Abstract
Covalent modifications of histone tails have a key role in regulating chromatin structure and controlling transcriptional activity. In eukaryotes, histone H3 trimethylated at lysine 4 (H3K4me3) is associated with active chromatin and gene expression1–4. We recently found that plant homeodomain (PHD) finger of tumour suppressor ING2 (inhibitor of growth 2) binds H3K4me3 and represents a new family of modules that target this epigenetic mark5. The molecular mechanism of H3K4me3 recognition, however, remains unknown. Here we report a 2.0 Å resolution structure of the mouse ING2 PHD finger in complex with a histone H3 peptide trimethylated at lysine 4. The H3K4me3 tail is bound in an extended conformation in a deep and extensive binding site consisting of elements that are conserved among the ING family of proteins. The trimethylammonium group of Lys 4 is recognized by the aromatic side chains of Y215 and W238 residues, whereas the intermolecular hydrogen-bonding and complementary surface interactions, involving Ala 1, Arg 2, Thr 3 and Thr 6 of the peptide, account for the PHD finger’s high specificity and affinity. Substitution of the binding site residues disrupts H3K4me3 interaction in vitro and impairs the ability of ING2 to induce apoptosis in vivo. Strong binding of other ING and YNG PHD fingers suggests that the recognition of H3K4me3 histone code is a general feature of the ING/YNG proteins. Elucidation of the mechanisms underlying this novel function of PHD fingers provides a basis for deciphering the role of the ING family of tumour suppressors in chromatin regulation and signalling.
Specific post-translational modifications of histone tails facilitate recruitment of distinct proteins to nucleosomes to either activate gene expression or condense the chromatin6. Despite a wide variety of known histone marks, only a few protein domains have been identified that recognize or ‘read’ the precise tail modifications. This includes bromodomain, which binds acetylated lysine residues of histone H4 (refs 7, 8), and chromodomain, which targets methylated lysine residues of histone H3 (refs 9–13). A number of recent reports suggest that PHD modules are involved in chromatin remodelling14,15. We found that the carboxy-terminal PHD finger of tumour suppressor ING2 directly associates with the histone H3 tail tri-methylated at Lys 4. To understand the molecular mechanism of this histone code recognition, we determined a 2 Å resolution crystal structure of the mouse ING2 PHD finger bound to the H3K4me3 peptide and established the determinants of specificity and functional significance of this interaction.
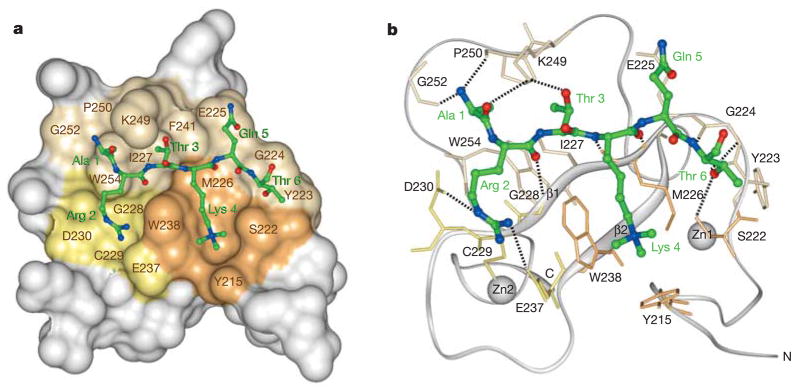
The structure of the complex indicates a conserved mode of methyl-lysine recognition by PHD fingers of ING proteins and reveals key elements that define the binding specificity. The overall fold of the ING2 PHD finger in the complex is similar to the fold previously found in ligand-free PHD modules16. The structure consists of three loops stabilized by two zinc-binding clusters and a small double-stranded anti-parallelβ-sheet (Fig. 1a, b). The structure of the ING2 PHD finger in complex with H3K4me3 peptide superimposes well with the structure of the free PHD finger of identical ING1L protein (Protein Data Bank 1WES) with a root-mean-squared deviation of 0.8 Å, thus indicating that binding of the peptide does not induce significant conformational changes in the protein.
Figure 1. Structure of ING2 PHD finger in complex with a histone H3 peptide trimethylated at Lys 4.
a, The PHD finger is shown as a solid surface with the binding site residues coloured and labelled. The Lys 4 and Arg 2 binding grooves are in brown and yellow, respectively. The histone peptide is shown as a ball-and-stick model with C, O and N atoms coloured green, red and blue, respectively. b, Ribbon diagram of the structure. Dashed lines represent intermolecular hydrogen bonds.
The histone H3K4me3 tail is bound in a deep and extensive binding site that engages nearly one-third of the PHD finger residues (Fig. 1a, b). The peptide adopts an extended β-strand-like conformation and resides in two grooves connected by a narrow channel (Fig. 1a). The fully extended side chain of Lys 4 occupies the major groove formed by the Y215, S222, M226 and W238 residues of the PHD finger, whereas Arg 2 is bound in the adjacent groove and is surrounded by G228, C229, D230 and E237. The aromatic rings of W238 and F241, which protrude orthogonally to the surface of the protein, separate the two grooves creating a narrow channel where Thr 3 is bound. The peptide lies anti-parallel to the protein’sβ1 strand and pairs with it, forming a third strand within the three-stranded β-sheet (Fig. 1b).
Numerous hydrogen bonds are formed between the H3K4me3 peptide and the PHD finger. Residues Arg 2, Lys 4 and Thr 6 of the peptide make characteristic β-sheet contacts with the G224, M226 and G228 residues of the PHD finger (Fig. 1b). The peptide–protein interaction is further stabilized by intermolecular hydrogen bonds involving residues S222, D230, E237, K249, P250 and G252 of the domain. Thus, the guanidino moiety of Arg 2 is restrained by the interactions with D230 and E237. The hydroxyl group of Thr 3 is hydrogen bonded to K249. The carbonyl and backbone group of Ala 1 form hydrogen bonds with K249, P250 and G252. The hydroxyl moiety of Thr 6 interacts with the amide group of G224 and with the hydroxyl group of S222.
In addition to the formation of intermolecular hydrogen bonds, the complementary surface interactions stabilize the complex. The side chains of each peptide residue precisely fit in the distinct binding pockets. Well-defined binding grooves for Lys 4 and Arg 2 residues are apparent. The methyl groups of Ala 1 and Thr 3 are buried in the deep hydrophobic pockets formed by the core residues W254 and I227. The side chain of Thr 6 is positioned in the site formed by S222, Y223 and G224. Thus, the H3K4me3 tail is bound by the PHD finger through an extensive network of hydrogen-bonding and complementary surface interactions. These contacts, which involve three residues preceding and Thr 6 following the methylated lysine 4, are responsible for the PHD finger specificity and unique recognition of the ARTK(me3)QT peptide sequence.
The trimethylammonium group of Lys 4 is recognized by two aromatic residues, Y215 and W238 (Fig. 1a, b). Aromatic side chains of Y215 and W238, positioned perpendicular to each other and orthogonal to the protein surface, make cation–π contacts with the trimethylammonium group of Lys 4. This mode of methyl-lysine recognition by two aromatic residues has been recently reported for double chromodomains of human CHD1 (ref. 13). Notably, both proteins, CHD1 and ING2, target methylated Lys 4 and use two aromatic residues, not the typical three-residue aromatic cage used by chromodomains of HP1 and Polycomb for binding to methylated H3K9 and H3K27, respectively11,12,17,18. Furthermore, although ING2 PHD finger and CHD1 chromodomains are structurally unrelated, further resemblance in the binding of H3K4me3 is evident. In both cases, the histone peptide is bound in an extended conformation with Lys 4 and Arg 2 occupying adjacent grooves separated by a tryptophan residue.
Alignment of amino acid sequences of PHD fingers is shown in Supplementary Fig. 1. Conservation of the binding site elements among all human ING and yeast YNG proteins suggests a similar mode of H3K4me3 recognition by their PHD fingers. However, the critical binding site residues are not found in many PHD sequences, indicating that the H3K4me3 interaction is likely to be a function of a subset of PHD modules. We found only 11 PHD finger-containing proteins outside of the ING family that contain residues important for the interaction. It remains to be tested whether these proteins are able to bind H3K4me3.
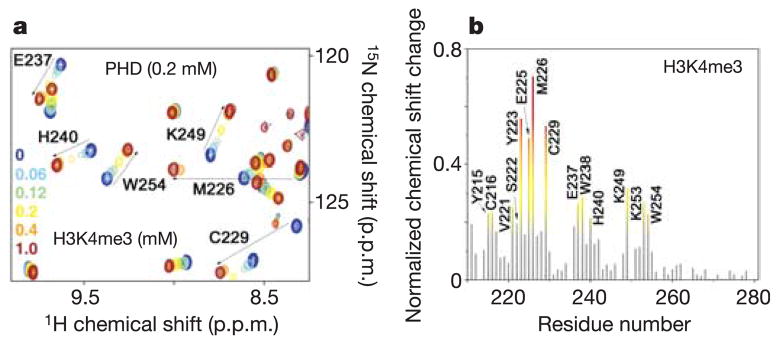
The ING2 PHD finger binds histone H3 trimethylated and dimethylated at Lys 4 but does not recognize histone H3 methylated at Lys 9 or histone H4 methylated at Lys 20. To establish the PHD finger specificity for naturally occurring histone modification, the peptides corresponding to methylated H3K4, H3K9 and H4K20 were tested by NMR and tryptophan fluorescence spectroscopy (Table 1 and Supplementary Fig. 2). Substantial chemical shift changes were observed in the residues comprising the H3K4me3 binding site when H3K4me3, H3K4me2 or H3K4me1 peptides were gradually added to the 15N-labelled ING2 PHD finger (Fig. 2 and Supplementary Fig. 2). An almost identical pattern of chemical shift perturbations indicated that these peptides were bound similarly; however, the PHD finger displayed one and two orders of magnitude higher affinity for H3K4me3 than for H3K4me2 and H3K4me1 (Table 1). Comparable affinities, in the range of 1–10μM, are exhibited by chromodomains and bromodomains for the correspondingly modified histone tails8,11,12, thus suggesting that the H3K4me3 interaction of the PHD finger is physiologically relevant. Yet, the ING2 PHD finger seems to be more specific for H3K4me3 than chromodomains, which show little preference for trimethylated over dimethylated histone peptides11–13. Chromatin immunoprecipitation assays in an accompanying paper corroborate this, showing strong in vivo association of ING2 with H3K4me3 and only weak association with H3K4me2 (ref. 5). The methylated H3K9 peptides or unmodified H3 were bound weakly, and no binding was detected for methylated H4K20 (Table 1). Thus, in agreement with our structural analysis, these data demonstrate that the ART/QT sequence around Lys 4 is critical for binding, and that substitution of these residues with either TAR/ST or RHR/VL disrupts the interaction. Furthermore, Thr 3 of the peptide is positioned in the narrow channel connecting two grooves. The small size of the channel, which is limited by two aromatic side chains of W238 and F141, would exclude any residue larger than threonine, such as arginine in TAR/ST or RHR/VL sequences. Other human ING and yeast YNG PHD fingers show comparable affinities for H3K4me3 (Table 1), indicating that the recognition of this histone code is a general function of the ING family proteins.
Table 1.
Affinities of the wild-type and mutant PHD fingers for the histone tail peptides
| PHD proteins | Ligand | Kd (μM) |
|---|---|---|
| ING2 wild type | H3K4me3 | 1.5 ± 1* |
| ING2 wild type | H3K4me2 | 15 ± 4* |
| ING2 wild type | H3K4me1 | 208 ± 80* |
| ING2 wild type | H3K9me3 | 2,000 ± 60† |
| ING2 wild type | H3K9me2 | 2,320 ± 300*† |
| ING2 wild type | H3K9me1 | 2,380 ± 800† |
| ING2 wild type | H3 unmodified | 2,240 ± 350*† |
| ING2 wild type | H4K20me3 | >10,000† |
| ING2 wild type | H4K20me2 | >10,000*† |
| ING2 wild type | H4K20me1 | >7,000*† |
| ING2 Y215A | H3K4me3 | >5,000* |
| ING2 S222A | H3K4me3 | 24 ± 10* |
| ING2 Y223A/E225A | H3K4me3 | 326 ± 169*† |
| ING2 D230A | H3K4me3 | 51 ± 15* |
| ING2 E237A/W238A | H3K4me3 | 726 ± 50* |
| ING2 K249A/K251A/K253A | H3K4me3 | 1.1 ± 0.6* |
| ING1 | H3K4me3 | 3.3 ± 1.6* |
| ING3 | H3K4me3 | 6.9 ± 1.1* |
| ING4 | H3K4me3 | 7.9 ± 2* |
| ING5 | H3K4me3 | 2.4 ± 1* |
| YNG1 | H3K4me3 | 2.3 ± 0.9* |
| YNG2 | H3K4me3 | 5.1 ± 1.8* |
| PHO23 | H3K4me3 | 4.5 ± 1.3* |
K d, dissociation constant.
Determined by tryptophan fluorescence.
Determined by NMR.
Figure 2. ING2 PHD finger recognizes H3K4me3.
a, Six superimposed 1H,15N heteronuclear single quantum coherence (HSQC) spectra of PHD (0.2 mM) collected during titration of H3K4me3 peptide are colour-coded according to the ligand concentration (inset). b, The histogram displays normalized 1H,15N chemical shift changes observed in the corresponding (a) spectra of the PHD finger. The normalized27 chemical shift change was calculated using the equation [(ΔδH)2 + (ΔδN/5)2]0.5, where δ is the chemical shift in parts per million (p.p.m.). Coloured bars indicate significant change being greater than average plus one-half standard deviation.
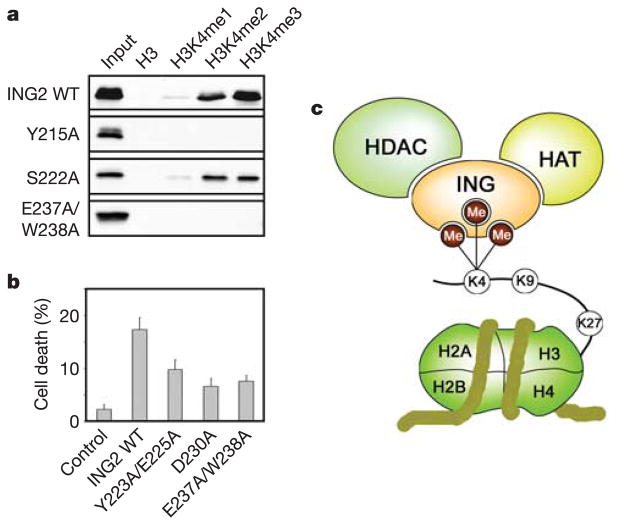
Mutation of the H3K4me3 active site residues disrupts binding. To determine the relative contributions of the binding site residues in the coordination of H3K4me3, they were substituted for Ala. Consequently, the mutant proteins were tested by pull-down experiments (Fig. 3a) and their binding affinities were measured by tryptophan fluorescence and NMR (Table 1). An Ala substitution of the Y215 residue abolished binding, whereas replacement of S222 reduced the binding affinity, suggesting a critical role of the aromatic residue and less significant role of serine of the major groove in the interaction (Table 1 and Fig. 3a). As expected, mutation of both W238 and E237 residues involved in the recognition of Lys 4 and Arg 2, respectively, disrupted the binding, whereas replacement of D230 resulted in a 30-fold decrease of the affinity, pointing to a significant contribution of Arg 2 recognition to the binding energetics. A substantial decrease of activity was observed for the Y223A/E225A mutant. This indicates the importance of surface complementarity at the PHD–H3K4me3 interface.
Figure 3. ING2 function requires its H3K4me3 binding activity.
a, Interactions of the GST fusion wild-type and mutant ING2 PHD fingers with biotinylated histone peptides examined by western blot analysis. b, Stimulation of apoptosis by overexpressed full-length wild-type and mutant ING2. Cell death was measured in HT1080 cells transfected with 10μg of the indicated plasmids. The error bars represent the mean ± s.e.m. from at least three independent experiments. c, A model of the association of HAT/HDAC chromatin-modifying complexes with the nucleosome through the binding of PHD fingers of ING proteins to H3K4me3.
To elucidate the functional significance of H3K4me3 binding, the ability of wild-type and mutant ING2 proteins to induce apoptosis was compared. Overexpression of ING2 is known to stimulate cell death19. We measured the apoptosis level by Trypan blue exclusion in HT1080 cells transfected with full-length wild-type ING2 and Y223A/E225A, D230A and E237A/W238A mutant proteins (Fig. 3b). These mutants were defective in H3K4me3 binding in vitro (Table 1). Furthermore, they substantially decreased the ability of the protein to induce apoptosis, demonstrating that this interaction is necessary for the function of ING2.
Our data reveal that the ING2 PHD finger directly and specifically binds histone H3 tail trimethylated at Lys 4. The family of ING tumour suppressors is implicated in chromatin remodelling, DNA damage repair and together with p53 induces apoptosis and senescence20,21. ING proteins associate with and modulate the activity of histone acetyl transferase (HAT) and histone deacetylase (HDAC) chromatin-modifying complexes20,22,23. Our results suggest that ING proteins may direct the HAT/HDAC complexes to chromatin through binding of their PHD fingers to histone H3 poly-methylated at Lys 4 (Fig. 3c). A number of PHD finger residues involved in H3K4me3 binding, including S222, G224 and M226, are found to be mutated in cancer cells (Supplementary Fig. 1), suggesting the vital role of this interaction in tumorigenesis21. Consequently, our results should help in elucidating the general principles by which ING proteins influence chromatin remodelling and transcriptional activation and may aid in the understanding of how the ING-dependent pathways can be therapeutically manipulated to prevent and treat cancer.
METHODS
See Supplementary Information for details of mutagenesis, expression and purification of proteins, NMR spectroscopy, cell-culture and western analyses, and fluorescence spectroscopy.
X-ray crystallography
The ING2 PHD finger (1.0 mM) was combined with H3K4me3 peptide (residues 1–12) in a 1:1.5 molar ratio before crystallization. Crystals of the complex were grown using the hanging-drop vapour diffusion method at 25 °C by mixing 1μl of the protein peptide solution with 1μl of a well solution containing 0.01 M NiCl2·6H2O, 0.1 M Tris (pH 8.0) and 22% poly-ethylene glycol monomethyl ether (PEGMME 2K). Crystals were soaked for 15 min in the same solution supplemented with 25% PEGMME 2K before being flash-cooled in liquid nitrogen. All data were collected at −180 °C. The native data set was collected on a RU-H3R generator with a Raxis IV++ detector at the University of Colorado Health Sciences Center’s X-ray Core Facility. The Zn MAD data set was collected at beamline X26C of the National Synchrotron Light Source. Data were processed with the HKL2000 package24 and statistics are shown in Supplementary Table 1. The molecular replacement method and Zn multi-wavelength anomalous dispersion (MAD) method were simultaneously pursued for structural determination. The molecular replacement solution was generated using the MolRep program in CCP4 and NMR structure of PHD finger of ING1L (Protein Data Bank 1WES) as a model. At the same time, two Zn ions were located using HKL2MAP and an initial experimental phase map was calculated using the Solve/Resolve program. This initial map is of excellent quality and shows clear density for main chains, side chains and main-chain carbonyl groups. Residues 212–263 of the PHD domain and 1–8 of the peptide can be built into the density using program O25 without any ambiguity. The remaining residues (205–211 and 264–265 of the PHD domain and residues 9–12 of the peptide) are disordered in the crystal structure. Refinement was performed using the program CNS26.
Supplementary Material
Acknowledgments
We thank D. Bentley and J. Tyler for discussions; M. Grunstein for providing GST fusion histone tails; B. Tripet and the Biophysics Core Facility for synthesis of the histone peptides; the University of Colorado Health Sciences Center’s X-ray Core Facility and NMR Core Facility, supported by the University of Colorado Cancer Center; and A. Heroux and the mail-in data collection service at the National Synchrotron Light Source (NSLS) for synchrotron data collection. Financial support for NSLS comes principally from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and from the National Center for Research Resources of the National Institutes of Health (NIH). This research was supported by grants from the NIH (V.V.V., O.G. and T.G.K.), Burroughs Welcome (O.G.), American Heart Association (T.G.K.) and University of Colorado Cancer Center (T.G.K.). T.G.K. is a NARSAD Young Investigator and an American Cancer Society Research Scholar.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
The coordinates have been deposited in the Protein Data Bank under accession number 2G6Q. Reprints and permissions information is available at npg.nature.com/reprintsandpermissions.
The authors declare no competing financial interests.
References
- 1.Santos-Rosa H, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 2.Schneider R, et al. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nature Cell Biol. 2004;6:73–77. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- 3.Briggs SD, et al. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev. 2001;15:3286–3295. doi: 10.1101/gad.940201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sims RJ, III, Nishioka K, Reinberg D. Histone lysine methylation: a signature for chromatin function. Trends Genet. 2003;19:629–639. doi: 10.1016/j.tig.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 5.Shi X, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006 doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 7.Dhalluin C, et al. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- 8.Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- 9.Bannister AJ, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 10.Lachner M, O’Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen PR, et al. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416:103–107. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- 12.Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 13.Flanagan JF, et al. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 14.Ragvin A, et al. Nucleosome binding by the bromodomain and PHD finger of the transcriptional cofactor p300. J Mol Biol. 2004;337:773–788. doi: 10.1016/j.jmb.2004.01.051. [DOI] [PubMed] [Google Scholar]
- 15.Eberharter A, Vetter I, Ferreira R, Becker PB. ACF1 improves the effectiveness of nucleosome mobilization by ISWI through PHD-histone contacts. EMBO J. 2004;23:4029–4039. doi: 10.1038/sj.emboj.7600382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pascual J, Martinez-Yamout M, Dyson HJ, Wright PE. Structure of the PHD zinc finger from human Williams-Beuren syndrome transcription factor. J Mol Biol. 2000;304:723–729. doi: 10.1006/jmbi.2000.4308. [DOI] [PubMed] [Google Scholar]
- 17.Fischle W, et al. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagashima M, et al. DNA damage-inducible gene p33ING2 negatively regulates cell proliferation through acetylation of p53. Proc Natl Acad Sci USA. 2001;98:9671–9676. doi: 10.1073/pnas.161151798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doyon Y, et al. ING tumor suppressor proteins are critical regulators of chromatin acetylation required for genome expression and perpetuation. Mol Cell. 2006;21:51–64. doi: 10.1016/j.molcel.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 21.Campos EI, Chin MY, Kuo WH, Li G. Biological functions of the ING family tumor suppressors. Cell Mol Life Sci. 2004;61:2597–2613. doi: 10.1007/s00018-004-4199-4. [DOI] [PubMed] [Google Scholar]
- 22.Loewith R, Meijer M, Lees-Miller SP, Riabowol K, Young D. Three yeast proteins related to the human candidate tumor suppressor p33(ING1) are associated with histone acetyltransferase activities. Mol Cell Biol. 2000;20:3807–3816. doi: 10.1128/mcb.20.11.3807-3816.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuzmichev A, Zhang Y, Erdjument-Bromage H, Tempst P, Reinberg D. Role of the Sin3-histone deacetylase complex in growth regulation by the candidate tumor suppressor p33(ING1) Mol Cell Biol. 2002;22:835–848. doi: 10.1128/MCB.22.3.835-848.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 25.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 26.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 27.Grzesiek S, Stahl SJ, Wingfield PT, Bax A. The CD4 determinant for downregulation by HIV-1 Nef directly binds to Nef. Mapping of the Nef binding surface by NMR. Biochemistry. 1996;35:10256–10261. doi: 10.1021/bi9611164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.