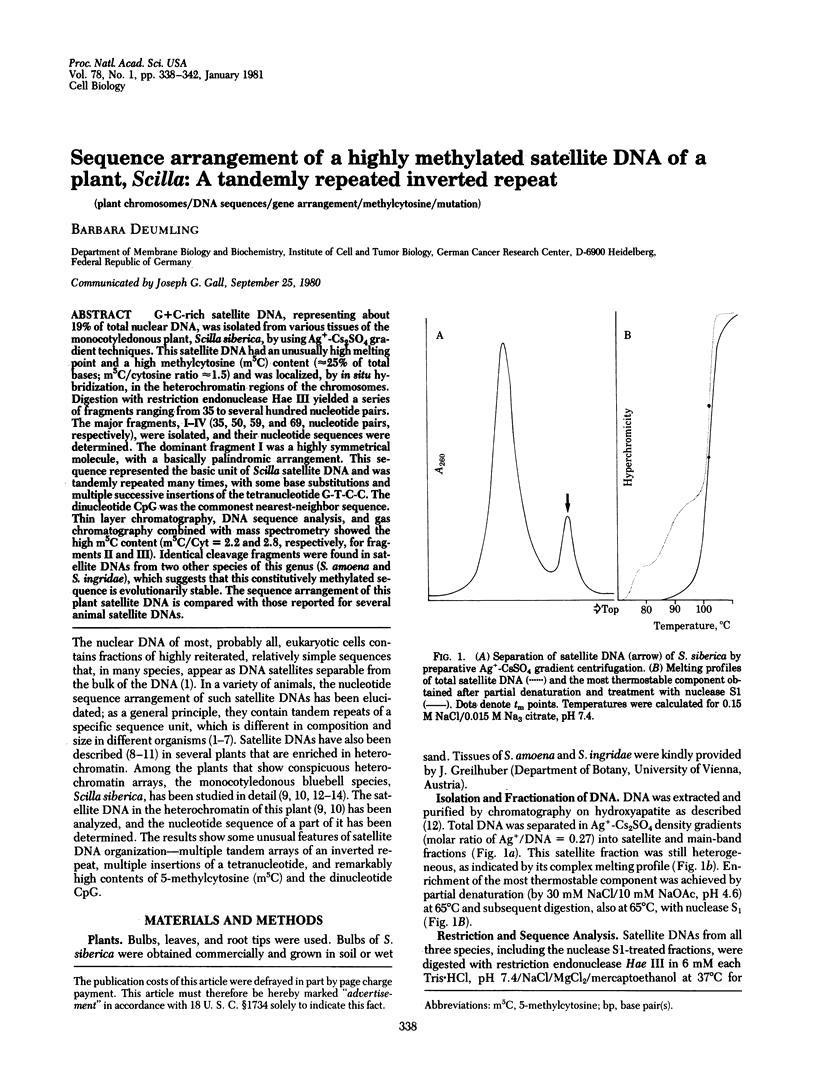



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Appels R., Peacock W. J. The arrangement and evolution of highly repeated (satellite) DNA sequences with special reference to Drosophila. Int Rev Cytol Suppl. 1978;Suppl 8:69–126. doi: 10.1016/s0074-7696(08)60472-6. [DOI] [PubMed] [Google Scholar]
- Baumann T. W. Heterochromatin und DNS-Replikation bei Scilla sibirica. Exp Cell Res. 1971 Feb;64(2):323–330. doi: 10.1016/0014-4827(71)90083-8. [DOI] [PubMed] [Google Scholar]
- Bedbrook J. R., Jones J., O'Dell M., Thompson R. D., Flavell R. B. A molecular description of telometic heterochromatin in secale species. Cell. 1980 Feb;19(2):545–560. doi: 10.1016/0092-8674(80)90529-2. [DOI] [PubMed] [Google Scholar]
- Bird A. P., Southern E. M. Use of restriction enzymes to study eukaryotic DNA methylation: I. The methylation pattern in ribosomal DNA from Xenopus laevis. J Mol Biol. 1978 Jan 5;118(1):27–47. doi: 10.1016/0022-2836(78)90242-5. [DOI] [PubMed] [Google Scholar]
- Chambers C. A., Schell M. P., Skinner D. M. The primary sequence of a crustacean satellite DNA containing a family of repeats. Cell. 1978 Jan;13(1):97–110. doi: 10.1016/0092-8674(78)90141-1. [DOI] [PubMed] [Google Scholar]
- Coulondre C., Miller J. H., Farabaugh P. J., Gilbert W. Molecular basis of base substitution hotspots in Escherichia coli. Nature. 1978 Aug 24;274(5673):775–780. doi: 10.1038/274775a0. [DOI] [PubMed] [Google Scholar]
- Deumling B. Localisation of foldback DNA sequences in nuclei chromosomes of Scilla, Secale, and of mouse. Nucleic Acids Res. 1978 Oct;5(10):3589–3602. doi: 10.1093/nar/5.10.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endow S. A., Polan M. L., Gall J. G. Satellite DNA sequences of Drosophila melanogaster. J Mol Biol. 1975 Aug 25;96(4):665–692. doi: 10.1016/0022-2836(75)90145-x. [DOI] [PubMed] [Google Scholar]
- Farabaugh P. J., Schmeissner U., Hofer M., Miller J. H. Genetic studies of the lac repressor. VII. On the molecular nature of spontaneous hotspots in the lacI gene of Escherichia coli. J Mol Biol. 1978 Dec 25;126(4):847–857. doi: 10.1016/0022-2836(78)90023-2. [DOI] [PubMed] [Google Scholar]
- Fry K., Salser W. Nucleotide sequences of HS-alpha satellite DNA from kangaroo rat Dipodomys ordii and characterization of similar sequences in other rodents. Cell. 1977 Dec;12(4):1069–1084. doi: 10.1016/0092-8674(77)90170-2. [DOI] [PubMed] [Google Scholar]
- Ingle J., Pearson G. G., Sinclair J. Species distribution and properties of nuclear satellite DNA in higher plants. Nat New Biol. 1973 Apr 18;242(120):193–197. doi: 10.1038/newbio242193a0. [DOI] [PubMed] [Google Scholar]
- Mandel J. L., Chambon P. DNA methylation: organ specific variations in the methylation pattern within and around ovalbumin and other chicken genes. Nucleic Acids Res. 1979 Dec 20;7(8):2081–2103. doi: 10.1093/nar/7.8.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miller J. R., Cartwright E. M., Brownlee G. G., Fedoroff N. V., Brown D. D. The nucleotide sequence of oocyte 5S DNA in Xenopus laevis. II. The GC-rich region. Cell. 1978 Apr;13(4):717–725. doi: 10.1016/0092-8674(78)90221-0. [DOI] [PubMed] [Google Scholar]
- Ohmori H., Tomizawa J. I., Maxam A. M. Detection of 5-methylcytosine in DNA sequences. Nucleic Acids Res. 1978 May;5(5):1479–1485. doi: 10.1093/nar/5.5.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pech M., Igo-Kemenes T., Zachau H. G. Nucleotide sequence of a highly repetitive component of rat DNA. Nucleic Acids Res. 1979 Sep 25;7(2):417–432. doi: 10.1093/nar/7.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pech M., Streeck R. E., Zachau H. G. Patchwork structure of a bovine satellite DNA. Cell. 1979 Nov;18(3):883–893. doi: 10.1016/0092-8674(79)90140-5. [DOI] [PubMed] [Google Scholar]
- Rae P. M., Steele R. E. Absence of cytosine methylation at C-C-G-G and G-C-G-C sites in the rDNA coding regions and intervening sequences of Drosophila and the rDNA of other insects. Nucleic Acids Res. 1979 Jul 11;6(9):2987–2995. doi: 10.1093/nar/6.9.2987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae P. M., Steele R. E. Modified bases in the DNAs of unicellular eukaryotes: an examination of distributions and possible roles, with emphasis on hydroxymethyluracil in dinoflagellates. Biosystems. 1978 Apr;10(1-2):37–53. doi: 10.1016/0303-2647(78)90027-8. [DOI] [PubMed] [Google Scholar]
- Ranjekar P. K., Pallotta D., Lafontaine J. G. Analysis of the genome of plants. II. Characterization of repetitive DNA in barley (Hordeum vulgare) and wheat (Triticum aestivum). Biochim Biophys Acta. 1976 Feb 18;425(1):30–40. doi: 10.1016/0005-2787(76)90213-6. [DOI] [PubMed] [Google Scholar]
- Razin A., Sedat J. Analysis of 5-methylcytosine in DNA. II. Gas chromatography. Anal Biochem. 1977 Feb;77(2):370–377. doi: 10.1016/0003-2697(77)90250-0. [DOI] [PubMed] [Google Scholar]
- Salomon R., Kaye A. M., Herzberg M. Mouse nuclear satellite DNA: 5-methylcytosine content, pyrimidine isoplith distribution and electron microscopic appearance. J Mol Biol. 1969 Aug 14;43(3):581–592. doi: 10.1016/0022-2836(69)90360-x. [DOI] [PubMed] [Google Scholar]
- Salser W. Globin mRNA sequences: analysis of base pairing and evolutionary implications. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 2):985–1002. doi: 10.1101/sqb.1978.042.01.099. [DOI] [PubMed] [Google Scholar]
- Shmookler Reis R. J., Biro P. A. Sequence and evolution of mouse satellite DNA. J Mol Biol. 1978 May 25;121(3):357–374. doi: 10.1016/0022-2836(78)90369-8. [DOI] [PubMed] [Google Scholar]
- Singer J., Roberts-Ems J., Luthardt F. W., Riggs A. D. Methylation of DNA in mouse early embryos, teratocarcinoma cells and adult tissues of mouse and rabbit. Nucleic Acids Res. 1979 Dec 20;7(8):2369–2385. doi: 10.1093/nar/7.8.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer J., Schnute W. C., Jr, Shively J. E., Todd C. W., Riggs A. D. Sensitive detection of 5-methylcytosine and quantitation of the 5-methylcytosine/cytosine ratio in DNA by gas chromatography--mass spectrometry using multiple specific ion monitoring. Anal Biochem. 1979 Apr 15;94(2):297–301. doi: 10.1016/0003-2697(79)90363-4. [DOI] [PubMed] [Google Scholar]
- Skinner D. M., Beattie W. G., Blattner F. R., Stark B. P., Dahlberg J. E. The repeat sequence of a hermit crab satellite deoxyribonucleic acid is (-T-A-G-G-)n-(-A-T-C-C-)n. Biochemistry. 1974 Sep 10;13(19):3930–3937. doi: 10.1021/bi00716a018. [DOI] [PubMed] [Google Scholar]
- Smith G. P. Evolution of repeated DNA sequences by unequal crossover. Science. 1976 Feb 13;191(4227):528–535. doi: 10.1126/science.1251186. [DOI] [PubMed] [Google Scholar]
- Sneider T. Methylation of mammalian deoxyribonucleic acid. 3. Terminal versus internal location of 5-methylcytosine in oligodeoxyribonucleotides from Novikoff hepatoma cell deoxyribonucleic acid. J Biol Chem. 1972 May 10;247(9):2872–2875. [PubMed] [Google Scholar]
- Sutter D., Doerfler W. Methylation of integrated adenovirus type 12 DNA sequences in transformed cells is inversely correlated with viral gene expression. Proc Natl Acad Sci U S A. 1980 Jan;77(1):253–256. doi: 10.1073/pnas.77.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmis J. N., Deumling B., Ingle J. Localisation of satellite DNA sequences in nuclei and chromosomes of two plants. Nature. 1975 Sep 11;257(5522):152–155. doi: 10.1038/257152a0. [DOI] [PubMed] [Google Scholar]
- van Ooyen A., van den Berg J., Mantei N., Weissmann C. Comparison of total sequence of a cloned rabbit beta-globin gene and its flanking regions with a homologous mouse sequence. Science. 1979 Oct 19;206(4416):337–344. doi: 10.1126/science.482942. [DOI] [PubMed] [Google Scholar]