Abstract
Sharks and skates are representatives of the earliest vertebrates with an immune system based on V(D)J rearrangement. They possess a unique Ig gene organization consisting of 15 to >50 individual IgM loci, each with one VH, two DH, one JH and one set of constant region exons. The present study attempts to understand how multiple Ig genes are regulated with respect to rearrangement initiation and to targeting during somatic hypermutation. The linkage of three single-copy IgH genes was determined, and single-cell genomic PCR studies in a neonatal animal was used to examine any relationship between relative gene position and likelihood of rearrangement. Our results show that 1–3 IgH genes are activated independently of linkage or allelic position and the data best fit with a probability model based on the hypothesis that V(D)J rearrangement occurs as a sequence of trials within the B cell. In the neonatal cell set two closely related IgH, G2A and G2B, rearranged at similar frequencies and their membrane forms were expressed at similar levels, like in other young animals. However, older animals displayed a bias in favor of the G2A isotype, which suggests that although rearrangement at G2A and G2B was randomly initiated during primary repertoire generation, the two very similar IgM sequences appear to be differentially expressed with age and exposure to antigen. We performed genomic single-cell PCR on B cells from an immunized individual to study AID targeting and found that hypermutation, like V(D)J rearrangement, occurred independently among the many shark IgH.
Keywords: evolution, V(D)J rearrangement, allelic exclusion
Introduction
In the mouse, V(D)J recombination at the immunoglobulin (Ig) heavy (H) chain locus proceeds in two distinct and ordered steps, D to JH followed by VH to DJ [1]. This is in part controlled by enhancer-blocking insulator sequences in the 96 kb V-D interval that prevent spreading of open chromatin and simultaneous activation of all elements [2]. The differential chromatin activation restricts recombination to DJ formation on one or both alleles before the second step, during which ~200 upstream VH occupying >2 Mb organize into multiple loops that gather in close spatial proximity to the DJ, to enable DJ rearrangement to one VH [3]. The two phenomena, sequential chromatin domain activation [4] and locus contraction [5, 6], are evolutionary adaptations for a highly complex and spatially extended IgH locus. All components, the VH, D and JH gene segments and constant (C) region exons, are brought under one set of cis-acting regulators that coordinate gene activation and transcription and accessibility to the recombinase during early B cell differentiation (for a recent review see 7). Both alleles are poised for rearrangement, but it is the asynchrony at the VH to DJ step that initiates allelic exclusion and events leading to a single type of antigen receptor expressed per lymphocyte. The underlying causes for one gene acting ahead of the other are the subject of some controversy [8–10].
Ig and V(D)J rearrangement exist in all jawed vertebrates; however, only in bony and cartilaginous fishes are the Ig genes present as multiple loci, or clusters [for reviews see 11, 12]. The existing cartilaginous fish subclasses, Elasmobranchii (sharks, rays and skates) and Holocephali (chimeras, ratfish), diverged more than 350 million years ago, and the IgH of all representative animals examined are arrayed as multiple miniloci. There can be 100–200 individual IgH and IgL genes, each with a few gene segments (V-D1-D2-JH and VL-JL, respectively) and one set of C region exons. This unit is considered ancestral to the classical Ig locus in other vertebrates. In this H chain gene system the close proximity of the rearranging gene segments (within 2 kb) do not require locus compaction, and they all are simultaneously available to RAG for the three recombination events generating the shark VDJ [13]. From the first report [14] of this alternative organization in the earliest of jawed vertebrates, the regulation of gene expression and allelic exclusion have been central questions of interest in an Ig system that is both ancient and conserved.
Our model is the nurse shark, where we have isolated and classified all of its IgM H chain genes, each of which must rearrange to be expressed [15]. There are 9–12 functional IgH genes whose VH are all members of one family [16], where the gene number in one subfamily (G4) differs between individuals [15]. Thus in any single differentiating shark B cell there are multiple potential targets for RAG (recombination activating gene) recombinase as there are for activation-induced cytidine deaminase (AID) after the B cell has been activated by antigen. AID is found in all vertebrates [17], and mutated shark Ig sequences are scarce in young animals (pups) but abundant in hyperimmunized adults [16, 18, 19].
The present study attempts to understand the regulation and activity of multiple copies of Ig genes within a cell. Pioneering work by Bengtén and coworkers [20] in the bony fish Atlantic cod demonstrated that the many L chain clusters did not possess individual enhancers. Since one set of regulators probably exerts influence over several gene clusters separated by distances of 2.1–4.8 kb, this casts some perplexity as to how cod L chain isotype exclusion is effected but raises interesting prospects for receptor editing [21]. We had previously demonstrated H chain exclusion in shark [13]; now our first task was to establish whether the shark IgH genes share regulatory elements and so discover if recombinase accessibility unfurled on the basis of linkage or allelism. Information on the spatial relationship of the IgM H chain genes is limited in nurse shark and unknown in other species. We linked and mapped three closely related IgH that are single-copy genes. This was followed by single-cell genomic PCR studies in a very young animal to examine the pattern of rearranged IgH relative to their gene position. The paucity of somatic mutations in neonates also affords the best opportunity for scoring VDJ, and based on the results, we constructed probability models to assess the nature of IgH activation in shark pre-pro B lymphocytes.
We went on to study AID targeting patterns among IgH in single B cells of an older, immunized shark and to ascertain whether genetic exchanges took place between the genes, rearranged or not. In the course of investigating whether an IgH gene in germline (GL) configuration was in fact quiescent in B cells, we found that targeting by somatic hypermutation (SHM) was largely conditional to the rearrangement status of the gene.
Materials and methods
Animals
Nurse sharks (Ginglymostoma cirratum) were captured off the coast of the Florida Keys. Blood was obtained from the caudal sinus. Peripheral blood leukocytes (PBL) were separated from erythrocytes (RBC) after centrifugation through Ficoll. Genomic DNA and total RNA were isolated using conventional techniques. Genomic DNA obtained from shark-GR PBL, after sorting with anti-IgM mAbs (CB5, CB11, CB16), was described previously where an aliquot was used for genomic Southern blotting [13]. Some animals were sacrificed after capture (shark-33) and others had been immunized with various antigens (shark-GR, shark-JS, shark-PI) [13, 15]. At the time of sample collection shark-GR was 7 years of age, shark-JS 5–6 years, shark-PI 3–4 years, and shark-33 2.5 years. Shark pups included shark-LA (est. <2 months of age); shark-AQ was one week old after caesarian section [22] and sharks-EC and -TH less than one week of age. All experiments involving animals were conducted in accordance with guidelines set by the Institutional Animal Care and Use Committees for the State University of New York Health Science Center at Brooklyn and the University of Maryland at Baltimore.
Libraries
The shark-Y BAC library [23] was screened with probes to IgM C region as described [15]. The 216 selected clones purchased from Arizona Genomics Institute (http://www.genome.arizona.edu) are identified by their grid positions (plate addresses). BAC DNA insert ends were sequenced using primers near the Hind III cloning site (T7, 5′-TAATACGACTCACTATAGGG-3′; BES_HR, 5′-CACTCATTAGGCACCCCA-3′). Pulse field electrophoresis was performed as previously described [15].
Germline IgH
Single-cell studies were performed with B cells from sharks-GR and – LA, whose germline IgH genes were individually identified. There are 9–12 functional IgH in nurse sharks, classified in five subfamilies called Groups 1–5 according to differences throughout the H chain sequence [15, 16]. The variation lies in number of members from one subfamily, G4, and these were identified in shark-LA in this study. All germline H chain genes from shark-GR were previously defined [13].
Single-cell PCR
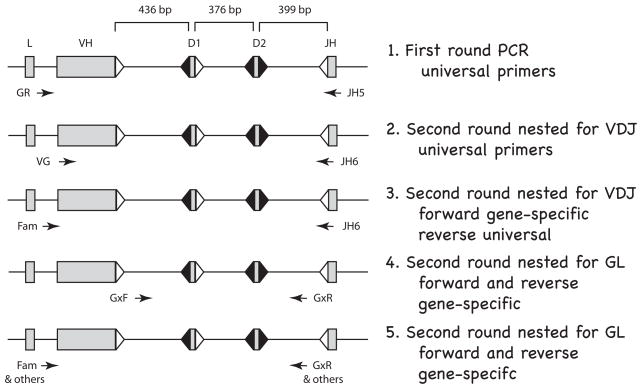
The surface Ig-positive (sIg+) cells were collected by magnetic cell (MAC) sorting. For shark-LA, the spleen cells were dissociated and incubated with a mix of anti-shark-IgM mAb (CB5, CB11, CB16 [24]) shown to collectively bind most, although not all, serum IgM. The MAC sorting and single-cell PCR procedures, including primer sequences and cycling parameters, were previously described in detail [13]. Single lymphocytes were picked by hand, alternating with single RBCs which served as negative (germline-only) control. The pipet was washed three times between cells. A flow diagram of the PCR reactions is shown in Fig. 1. First round PCR was performed with universal primers in the leader intron (primers GR1, GR2-5) and in JH (primer JH5) [unless specified, oligonucleotide sequences are in ref. 13]. These PCR products were treated with ExoSAP-IT (USB) to remove remaining primers and used separately in four different second round PCR reactions, as depicted. The first nested reaction (Fig. 1, line 2) was primarily used to see whether any VDJ could be amplified. The universal primers (VG1, VG2-5 and JH6) generate only nonrearranged GL sequences of 1.6 kb bp in the RBC but in lymphocytes both the GL as well as rearranged VDJ of 350 bp. This reaction allowed us to select positive lymphocyte samples suitable for further analysis, that is, those VDJ-containing samples that were flanked by GL-only signals in the adjacent RBC reactions. This criterion was used to avoid carryover contamination between lymphocytes during picking. For isolation and identification of the VDJ another set of PCRs (Fig. 1, line 3) was performed on the selected lymphocytes: five reactions each using a forward primer specific for the VH subfamily (Fam1 for G1, Int for G2, GR3N2 for G3, Fam4 for G4, or Fam5 for G5 each in combination with JH6). Examples of such reactions for shark-LA B cells are shown in Supplemental Materials, Fig. S1, top. The VDJ of 400–420 bp was directly sequenced (Genewiz, Inc.) or cloned into pGEM (Invitrogen). To determine which genes remained nonrecombined, those in GL configuration were amplified in another reaction (Fig. 1, line 4) where the primers targeted the intersegmental sequence in V-D1 and in D2-JH. These primers were subfamily specific (G1DF/G1JR for G1, FD2-1/RD2 for G2, G3DF/G3JR for G3, G4DF/DR3/4 for G4, G5DF/G5JR for G5). Examples of such reactions are shown in Fig. S1, bottom. The identity of the 1.1–1.2 kb PCR products was ascertained by restriction enzyme analyses (Supplemental Fig. S2).
Figure 1.
Flow diagram for single-cell PCR in shark B cells. The rearranging gene segments (VH, D1, D2, JH) are separated by ~400 bp; the specific distances are from the gene G2A; L is leader. The flanking recombination signal sequences (RSS) with 23 bp spacers are represented as open triangles and RSS with 12 bp spacers filled triangles. Arrows indicate position and orientation of PCR primers. Line 1. First round PCR. A 40-cycle PCR was performed on B cells and on RBC, using “universal” primers detecting all functional IgH. Thereafter, the PCR products were depleted of oligonucleotides and used in the following four kinds of nested second-round reactions (25–30 cycles). Line 2. Second round nested for VDJ. Universal primers targeting FR1 and JH can amplify VDJ and/or GL fragments in B cells but only GL in the RBC samples. Line 3. Second round nested for VDJ. Specific forward primers reveal which subfamily (G1, G2, G3, G4, G5) the VDJ belongs to. VDJ is isolated and sequenced. Line 4. Second round nested for GL. Subfamily specific primers in the V-D1 and D2-J intersegmental regions amplify the non-rearranged genes. The five subfamily reactions are analyzed by restriction enzyme analyses detailed in Fig. S2, Supplementary Materials. Line 5. Second round nested for GL. The unrearranged VH is amplified for sequencing by using subfamily or gene-specific primers. Primer information is supplied in Materials and Methods.
To characterize the individual nonrearranged VH gene segment (Fig. 1, line 5), specific primers in the leader intron and D2-JH sequence were used. First-round samples from G1, G3, and G5 are amplified by using subfamily-specific primers in the leader intron and the D2-J region. Since there are two members of G2 and four of G4, additional gene-specific primers were generated for these studies. These included reverse primers that distinguished G2A from G2B (G2A-DJ, 5′-CGTCATCAAATTATAGTTGTGCAAA-3′; G2B-DJ, 5′-TTCATCAAATTATAGCTGTGCAAG-3′) in combination with forward G2 primer Int.
To determine the status of individual G4 members in shark-GR (G4A, G4CG, G4D, G4E), primers specific for sequence differences in V and in the D-J region were used to amplify the germline genes for direct sequencing (G4A-5′, 5′-TTTCATCAGTAACACTGG-3′; G4A-3′, 5′-GTCCTTGTCCCCAGTTAT-3′; G 4 D-5′, 5′-CATTTCATCAGTAACAC-3′; G 4 D-3′, 5′-TTCCTTGTCCCCAGTTAC-3′; G4E-V, 5′-GAAACTAGCGGGTTCA-3′; G4E-JR, 5′-CTTATTCTAATGTTTTC-3′; G4G-5′, 5′-CATTTCATCAGTAACAT-3′; G4G-3′, 5′-TTCCTTGTCCCCAGTTAG-3′).
The CDR3 of all the VDJ isolated are provided in the Figures. Mutant VDJ sequences were submitted to GenBank (http://www.ncbi.nlm.nih.gov/sites/entrez?db=nucleotide) as JN087503-JN087509.
RT-PCR, PCR and probes
First strand cDNA was primed in total RNA using an oligonucleotide in the G2 membrane sequence (G2mem4, 5′-GACGGCAGCGCTGTAGAA-3′) according to manufacturer’s instruction for use of the SuperScript III Reverse Transcriptase kit (Invitrogen). Subsequent PCR was performed with primers specific for G2 leader (G2L, 5′-ACCAGAATGACKACGATG-3′) and G2 CH1 (G2CH1R, 5′-CGCATCTGGCTCGTTGAG-3′); the optimal annealing temperature was determined by a T-Gradient thermocycler (Biometra) and the elongation time was usually set at 30 seconds for <500 bp products and one minute for 800–1200 bp products. After 30 cycles the ~660 bp PCR product was digested with BstE II, a restriction enzyme site in the G2A JH that distinguishes it from G2B sequences.
RT-PCR was performed on shark-GR spleen RNA to detect pseudogene G8 transcripts. Primers targeting FR1 (V2–5: 5′-TGAYTCAACCAGAGGCA-3′) and the G8 CH2 (G8CH2R, 5′-GAATGACCTCTTCATGAG-3′) amplified DNA fragments of about 955 bp from oligo dT-primed cDNA. Sequences were cloned into pGEM-T-Easy and their identity ascertained by the unique 9-bp gap in FR1 and the G8 C region (EU106187).
Long template PCR (Roche) was performed according to manufacturer’s instructions for a total of 35 cycles, and optimized for the amount of input genomic DNA. Primers for G2B in FR1 (S1, 5′-AGAGGAGGTTACTTTGATC-3′) and in J-C intron (J-C3, 5′-ATGGTTAACTGCGATAC-3′) were used to raise rearranged products of about 6.2 kb (GL) to 5 kb (fully rearranged VDJ) from B cell genomic DNA.
Amplification of G4 sequences from shark-LA RBC DNA was performed with G4-specific primers in leader (G4L, 5′-TTCTGACTTTCTTATCCC-3′) and in the D2-JH interval (DR3/4 [13]), generating 1772 bp fragments that were cloned into pGEM (Promega). Bacterial colonies were picked with pipet tips and resuspended in 50 microliters of LB broth with ampicillin. The suspension was allowed to settle for an hour and 1.5 microliter was added to 25–50 microliter PCR reactant mixture that contained primers to the region bordering the pGEM polylinker site, T7 and Sp6. All PCR products of the anticipated size were selected for restriction enzyme analyses.
For Southern blotting, electrophoresed DNA samples were transferred to nylon filters (HyBond-N, GE Healthcare), and the membranes hybridized with radioactive probe under routine stringency conditions. Probes to H chain derived from Group 2 cDNA (vh, cμ1, cμ2, cμ3-cμ4) have been described elsewhere [15]. The blots were subjected to autoradiography, and signal intensities of bands were quantified using a Storm 860 phosphorimaging system with ImageQuant software (Molecular Dynamics, Sunnyvale, CA).
Statistical methods
We considered two models where we asked, is the total number of VDJ per cell fixed or constrained, and if so, what value might it be fixed at or have as a maximum? Each probability model was based the following assumptions: (a) in each cell there are 13 genes of interest; (b) four of these 13 genes are non-functional and never evaluated for VDJ status; (c) in each cell, the number of genes whose VDJ status can be evaluated is a random “nuisance” statistic; (d) VDJ events are equiprobable across the 13 genes; (e) ascertainability of a VDJ event is equiprobable across the 9 functional genes; (f) the presence or absence of a VDJ event, where ascertainable, is established without error; (g) partially rearranged genes such as 1R or 2R should be treated as nonproductive VDJ events.
For the “fixed genes” model there is the assumption (h) the total number of VDJ events in each cell is fixed across cells and has value ϑ. For the ith shark cell (i = 1 … n), let Yi denote the number of genes evaluated and Xi the number of observed VDJ events. There are 13Cϑ ways in which the ϑ VDJ events can be arranged in the 13 genes. Since the sample includes an observation with data (Xi = 3, Yi = 9), ϑ is constrained to be an integer within the interval [3, 7]; Y is similarly constrained within the interval [1, 9]. For each (ϑ, Y) combination we calculated conditional probabilities under the model CP ϑ,Y(x) = Prob(X = x | ϑ, Y), x = 0 … ϑ. From these probabilities we calculated the conditional likelihood (CL) of the sample data as CL ϑ,Y = Πi=1…n Prob(Xi | ϑ, Yi); we used these numbers to construct a conditional maximum likelihood estimate (CMLE) of ϑ.
We asked, how well do the sample data fit an “optimal” (i.e., with ϑ estimated by CMLE) ϑ-VDJ model? In an initial approach we conducted an exact chi-square goodness of fit test [25] separately for each Y stratum, using as expected probabilities the CP values under the optimal model. As a second analysis, we calculated a single global chi-square goodness of fit statistic G, conditioned on the marginal distribution of Y, using as expected probabilities the CP values under the optimal model multiplied by the ratio (Y total frequency/n). Since the number of permutations needed to calculate an exact p-value for this test is extremely large, we conducted a Monte Carlo simulation in which a dataset constituting the 13Cϑ equiprobable outcomes under the probability model (i.e., not the sample dataset) was resampled with replacement k = 1 … 20,000 times, each pseudo-set having both the same n as the sample dataset and the same marginal distribution of Y. For the kth pseudo-set a goodness of fit statistic Gk was calculated; a p-value for G in the sample set was estimated by determining its ordinality relative to the distribution of the simulated Gk values.
For the “sequential activation” model assumptions (a)-(g) above remain, but this time VDJ events occur in each cell as a sequence of trials. For the jth trial, Prob(VDJ+) = 1/(ϑ−j+1); the process stops as soon as the first VDJ+ event has occurred. If the VDJ+ event occurs in one of the four non-functional genes then the cell is considered dead and therefore unobservable (i.e., excluded from the probability model). This time there are (9/13) Σj=1 … ϑ 13Pj observable cell outcomes. It follows from this model that a maximum of ϑ VDJ events can occur in any cell, with exactly one of them being VDJ+. Let represent the number of VDJ+ & VDJ− events respectively in a cell having Yi ascertainable genes. From the probability model we calculated the joint conditional probabilities CP = Prob(X+ = x1, X− = x2 | ϑ, Y), for x1 = 0 or 1, x2 = 0 … ϑ – 1.
Estimation of ϑ and fit of data to an optimal model was conducted in a manner similar to that used for the fixed-genes model; the Monte Carlo simulation of the probability model data took into account the fact that under this model the observable outcomes are not all equally probable.
We conducted exact chi-square goodness of fit tests of our data to assumptions (d) & (e). Statistical software used were SAS (SAS Institute, Cary NC) Release 9.2, and StatXact (Cytel Corp., Cambridge MA) PROCS for SAS, version 9.
Results
Experimental overview
The linkage of three IgM H chain genes was established, and experiments focused on whether there existed any regulation shared between the adjacent or allelic IgH genes. V(D)J recombination and SHM patterns among the functional IgH were studied in single B cells from two individuals, pup shark-LA and adult shark-GR. After discovering that pseudogenes also rearranged we tested probability models that included all IgH in order to obtain a global picture of IgH rearrangement initiation in a B lymphocyte. Since our results suggested that the IgH genes operated autonomously, we went on to investigate whether the different IgM isotypes were of the same value to the animal. Two closely related IgH (G2A, G2B) that rearranged at similar frequency in the neonate were examined for expression. The membrane forms were found at similar levels in young animals but a distinct bias for one (G2A) was observed in adult animals.
Mapping of three IgM genes
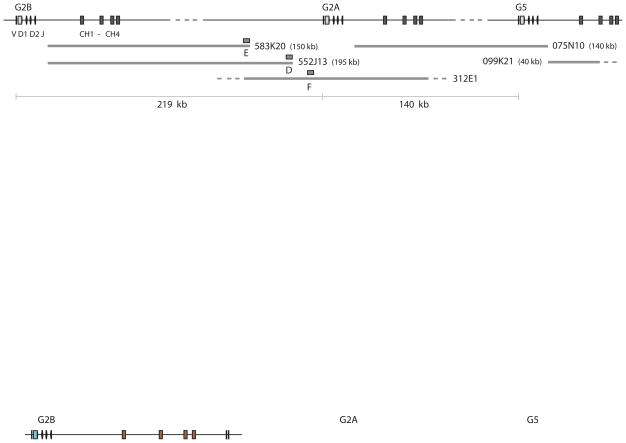
The shark-Y BAC library was screened with Cμ3–4 sequence, as previously described [15], and 216 candidate clones were analyzed. Hybridization of dot blots showed that 156 contained V and C components and 21 carried only C; the first 78 clones were characterized and reported while the remaining 99 were completed at a later time using the same techniques (V. Lee, K. Malecek and E. Hsu, unpublished results). The classification of all clones according to IgM subfamily (G1, G2A, G2B, G3, G4, G5, and pseudogenes G2C, G7, G8) is available in Table S1, Supplemental Materials. Clone 075N10 [15] carried the C exons of gene G2A and the VH gene segments of G5 (Fig. 2), ~120 kb apart. Because G2B is very closely related to G2A, four BAC clones carrying the G2B C region alone (Table S1, 399C2, 583K20, 552J13, 639A12) were analyzed for linkage to any downstream IgH. Their insert ends were sequenced. Each of the four contained one insert end in the G2B J-C intron, at the Hind III site 1476 bp downstream from JH; probes were generated for the opposite ends in two (Fig. 2, boxes E and D) where non-repetitive sequence was available.
Figure 2.
Mapping of IgH genes G2B, G2A, G5. The orientation and order of the IgM H chain genes G2B, G2A and G5 are shown at top, with the VH gene segments and C exons, labeled at left. The distance between G2B leader and its transmembrane exons is 17.9 kb; dotted line indicates map is not drawn to scale. BAC clones, drawn as thick gray lines, include two clones carrying only the C regions of G2B (583K20, 552J13), whose insert-end sequences (box E and D) hybridized to BAC clone 312E1. The F sequence from 312E1 (box F) was located 4.4 kb upstream of the G2A leader and 16 kb downstream of the D sequence. BAC clone 075N10 bridged the C exons of G2A and the VH gene segments of G5 [15]. BAC clone 099K21 carried only the G5 C exons and extended 35 kb beyond CH1. The distances are shown at the bottom, as determined between the genes. The entire area includes: distance from G2B to G2A (219 kb), from G2A to G5 (140 kb), and the distance G5 to the end of 099K21 (43 kb), a total of 402 kb. Except for 312E1, the BAC clone insert sizes were determined by pulsed field electrophoresis. Boxes labeled D, E, F represent sequences from which probes were generated.
Probes E and D were hybridized to the dot blots that carried DNA from all IgM BAC clones, and signals were observed only from some G2A and G2B clones. Since neither hybridized to the single BAC clone that carried all the sequence downstream of G2A C region (075N10), the area that E and D detected must be located upstream of the G2A V gene segments. The distance between D and the G2A leader was determined on one of the G2A BAC clones (312E1), allowing us to conclude that the G2B and G2A leaders were separated by 219 kb (see Fig. 2 legend). A probe to the non-J-C intron insert end of 099K21, the G5 clone, detected only other G5-containing BAC clones (Fig. 2 and unpub. results), probably because the insert is short, 40 kb. The order and orientation of G2B-G2A-G5 was established over a distance of 402 kb.
Germline IgH determined in shark-LA
The G4 subfamily members vary among individual sharks, but all other IgH (G1, G2A, G2B, G3, G5) are single-copy genes [15]. The germline G4 genes in shark-LA RBC DNA were amplified by G4-specific primers and cloned into pGEM (see Materials and Methods). Restriction enzyme analyses of 156 bacterial colony PCR fragments, supported by sequencing, showed there were four different G4 genes. The novel G4D2 (accession number JN087510) closely resembles G4D, differing only by 8 substitutions; however, one difference in CDR1 in the VH and another the loss of a Hinc II site in the V-D interval permitted G4D2 to be distinguished in the single-cell studies described below. Shark-LA carried G4A, G4CG, G4D, G4D2; the other H chain genes (G1, G2A, G2B, G3, G5) are single copy genes, and the shark-LA sequences obtained from RBC matched those previously submitted to GenBank. The restriction enzyme analyses for non-rearranged shark-LA IgH is shown in Supplemental Fig. S2.
Single-cell genomic PCR in neonatal shark
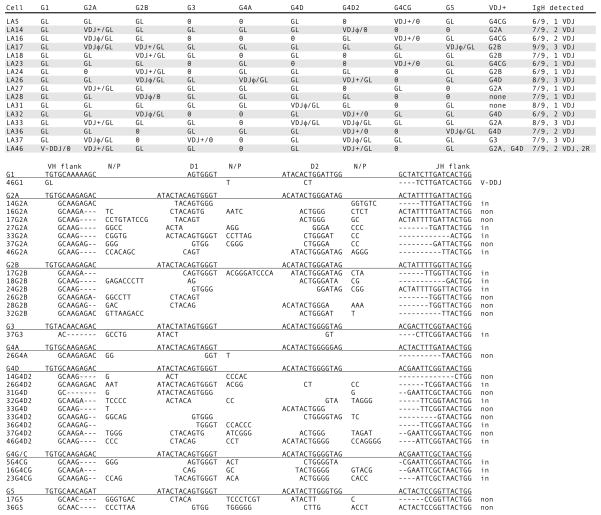
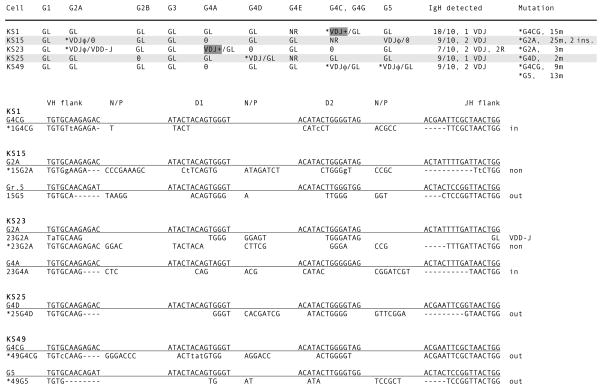
Surface IgM-positive (sIgM+) splenocytes from nurse shark pup LA were isolated and 54 single cells were picked by hand, alternating with 54 erythrocytes. In the course of the first nested reactions (Fig. 1, line 2) 28 lymphocytes with VDJ were selected after GL-only signals were found in the flanking RBC samples. In Fig. 3 (top) 16 B cells are shown where at least 6 out of 9 IgH genes could be detected. The rearranged IgH were identified as VDJ (Fig. 1, line 3) and their CDR3 shown in Fig. 3 (bottom). In each cell 1–3 rearrangements are present, and in one case (LA46) there are two in-frame VDJ with CDR3 of viable sizes. In the same cell a partially rearranged gene (V-DDJ, called 2R for two rearrangement events) was found; the reason for its noncompletion is that D1 and its 5′ RSS were deleted. The paucity of junctional sequence (Fig. 3, bottom, 46G1) does not permit analysis as how this was generated.
Figure 3.
Single-cell PCR results from shark-LA sIgM+ cells. Top. Rearrangement configurations of the nine IgH in each cell. The IgH genes present in shark-LA were: G1, G2A, G2B, G3, G4A, G4D, G4D2, G4CG, and G5. The status of the Ig gene segments is indicated as fully rearranged (VDJ), partly rearranged as V-DDJ, or not rearranged (GL). In-frame, potentially functional VDJ are indicated by plus (+) sign; nonproductive VDJ have φ sign. Zero indicates that the gene could not be detected. The column VDJ+ shows the probable IgH gene encoding the receptor. IgH detected indicates tally of the genes characterized per B cell and the number of unique VDJ. Bottom. CDR3 of VDJ. Their junctions are shown according to assignment to VH flank, D1 gene, D2 gene, and JH flank, classified according to subfamily. N/P, nontemplated or palindromic sequence. Dashes denote nucleotide deletions at the flanks. At the right, the VDJ are indicated as in-frame or nonproductive (non). V-DDJ is sequence with only two rearrangement (2R) events, where the V-D intersegmental region was not been deleted by recombination.
In the established single-copy genes G1, G2A, G2B, G3 and G5, the nonrearranged allele can be detected in GL configuration in 14 out of 17 instances in 12 B cells. For example, in LA16 the G2A VDJ was out-of-frame and presumed to be the first IgH to recombine; the second rearrangement event did not take place at its allele but at G4CG, which recombined to an in-frame VDJ. Such instances suggest that activation of IgH is allele-independent. Moreover, since G2A is rearranged as often as and independently of the adjacent G2B, it appears that there is no connection between their accessibility to RAG and their chromosomal positions.
Rearrangement occurs at pseudogenes
The single-cell experiments monitored only the functional IgH. Transcripts from four pseudogenes in nonrearranged, non-mutated form have been described [15 and references therein] but it was not clear whether RAG or AID would act upon them. RT-PCR was performed using shark-GR spleen RNA to amplify pseudogene G8 transcripts. Among 23 clones were three partially rearranged V-DDJ as well as six VDDJ with the J spliced to C region (not shown). Most of these carried substitutions, as did half of the 14 unrearranged VH (G8-15, -28, -31, Fig. 4); it is not known whether the latter sequences were transcribed from a GL or V-DDJ configuration. In other experiments we have found that the pseudogene G2C can also rearrange and hypermutate (C. Zhu and E. Hsu, unpub. results). We assume that the four pseudogenes (G2C, G6, G7, and G8) are potentially accessible to RAG as they are transcribed and carry canonical RSS [15].
Figure 4.
Mutated pseudogene G8 sequences. The G8 cDNA sequence, consisting of leader, leader intron, germline VH gene segment directly spliced to C region, is aligned to the most closely related functional gene, G1 (top). The genomic germline G1 sequence shows leader, leader intron with splice sites (indicated in lower case) and the VH gene segment demarcated into framework (FR) and complementarity-determining region (CDR, underlined). The recombination signal sequences (heptamer, spacer, nonamer) are labeled. G8 is nonfunctional, due to a mutation disabling the leader intron splice acceptor site. G8 mutants 15, 28 and 31 are compared with the G8 cDNA, with dots representing identity and dashes gaps. The PCR primer is shown in italics.
Sequential activation of IgH
Since the four pseudogenes are potential targets of RAG, there are 13 IgH loci in shark-LA pre-pro B cells that can become accessible for rearrangement. In order to obtain the full picture of all IgH in a shark B lymphocyte we used data of varying completeness from 36 samples (Supplemental Fig. S3), thereby taking into account missed information. We asked, what is the potential number of VDJs in each cell and is this number fixed across cells?
Table I shows the results obtained from two kinds of probability models. In the first, the total number of VDJ events in each cell is fixed and has a value of ϑ to be estimated. In the second, VDJ events occur as a sequence of trials that stop when the first in-frame VDJ occurs. If this happened at a pseudogene, it is considered unobservable. A maximum of ϑ VDJ events can occur and only one is in-frame. Results of the statistical calculations are detailed in footnotes to Table I. In the first model the best ϑ estimate was three (Table I, Fixed column), and although most categories showed good fit with the model, the global goodness of fit (Gk = 38.0 with p = 0.076) did not.
Table I.
Joint frequency of number of evaluated genes (Y) with number of observed VDJ events (X)a
| total IgH per cell (Y) | Number of VDJ (X) | p-valueb | ||||||
|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | total | Fixedc ϑ = 3 |
Sequentiald ϑ = 3 |
Sequentiald ϑ = 4 |
|
| 2 | 2 | 2 | 0 | 0 | 4 | 1.000 | 0.352 | 0.409 |
| 3 | 7 | 0 | 0 | 0 | 7 | 0.033 | 0.186 | 0.114 |
| 4 | 2 | 1 | 1 | 1 | 5 | 0.056 | 0.010 | 0.065 |
| 5 | 0 | 3 | 0 | 0 | 3 | 0.382 | 0.238 | 0.224 |
| 6 | 1 | 3 | 1 | 0 | 5 | 0.904 | 0.954 | 0.947 |
| 7 | 0 | 3 | 3 | 2 | 8 | 0.698 | 0.333 | 0.682 |
| 8 | 0 | 1 | 0 | 2 | 3 | 0.163 | 0.059 | 0.078 |
| 9 | 0 | 0 | 0 | 1 | 1 | 0.497 | 0.189 | 0.289 |
| Total | 12 | 13 | 5 | 6 | 36 | 0.076 | 0.041 | 0.209 |
Y is the total number of IgH, GL or rearranged, detected per cell; X includes VDJ (3R), 2R, and 1R.
for an exact test of the null hypothesis that data fit the ϑ-VDJ model. Total designates global goodness of fit.
Logarithms of the conditional likelihoods (CLs) were computed as CL3,Y : −41.3; CL4,Y : −45.0; CL5,Y : −56.6; CL6,Y : −75.5; CL7,Y : −95.7. Our conditional maximum likelihood estimate (CMLE) of discrete parameter ϑ is 3.
Logarithms of the CLs were computed as CL3,Y : −53.6; CL4,Y : −53.0; CL5,Y : −54.0; CL6,Y : −55.0; CL7,Y : −55.6. Our CMLE of ϑ is 4.
The sequential model ϑ = 4 fares better than the ϑ = 3 model; in the former, no p-value is below the customary 0.05 threshold (Table I, Sequential, ϑ = 4 column). The ϑ = 3 model overall goodness of fit statistic G = 72.5 with p = 0.041, suggesting poor model fit, but for the ϑ = 4 model G = 56.5 with p = 0.209. Thus of the two types of model examined, the one that fits the data more satisfactorily states that the events (gene activation to rearrangement) occur sequentially and features a maximum of 4 VDJ events per cell.
G2A is different from G2B
Rearrangement, in-frame as well as nonproductive, is evenly distributed between G2A and G2B among the shark-LA cells (Fig. 3, top). This result suggests that the two closely related H chain isotypes are activated for recombination at the same frequency and in this animal are expressed on the cell surface at similar levels. We investigated G2A/G2B expression in pups like shark-LA as well as in adults. First strand cDNA was primed with the G2 transmembrane sequence, so that the PCR products have a greater likelihood of reflecting the resting cell population [26].
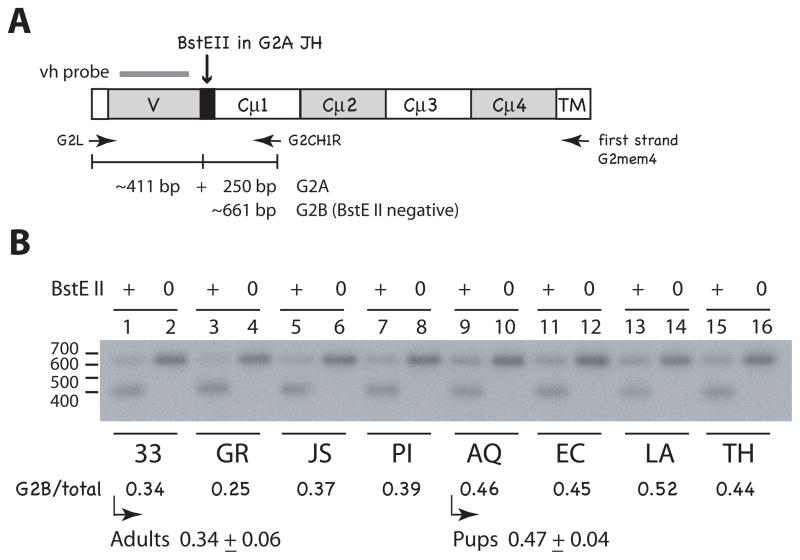
The RT-PCR products were blotted, hybridized with vh probe and the signals analyzed by phosphorimaging. Comparable amounts of DNA were digested with BstE II, a site present only in the JH of G2A (Fig. 5A). The intensity of the ~661 bp BstE II-negative band (G2B) is calculated as a fraction of the total G2 DNA (Fig. 5B). The pups express similar amounts of G2A and G2B (0.47 ± 0.04 S.D.) whereas the adults overall express significantly less G2B (0.34 ± 0.06 S.D.). The Mann-Whitney U test gave a value of p = 0.021, disproving the null hypothesis that the distribution of values is the same across the categories. The adults showed varied skewing in favor of G2A, whether the shark was injected with hen egg lysozyme (shark-JS), Ebola virus (shark-GR), DNP-LPS (shark-PI) or was unimmunized (shark-33).
Figure 5.
Comparing G2A and G2B expression in individuals. A. Representation of G2 PCR products and BstE II-digested fragments. The G2 H chain is shown with labeled domains; first strand cDNA primer G2mem4 targets the transmembrane sequence (TM) of both G2A and G2B. PCR primers G2L and G2CH1R generate DNA fragments of ~661 bp (calculated average CDR3 size of 11 codons). G2A sequences can be differentiated from G2B by the BstE II site in G2A JH (black box). Bar over V region represents the vh probe. B. Relative G2A and G2B amounts in pup and adult spleen RNA. RT-PCR products from adult (shark-33 epigonal organ, GR, JS, and PI spleen) and pup RNA (shark-AQ, EC, LA, and TH spleen) were incubated with (+ lanes) or without endonuclease (0 lanes), electrophoresed on a 1.5% TBE gel, and transferred onto a nylon filter (Hybond N+, GE Healthcare). The blot was hybridized to vh probe and submitted for scanning and autoradiography (x-ray film shown). Phosphorimager values obtained: (lane 1) 9759 at 661 bp; 18,497 at 411 bp, (lane 2) 28,405 at 661 bp, (lane 3) 6930; 19,296, (lane 4) 27,624, (lane 5) 9374; 15,764, (lane 6) 24,960, (lane 7) 12,430; 19,732, (lane 8) 31,166, (lane 9) 12,997; 13,720, (lane 10) 27,911, (lane 11) 13,567; 16,951, (lane 12) 30,049, (lane 13) 14,587; 13,441, (lane 14) 27,663, (lane 15) 14,348; 18,677, (lane 16) 32,019. G2B/total is the BstE II-negative band value divided by the value of the untreated sample. For shark-33 (lanes 1 and 2) it would be 9759/18,497 or 0.34 obtained as the proportion of G2B-containing sequences. The mean value with standard deviation is given for adult and pup groups.
Hypermutation at individual IgH genes in a B cell
The criterion for scoring mutant Ig is the presence of adjacent substitutions that are the hallmark of SHM in sharks [18, 27].
Neonatal Ig sequences are hardly ever mutated, and only one shark-LA rearrangement (16G2A) carried substitutions; these consisted of three point mutations and one doublet. The in-frame VDJ in the same cell (16G4CG) contained one point mutation in CDR1. To study patterns of hypermutation among the IgH genes in one cell, sIg+ cells from the immunized adult shark-GR were analyzed. The VDJ had been sequenced from 13 single cells [13], eight of which contained mutants (62%). We re-analyzed these eight cells, as well as eight new ones of this series selected according to the criteria described in Material and Methods. The five cells in Fig. 6 were chosen for presentation because most of their IgH genes (≥7/10) could be characterized. The VDJ were isolated as described for Fig. 1, reaction 3, as were the GL VH per Fig. 1, reaction 5.
Figure 6.
Single-cell PCR results from mutant shark-GR sIg+ cells. Top. Rearrangement configurations of the ten IgH in each cell. The IgH genes present in shark-GR were: G1, G2A, G2B, G3, G4A, G4D, G4E, G4C, G4G, and G5. The status of the Ig gene segments is indicated as fully rearranged (VDJ), partly rearranged as VDD-J, or not rearranged (GL). The plus or φ sign indicate respectively in-frame (highlighted) or nonproductive. The mutated VDJ are marked with asterisk. The GL VH has been sequenced in every case except where marked as NR; this means that the GL configuration was detected but a sequence with the VH could not be isolated. Bottom. CDR3 of the VDJ. Six out of nine rearrangements in 5 B cells were mutated. Substitutions indicated in lower case in the CDR3. See Fig. 3 legend. Genbank accession numbers for mutants are: 1G4CG, 15G2A, 23G2A, 25G4D, 49G4CG, and 49G5 (JN087503-JN087508).
Only rearranged VDJ were mutated (asterisked in Fig. 6), irrespective of functionality. None of the 34 GL VH was changed; in other B cells with mutants (KM5, KM13, KM17, KS53, not shown) 11 GL VH also were unchanged. There was no sharing of substitutions in the two mutant VDJ in cell KS49. In KS15, one VDJ was extensively mutated while the other was unchanged.
Screening for SHM at nonrearranged genes
In the previous experiment the GL sample size may have been too small to detect SHM occurring at low frequency. GL VH sequences were therefore amplified from genomic DNA from shark-GR sIgM-positive cells [13]. G2B was chosen because G2B genes in GL configuration are expected to be abundant in shark-GR (see “G2A is different from G2B”). Sequences in GL and various stages of recombination were analyzed. Of 88 GL sequences, 55 carried 1–4 substitutions, a frequency of 0.04% (33/76,912 bp) and 13 independent 1R (VD-D-J) sequences 0.05% (6/11,362 bp). In no case were any tandem substitutions observed. These frequencies were indistinguishable from that obtained from 12 GL sequences from shark-GR erythrocyte DNA, where the polymerase misincorporation frequency was 0.04% (3/8474 bp). Of the 11 2R (VDD-J) sequences eight of them contained one substitution (1/3768 or 0.03%) but three other sequences were mutated throughout VH and the intersegmental region (8–48 changes).
Discussion
A limited window for rearrangement
Using monoclonal antibodies against nurse shark L chain and nurse shark IgM H chain, B cells were respectively isolated from PBL of an immunized shark and spleen of a non-immunized neonate. In each case not more than three VDJ were isolated per cell [Fig. 3 and ref. 13] although 9–10 functional μ genes and their alleles were available. As few cells carried more than one in-frame VDJ, H chain exclusion holds throughout ontogeny. Assuming that it is primarily rearranged genes that are transcribed, these results are comparable to what was found by single-cell RT-PCR in clearnose skate, where only a few μ transcripts were isolated per cell [28]. Since the number of IgM H chain genes can vary considerably among cartilaginous fishes, est. 50–100 in horned shark [29], it is the restriction of gene activation that the two systems display in common.
In nurse shark every μ gene must rearrange to be expressed, and RAG acts upon only a few IgH per B lymphocyte [15]. The chromatin accessibility appears to affect genes individually and at random, since the pattern of rearranged genes has no correlation with their linkage relationship. Nor was the allele more likely to be activated, since it was usually in GL configuration. These observations suggest that shark IgH gene clusters operate autonomously. Although it would have seemed that multiple loci offer an advantage for multiple rounds of rearrangement, viz. until a productive VDJ is generated, this in fact does not occur since we have so far not detected >3 VDJ in any sIgM+ cell. Moreover it appears that the pseudogenes could compete for nuclear factors that bring about chromatin activation.
To obtain some idea of the manner in which the IgH are activated two scenarios were tested, the first being an instructive model where a set number of genes/chromosome locations become activated, in this way directing suppression of other locations. The second model was one where gene activation occurred in sequence and stopped with in-frame VDJ in the functional IgH. Our data showed the most satisfactory overall goodness of fit with the latter model, with up to a maximum of four activated IgH per B cell. We have not cloned four VDJ in a B cell so far, but the sample size was small and we have not included detecting the pseudogenes (31% of gene pool) in the single-cell assay.
In the mouse, Hewitt and coworkers [30] suggested that a direct interaction between the two IgH alleles leads to the relocation of one to suppressive nuclear compartment after cleavage by RAG on the other. However, interallelic crosstalk cannot fully explain H chain exclusion in the shark since linked IgH and their alleles rearrange independently of each other. Perhaps more relevant to the shark observations is a model recently generated for the TCRβ locus, which proposes that if the two alleles are autonomous and activated stochastically they are unlikely to initiate recombination at the same time [10]. A sufficient time lag between the two steps, D to J and V to DJ, would permit a productive VDJ at the first allele to effect inhibition on the second, rearranging allele. In shark a few 2R/1R, like DJ, were found in 36 B cells, suggesting feedback was possible. The feedback signal could come from a surface receptor consisting of H and L chains, if IgL rearranged at the same time, as we have proposed [31], or with an as-yet unidentified shark surrogate L chain that might be a pre-joined (VJ) L chain type like “NS3” [18].
Because shark VDJ (VDDJ) is formed in one step, feedback inhibition could be expected to occur during a time lag between the activation of individual H chain genes. If rearrangement initiation is stochastic, this reduces the likelihood that more than one IgH is poised for rearrangement in a cell. Coupled to a time window, the number of opportunities for attempting rearrangement would be limited; our data fit a model with a maximum of four activated IgH. Taken together, IgH autonomy, stochastic activation of IgH, and a restricted time window could explain much of the basis for H chain exclusion in a system with multiple Ig loci. These conclusions might be applied to the amphibian Xenopus with tetraploid IgH [32], and the Atlantic salmon with its two independently expressed IgM loci [33, 34].
RAG accessibility is regulated
Some insight into what is involved in regulated activation at a shark IgH gene may be gained by re-examining IgH recombination events in thymocytes [13]. Two cell populations are compared in Table II, one where the sIg+ cells were depleted (thymocytes) and the other positively selected (this study). In single thymocytes, up to 7 rearranged genes can be observed although 82% (46/56) were not fully recombined (1R and 2R). In contrast, only 1–3 IgH genes were recombined in B cells, but 29/30 were VDJ (3R). There were more recombination events in thymocytes: 109 events in 12 cells in Table II (13 in 1R + 66 in 2R + 30 in 3R) or an average of 9 per cell, compared to 5.6 per B cell (89 events in 16 cells, 2 in 2R + 87 in 3R). It is not that there is less recombinase activity in thymocytes but that RAG might be described as not acting processively at most genes.
Table II.
Comparison of IgH gene rearrangement configurations in shark T and B cells
| Thymocyte | sIgM+ cell | |
|---|---|---|
| Single cella: | Up to 8 IgH recombined | Up to 3 IgH recombined |
| Average 5 | Average 2 | |
| 56 IgH in 12 cells | 30 IgH in 16 cells | |
| 13 1R | 0 1R | |
| 33 2R | 1 2R | |
| 10 3R | 29 3R | |
| Genomic DNAb: | IgH rearrangements | IgH rearrangements |
| no IgL rearrangements | IgL rearrangements | |
| RNA studiesc: | no H chain mRNA detected | H chain mRNA |
| no L chain mRNA detected | L chain mRNA | |
| no VDD-J detected | VDD-J mutants cloned |
Single thymocyte data are from ref. 13; the sIgM+ column is from present study. 1R is one rearrangement event: VD-D-J or V-DD-J or V-D-DJ. 2R is two events: VDD-J or V-DDJ or VD-DJ. 3R is three events or VDJ.
Rearranged IgH in lymphocyte DNA, as detected by Southern blotting and PCR, were obtained in refs. 13 and 41. Rearranged IgL in sIgM+ genomic DNA were obtained in ref.41; the absence of rearranged IgL in thymus DNA was reported in ref.13.
Northern blotting and RT-PCR [13].
With such striking differences between B and T cells, the recombination events occurring in thymocytes could not have taken place in a common lymphoid progenitor. Since RAG can be recruited to perform VH to D1 rearrangement in G2 genes of any developing lymphocyte, the difference arises from its ability to continue VD to D2 in B cells. Recent findings indicate that RAG activity is regulated by the nature of chromatin structure, or the epigenetic environment of the RSS [35, 36].
Chromatin immunoprecipitation assays have revealed that whereas RAG1 binds RSS at active J gene segments RAG2 is separately brought to the site by binding of modified histone (trimethylated lysine 4 of histone 3), an activating histone mark [36, 37]. The RAG proteins recruited at this “recombination center” function to capture DNA with RSS (e.g. D gene segments) and bring about rearrangement. In thymocytes we speculate that some kind of recombination centers can form but they are not stable because the epigenetic environment is not optimal for recombinase catalytic activity. The fact that we could not detect H chain transcription in thymocytes furthermore suggests that the nature of the chromatin activation is different from B cells [13, 38].
SHM is not uniform
We investigated SHM among the IgH genes in single cells, to determine which genes were targeted for change and whether gene conversion/sequence exchange could be detected. Because we found occasional GL Ig transcripts in cDNA libraries (E.H., unpub. obs.), there was the possibility that transcribing, non-rearranged genes could be bystanders in a hypermutating B cell. Although we have comprehensive information for only five B cells, the results are clear. SHM targeted rearranged genes and its effect was not necessarily uniform within a cell. The extreme instance is in KS15, where only one of the two nonproductive VDJ was targeted and carried 34 substitutions and 2 insertions. If tandem changes are considered single events, then there were 15 “hits” on one gene and none on the other VDJ or in any VH gene segments of the non-rearranged genes. In support of these observations we found that genomic Ig sequence from sIgM+ cells in GL and 1R configurations have a substitution frequency not significantly different from Taq misincorporation, in contrast to some genes in 2R (and 3R, not shown) configurations that can be highly mutated.
However, the situation is not quite so clear-cut. In a recent course of experiments aimed at cloning precursor Ig transcripts in shark-JS as well as GR we found some GL Ig sequences in which mutations occurred throughout VH and the intersegmental regions (P. Hua and E. Hsu, unpub. results); this means that SHM at non-rearranged IgH can take place. The fact that we did not detect any in genomic B cell DNA in the current studies means that this phenomenon is at low frequency in the general sIgM+ cell population in shark-GR. The difference between results from genomic and cDNA sources suggests that SHM at IgH in GL configuration occurs in a minority of cells and under circumstances that are at present not clear.
IgM isotypes may not be all the same
Whether all the IgMs in a cartilaginous fish serve the same function has not been investigated. G2A and G2B rearranged at the same rate in pup cells (Fig. 3) and the transmembrane receptors they encode were expressed at similar levels in shark-LA as well as three other pup samples (Fig. 5B). However, in all the older animals G2A predominated. It is difficult to understand why such a skewing came about based on the V regions, since the few differences are Met/Leu in FR1, Ala/Thr in CDR1, Gly/Ala in CDR2 and Ile/Lys in FR3; the Asp/Gly in JH is often deleted during rearrangement. G2A and G2B differ by 24 amino acids in the 415 residue C region, and of 17 nonconservative differences 10 are in the 104-residue CH2 (3/99 in CH1, 3/102 in CH3 and 1/110 in CH4). These differences reflect our previous phylogenetic analyses [15], that the domains in all functional μ genes evolved at comparable rates except for VH and CH2, which are under strong positive selection for sequence diversity. There is little information on shark antibody effector function and none on whether the various IgM isotypes are specialized. The IgM are secreted as pentamers and switch to monomers during an antibody response [39–40], but there is no data on whether all the H chains are equally capable of doing so. Since there appears to be G2A prominence in every adult, we propose that not all IgM isotypes are equal. But the basis for expression differences, in the absence of information on effector function or RNA half-life, requires further investigation.
Summary
We propose that the stochastic formation of a few stable recombination centers allows rearrangement in B cells to proceed by the RAG efficiently capturing nearby RSS that flank the four gene segments. The neighboring IgH genes are too distant for frequent intergenic rearrangement events, even if all are simultaneously accessible to RAG; this partly explains the recombinational autonomy observed. The nurse shark IgH are regulated independently of allele or chromosomal position. The scoring of VDJ in single B cells best fits a probability model based on sequential IgH activation, which implies possibility for feedback inhibition once a productive VDJ has been generated.
Supplementary Material
Acknowledgments
We thank Victor Lee, Karolina Malecek, and Anita Lui for their work characterizing the nurse shark BAC library.
Abbreviations used in this paper
- RAG
recombination-activating genes
- AID
activation-induced cytidine deaminase
- SHM
somatic hypermutation
- GL
germline
- 3R
three recombination events
- 2R
two recombination events
- 1R
one recombination event
- VD-D-J
IgH gene where one recombination took place between V and D
Footnotes
This research was supported by grants from the National Institutes of Health (GM068095 to E.H., RR006603 to M.F.F.).
References
- 1.Alt FW, Yancopoulos GD, Blackwell TK, Wood C, Thomas E, Boss M, Coffman R, Rosenberg N, Tonegawa S, Baltimore D. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J. 1984;3:1209–1219. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Featherstone K, Wood AL, Bowen AJ, Corcoran AE. The mouse immunoglobulin heavy chain V-D intergenic sequence contains insulators that may regulate ordered V(D)J recombination. J Biol Chem. 2010;285:9327–9338. doi: 10.1074/jbc.M109.098251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jhunjhunwala S, van Zelm MC, Peak MM, Cutchin S, Riblet R, van Dongen JJ, Grosveld FG, Knoch TA, Murre C. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell. 2008;133:265–279. doi: 10.1016/j.cell.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chowdhury D, Sen R. Stepwise activation of the immunoglobulin μ heavy chain gene locus. EMBO J. 2001;20:6394–6403. doi: 10.1093/emboj/20.22.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fuxa M, Skok J, Souabni A, Salvagiotto G, Roldán E, Busslinger M. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 2004;18:411–422. doi: 10.1101/gad.291504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- 7.Feeney AJ. Epigenetic regulation of antigen receptor gene rearrangement. Curr Opin Immunol. 2011;23:171–177. doi: 10.1016/j.coi.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hewitt SL, Chaumeil J, Skok JA. Chromosome dynamics and the regulation of V(D)J recombination. Immunol Rev. 2010;237:43–54. doi: 10.1111/j.1600-065X.2010.00931.x. [DOI] [PubMed] [Google Scholar]
- 9.Vettermann C, Schlissel MS. Allelic exclusion of immunoglobulin genes: models and mechanisms. Immunol Rev. 2010;237:22–42. doi: 10.1111/j.1600-065X.2010.00935.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farcot E, Bonnet M, Jaeger S, Spicuglia S, Fernandez B, Ferrier P. TCR beta allelic exclusion in dynamical models of V(D)J recombination based on allele independence. J Immunol. 2010;185:1622–1632. doi: 10.4049/jimmunol.0904182. [DOI] [PubMed] [Google Scholar]
- 11.Edholm ES, Wilson M, Bengtén E. Immunoglobulin light (IgL) chains in ectothermic vertebrates. Dev Comp Immunol. 2011 doi: 10.1016/j.dci.2011.01.012. [DOI] [PubMed] [Google Scholar]
- 12.Rast JP, Litman GW. Towards understanding the evolutionary origins and early diversification of rearranging antigen receptors. Immunol Rev. 1998;166:79–86. doi: 10.1111/j.1600-065x.1998.tb01254.x. [DOI] [PubMed] [Google Scholar]
- 13.Malecek K, Lee V, Feng W, Huang JL, Flajnik MF, Ohta Y, Hsu E. Immunoglobulin heavy chain exclusion in the shark. PLoS Biol. 2008;6:e157. doi: 10.1371/journal.pbio.0060157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hinds KR, Litman GW. Major reorganization of immunoglobulin VH segmental elements during vertebrate evolution. Nature. 1986;320:546–549. doi: 10.1038/320546a0. [DOI] [PubMed] [Google Scholar]
- 15.Lee V, Huang JL, Lui MF, Malecek K, Ohta Y, Mooers A, Hsu E. The evolution of multiple isotypic IgM heavy chains in the shark. J Immunol. 2008;180:7461–7470. doi: 10.4049/jimmunol.180.11.7461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rumfelt LL, Lohr RL, Dooley H, Flajnik MF. Diversity and repertoire of IgW and IgM VH families in the newborn nurse shark. BMC Immunol. 2004;5:8. doi: 10.1186/1471-2172-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conticello SG, Thomas CJ, Petersen-Mahrt SK, Neuberger MS. Evolution of the AID/APOBEC family of polynucleotide (deoxy)cytidine deaminases. Mol Biol Evol. 2005;22:367–377. doi: 10.1093/molbev/msi026. [DOI] [PubMed] [Google Scholar]
- 18.Lee SS, Tranchina D, Ohta Y, Flajnik MF, Hsu E. Hypermutation in shark immunoglobulin light chain genes results in contiguous substitutions. Immunity. 2002;16:571–582. doi: 10.1016/s1074-7613(02)00300-x. [DOI] [PubMed] [Google Scholar]
- 19.Diaz M, Stanfield RL, Greenberg AS, Flajnik MF. Structural analysis, selection, and ontogeny of the shark new antigen receptor (IgNAR): identification of a new locus preferentially expressed in early development. Immunogenetics. 2002;54:501–512. doi: 10.1007/s00251-002-0479-z. [DOI] [PubMed] [Google Scholar]
- 20.Bengtén E, Strömberg S, Daggfeldt A, Magor BG, Pilström L. Transcriptional enhancers of immunoglobulin light chain genes in Atlantic cod (Gadus morhua) Immunogenetics. 2000;51:647–658. doi: 10.1007/s002510000176. [DOI] [PubMed] [Google Scholar]
- 21.Hsu E, Criscitiello MF. Diverse immunoglobulin light chain organizations in fish retain potential to revise B cell receptor specificities. J Immunol. 2006;177:2452–2462. doi: 10.4049/jimmunol.177.4.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malecek K, Brandman J, Brodsky JE, Ohta Y, Flajnik MF, Hsu E. Somatic hypermutation and junctional diversification at Ig heavy chain loci in the nurse shark. J Immunol. 2005;175:8105–8115. doi: 10.4049/jimmunol.175.12.8105. [DOI] [PubMed] [Google Scholar]
- 23.Luo M, Kim H, Kudrna D, Sisneros NB, Lee S-J, Mueller C, Collura K, Zuccolo A, Buckingham EB, Grim SM, Yanagiya K, Inoko H, Shiina T, Flajnik MF, Wing RA, Ohta Y. Construction of a nurse shark (Ginglymostoma cirratum) bacterial artificial chromosome (BAC) library and a preliminary genome survey. BMC Genomics. 2006;7:106. doi: 10.1186/1471-2164-7-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greenberg AS, Avila D, Hughes M, Hughes A, McKinney EC, Flajnik MF. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature. 1995;374:168–173. doi: 10.1038/374168a0. [DOI] [PubMed] [Google Scholar]
- 25.Radlow R, Alf EF. An alternate multinomial assessment of the accuracy of the χ2 test of goodness of fit. J Am Stat Assoc. 1975;70:811–813. [Google Scholar]
- 26.Diaz M, Greenberg AS, Flajnik MF. Somatic hypermutation of the new antigen receptor (NAR) in the nurse shark does not generate the repertoire: possible role in antigen-driven reactions in the absence of germinal centers. Proc Natl Acad Sci USA. 1998;95:4343–14348. doi: 10.1073/pnas.95.24.14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diaz M, Velez J, Singh M, Cerny J, Flajnik MF. Mutational pattern of the nurse shark antigen receptor gene (NAR) is similar to that of mammalian Ig genes and to spontaneous mutations in evolution: the translesion synthesis model of somatic hypermutation. Int Immunol. 1999;11:825–833. doi: 10.1093/intimm/11.5.825. [DOI] [PubMed] [Google Scholar]
- 28.Eason DD, Litman RT, Luer CA, Kerr W, Litman GW. Expression of individual immunoglobulin genes occurs in an unusual system consisting of multiple independent loci. Eur J Immunol. 2004;34:2551–2558. doi: 10.1002/eji.200425224. [DOI] [PubMed] [Google Scholar]
- 29.Kokubu F, Hinds K, Litman R, Shamblott MJ, Litman GW. Extensive families of constant region genes in a phylogenetically primitive vertebrate indicate an additional level of immunoglobulin complexity. Proc Natl Acad Sci USA. 1987;84:5868–5872. doi: 10.1073/pnas.84.16.5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hewitt SL, Yin B, Ji Y, Chaumeil J, Marszalek K, Tenthorey J, Salvagiotto G, Steinel N, Ramsey LB, Ghysdael J, Farrar MA, Sleckman BP, Schatz DG, Busslinger M, Bassing CH, Skok JA. RAG-1 and ATM coordinate monoallelic recombination and nuclear positioning of immunoglobulin loci. Nat Immunol. 2009;10:655–664. doi: 10.1038/ni.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fleurant M, Changchien L, Chen CT, Flajnik MF, Hsu E. Shark Ig light chain junctions are as diverse as in heavy chains. J Immunol. 2004;173:5574–5582. doi: 10.4049/jimmunol.173.9.5574. [DOI] [PubMed] [Google Scholar]
- 32.Du Pasquier L, Hsu E. Immunoglobulin expression in diploid and polyploid interspecies hybrids of Xenopus: evidence for allelic exclusion. Eur J Immunol. 1983;13:585–590. doi: 10.1002/eji.1830130714. [DOI] [PubMed] [Google Scholar]
- 33.Hordvik I, Berven FS, Solem ST, Hatten F, Endresen C. Analysis of two IgM isotypes in Atlantic salmon and brown trout. Mol Immunol. 2002;39:313–321. doi: 10.1016/s0161-5890(02)00114-1. [DOI] [PubMed] [Google Scholar]
- 34.Yasuike M, de Boer J, von Schalburg KR, Cooper GA, McKinnel L, Messmer A, So S, Davidson WS, Koop BF. Evolution of duplicated IgH loci in Atlantic salmon, Salmo salar. BMC Genomics. 2010;11:486. doi: 10.1186/1471-2164-11-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y, Subrahmanyam R, Chakraborty T, Sen R, Desiderio S. A plant homeodomain in RAG-2 that binds Hypermethylated lysine 4 of histone H3 is necessary for efficient antigen-receptor-gene rearrangement. Immunity. 2007;27:561–571. doi: 10.1016/j.immuni.2007.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matthews AG, Kuo AJ, Ramón-Maiques S, Han S, Champagne KS, Ivanov D, Gallardo M, Carney D, Cheung P, Ciccone DN, Walter KL, Utz PJ, Shi Y, Kutateladze TG, Yang W, Gozani O, Oettinger MA. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ji Y, Resch W, Corbett E, Yamane A, Casellas R, Schatz DG. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 2010;141:419–431. doi: 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ji Y, Little AJ, Banerjee JK, Hao B, Oltz EM, Krangel MS, Schatz DG. Promoter, enhancers, and transcription target RAG1 binding during V(D)J recombination. J Exp Med. 2010;207:2809–2816. doi: 10.1084/jem.20101136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Voss EW, Sigel MM. Distribution of 19S and 7S IgM antibodies during the immune response in the nurse shark. J Immunol. 1971;106:1323–1329. [PubMed] [Google Scholar]
- 40.Dooley H, Flajnik MF. Shark immunity bites back: affinity maturation and memory response in the nurse shark. Ginglymostoma cirratum. Eu r J Immunol. 2005;35:936–45. doi: 10.1002/eji.200425760. [DOI] [PubMed] [Google Scholar]
- 41.Zhu C, Hsu E. Error-prone DNA repair activity during somatic hypermutation in shark B lymphocytes. J Immunol. 2010;185:5336–5347. doi: 10.4049/jimmunol.1000779. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.