Abstract
Helicases are vital enzymes that carry out strand separation of duplex nucleic acids during replication, repair, and recombination1,2. Bacteriophage T7 gene product 4 is a model hexameric helicase which has been observed to utilize dTTP, but not ATP, to unwind dsDNA as it translocates from 5′ to 3′ along ssDNA2–6. Whether and how different subunits of the helicase coordinate their chemo-mechanical activities and DNA binding during translocation is still under debate1,7. Here we address this question using a single molecule approach to monitor helicase unwinding. We discovered that T7 helicase does in fact unwind dsDNA in the presence of ATP and the unwinding rate is even faster than that with dTTP. However unwinding traces showed a remarkable sawtooth pattern where processive unwinding was repeatedly interrupted by sudden slippage events, ultimately preventing unwinding over a substantial distance. This behavior was not observed with dTTP alone and was greatly reduced when ATP solution was supplemented with a small amount of dTTP. These findings presented an opportunity to use nucleotide mixtures to investigate helicase subunit coordination. We found T7 helicase binds and hydrolyzes ATP and dTTP by competitive kinetics such that the unwinding rate is dictated simply by their respective Vmax, KM, and concentrations. In contrast, processivity does not follow a simple competitive behavior and shows a cooperative dependence on nucleotide concentrations. This does not agree with an uncoordinated mechanism where each subunit functions independently, but supports a model where nearly all subunits coordinate their chemo-mechanical activities and DNA binding. Our data indicate that only one subunit at a time can accept a nucleotide while other subunits are nucleotide-ligated and thus interact with the DNA to ensure processivity. Such subunit coordination may be general to many ring-shaped helicases and reveals a potential mechanism for regulation of DNA unwinding during replication.
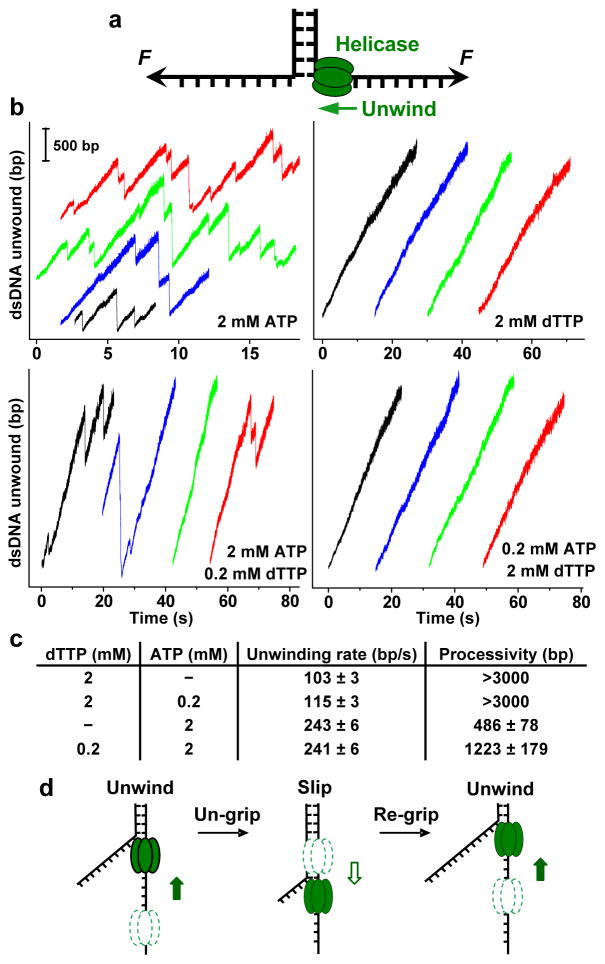
Despite the fact that most motor proteins use ATP as a fuel source, previous bulk studies have shown T7 helicase does not unwind DNA efficiently in the presence of ATP, although it is capable of ATP hydrolysis5,6,8. To investigate why ATP appeared not to support T7 helicase unwinding, we employed a single molecule optical trapping assay that we previously developed to measure unwinding of double-stranded DNA (dsDNA) or translocation on single-stranded DNA (ssDNA) (Fig. 1a; Supplementary Fig. 1)9. Briefly, two strands of a DNA fork junction were held under tension which was not sufficient to mechanically unwind the junction without a helicase. Helicase unwinding of the junction resulted in an increase in the ssDNA length, permitting tracking of the helicase location. When experiments were conducted with 2 mM ATP, we were surprised to find that ATP supported not only dsDNA unwinding but also a significantly faster rate than that with dTTP (Fig. 1b-c). However, processive unwinding was interrupted by slippage events, resulting in a remarkable sawtooth pattern in the unwinding trace (Fig. 1b). Control experiments verified that each trace was the action of a single helicase (Supplementary Fig. 2). We attribute this pattern to helicase losing its grip on the ssDNA, sliding backwards under the influence of the reannealing DNA fork, and then regaining its grip and resuming unwinding (Fig. 1d). In contrast, slippage behavior was essentially absent with 2 mM dTTP alone (Fig. 1b). These results resolve the mystery of the apparent lack of significant unwinding activity seen in bulk studies4–6,8; unwinding and slippage could not be separated, so unwinding was masked by unobservable slips that prevented helicase from moving over a substantial distance. The current work is the first direct observation of helicase nucleotide-specific slippage. Previous studies of non-ring-shaped helicases have reported reverse motions of the unwinding fork attributable to helicase reaching the end of the DNA or encountering a barrier10,11, dissociating from the DNA12,13, or moving in the reverse direction9,12,13. These are of a somewhat different nature than what we have observed. The only slippage behavior that may resemble ours is from non-helicase bacteriophage motors14,15, but their slippage is not a result of the use of a specific nucleotide.
Figure 1. Comparison of helicase unwinding behaviors with different nucleotides.
a, Schematic of the single molecule configuration (not to scale). The single-stranded ends of a dsDNA were held at a constant unzipping force of 8 pN while T7 helicase unwound the dsDNA by translocating on ssDNA. b, Representative traces showing the number of unwound base pairs versus time in the presence of various concentrations of nucleotides. For clarity, traces have been arbitrarily shifted along both axes. c, A summary of unwinding rates and processivities. Uncertainties are s.e.m. d, Cartoon illustrating slippage behavior. Helicase unwinds, loses grip, slips, re-grips, and resumes unwinding. Dotted helicase indicates a previous location of the helicase.
Slippage was not observed with dTTP alone (Fig. 1b) and therefore appears to be sensitive either to the base composition of the bound nucleotide (e.g., adenosine versus thymidine) or the type of sugar (ribose versus deoxyribose). We compared slippage for all four NTPs and their dNTP counterparts (Supplementary Fig. 3). For each nucleotide we measured processivity, defined as the mean distance between slips (Supplementary Fig. 4). The results indicate that the additional 2′-OH group on the ribose sugar makes the helicase more prone to slipping. Examination of the helicase structure at the nucleotide binding pocket16 reveals the 2′-OH group of a bound nucleotide may displace the -OH group on the side chain of residue Y535 (Supplementary Fig. 5a). We thus generated a Y535F mutant to remove the -OH group and it showed significantly increased processivity in the presence of ATP, albeit still less than that seen for dATP (Supplementary Fig. 5b).
Although ATP caused helicase to slip more frequently, it supported a much faster unwinding rate between slips, consistent with an earlier finding of a faster rate of ATP hydrolysis17. Because ATP and dTTP support different unwinding rates and processivities, we used nucleotide mixtures to understand how multiple subunits of the helicase coordinate unwinding activity. We approximated the in vivo concentrations of ATP and dTTP of E. coli18, by utilizing 2.0 mM ATP and a small amount of dTTP, 0.2 mM (Fig. 1b-c). Although the unwinding rate between slips was close to the value observed with 2 mM ATP alone, the processivity increased by ~ 3-fold. When the converse experiment was performed (0.2 mM ATP and 2.0 mM dTTP), the unwinding rate was comparable to that with 2 mM dTTP alone and minimal slippage was observed (Fig. 1b-c). These results imply that even a small fraction of helicase subunits, when bound with dTTP, reduce slippage and substantially increase processivity. This finding was further substantiated by bulk experiments using ATP alone, and an ATP/dTTP mixture (Supplementary Fig. 6). To determine if T7 helicase binds DNA with different affinities in the presence of dTTP and ATP, bulk binding studies were carried out using fluorescence anisotropy with dTTP and ATP analogs (Supplementary Fig. 7). The results show that T7 helicase binds ssDNA 100-fold more tightly with dTMPPCP than with AMPPCP, and indicate that the greater slippage in the presence of ATP is likely due to weaker binding to DNA.
The discovery of helicase slippage and the ability to directly measure helicase processivity provided a unique opportunity to address the following questions. How do ATP and dTTP compete for binding to helicase subunits? How does nucleotide binding regulate helicase affinity to DNA? How do multiple subunits of helicase coordinate their activities?
To understand how ATP and dTTP compete for binding to helicase subunits, we determined the unwinding rates between slippage events (Fig. 2a) as a function of nucleotide concentration. For each nucleotide alone, the unwinding rate followed Michaelis-Menten-like kinetics, yielding Vmax and KM values, both higher for ATP than for dTTP (Fig. 2b). This kinetics indicated that there was no cooperativity in NTP binding and hydrolysis. Next, we conducted experiments in which the concentration of one nucleotide was fixed while that of the other nucleotide was varied. The resulting unwinding rates could be explained by competitive kinetics: ATP and dTTP compete for binding based on their respective affinities and the resulting reaction rate is determined by their concentrations, Vmax, and KM (Fig. 2c-d; Methods Summary and Supplementary Discussion). A comparison of unwinding rates with mixed nucleotides and direct predictions (not fits) by the competitive binding kinetics showed excellent agreement. These results were further substantiated by ssDNA translocation rate experiments (Supplementary Fig. 8). This also explains why in Fig. 1b-c the unwinding rate was minimally altered when 0.2 mM of dTTP was added to 2 mM ATP. Under those conditions, only about 16% of the nucleotide bound to the helicase hexamer was dTTP.
Figure 2. Helicase unwinding kinetics.
a, Example of unwinding with ATP to illustrate the method of determining unwinding rate by analyzing data between slips. b, Kinetic constants for unwinding under a constant unzipping tension of 8 pN in the presence of either ATP or dTTP. For each nucleotide, KM and Vmax were obtained by fitting the unwinding rates as a function of NTP concentration to the Michaelis-Menten equation. c, Measured unwinding rates at either fixed [dTTP] and varying [ATP], or fixed [ATP] and varying [dTTP], and comparison with direct predictions (not fits) from the competitive nucleotide binding model using kinetic constants KM and Vmax shown in b. Error bars indicate s.e.m. d, Kinetic pathway of a competitive binding model where ATP and dTTP compete for binding and hydrolysis by the helicase (denoted by E here).
The competitive binding kinetics for nucleotides, however, does not explain the observed slippage behavior with mixed nucleotides (Fig. 1b-c). That is, how did the 16% bound dTTP result in a 3-fold increase in processivity? If only a single nucleotide can be bound by the helicase at a time and the type of the bound nucleotide determines the helicase’s affinity to the DNA, then processivity should only increase by 7% (Supplementary Discussion). In addition, it has previously been shown that the helicase subunits do not bind to ssDNA in the absence of a nucleotide19. However, we found minimal slippage even at [dTTP] much below its KM. These observations indicate participation of multiple subunits in both nucleotide and DNA binding, where each subunit would have a nucleotide-specific DNA binding affinity. Our data suggest that helicase may not slip if at least one subunit of the hexamer is in a deoxythymidine-ligated state, which has a higher affinity for the DNA.
Two models may be consistent with this idea. In an uncoordinated model1,2,7, each helicase subunit functions independently in its nucleotide binding/hydrolysis, and DNA binding/release (Supplementary Discussion). Conversely, coordinated models have been proposed for T7 helicase1,2,7, but details of the coordination remain unclear. Biochemical and structural studies suggest that nucleotide hydrolysis may occur sequentially around the hexameric ring16,20,21, that roughly four subunits are nucleotide-ligated at any given time20, and that DNA binding to the helicase might involve one to two helicase subunits16,20–22. A model based on structural studies has been proposed for ring-shaped helicases E1 (ref. 23) and Rho24 where all or some of the subunits coordinate their chemo-mechanical activities (Fig. 3d). Coordination could occur sequentially around the hexameric ring with the leading subunit poised for NTP binding and each successive subunit having a bound nucleotide in states of progression along the chemical reaction pathway (NTP, NDP + Pi, NDP, etc.). Depending on the state and type of nucleotide bound each subunit may have a different affinity to DNA. Once the leading subunit binds to an NTP and reels in the DNA, the remaining subunits progress to their next reaction states. Product release by the last participating subunit results in release of DNA from that subunit, and thus completes a single cycle.
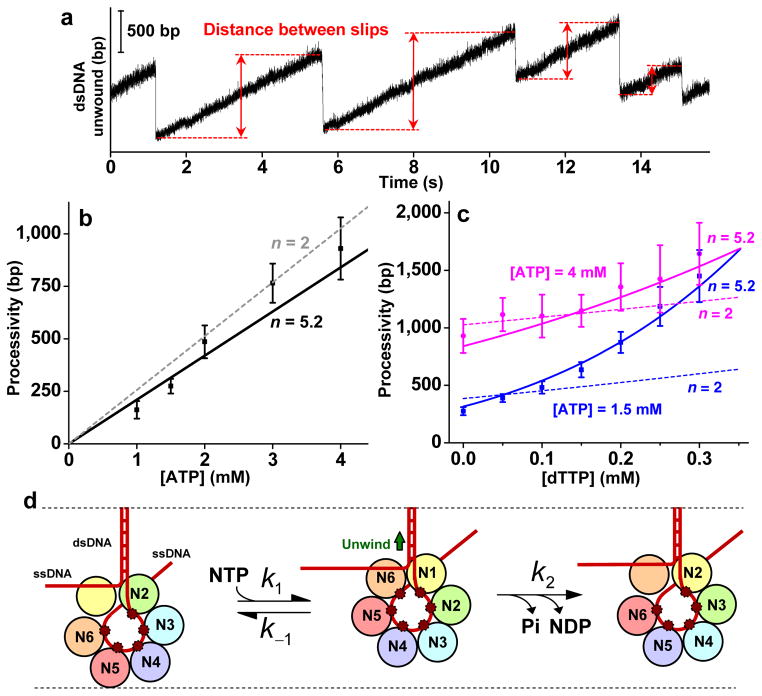
Figure 3. Processivity dependence on nucleotides and a proposed coordinated model.
a, An example of unwinding with ATP to illustrate the method of determining distance between slips. b-c, Measured processivity (mean distance between slipping events) as a function of [ATP] alone, and as functions of [dTTP] at two fixed concentrations of ATP. Note processivity increased substantially when a small amount of dTTP was added to the reaction. Solid lines are global fits using the coordinated model, yielding n = 5.2 ± 0.4 . For comparison, fits using n = 2 are also shown. Error bars indicate s.e.m. d, An interpretation of the proposed coordinated model. Each subunit is uniquely labeled with a different color and has a potential ssDNA binding site (small red dot). Nucleotide binding and subsequent hydrolysis occur sequentially around the ring. If a subunit is nucleotide-ligated (the state of hydrolysis indicated by Ni), it has a nonzero probability of being bound to ssDNA. During unwinding, the leading subunit can bind to a nucleotide (N) and thus acquire affinity to the upstream ssDNA. This stimulates the last nucleotide-bound subunit to release its nucleotide and ssDNA. Then the cycle proceeds again around the ring. Slippage occurs when all subunits simultaneously release ssDNA, as determined by the joint probability of detachment for all subunits (Supplementary Discussion).
We formulated quantitative descriptions for the uncoordinated and coordinated models (Supplementary Discussion). The observed rate of unwinding as a function of [ATP] or [dTTP] is consistent with both models which predict an apparent Michaelis-Menten-like kinetics. The observed unwinding rate with ATP and dTTP mixtures is also consistent with the competitive binding kinetics for both models as long as, in the case of the coordinated model, the rates are treated as averages over time (Supplementary Discussion). Although the two models cannot be distinguished based on rate measurement studies, they do yield different predictions for the DNA slippage behavior. The uncoordinated model (Supplementary Discussion) requires that each subunit binds and hydrolyzes nucleotides independently with an affinity to DNA dependent on the state and the type of nucleotide bound. This model is not consistent with the processivity data taken with mixed nucleotides at concentrations near or lower than their respective KM (Supplementary Fig. 9).
On the other hand, the coordinated model requires that subunits participating in coordination bind and hydrolyze nucleotide in coordination, with only one subunit poised to bind a nucleotide at a time and with each subunit having an affinity to DNA dependent on the state and type of nucleotide bound. This model predicts that processivity should increase linearly with [NTP] in the presence of a single type of NTP. Indeed, our data show that the processivity increases linearly with increasing [ATP] (Fig. 3a-b). If multiple helicase subunits coordinate in their chemo-mechanical activities, what is the degree of coordination as measured by the number of participating subunits at any given time (n)? This is a key parameter that characterizes the mechanism of the helicase. Previous studies indicate that only one or two subunits are involved in significant DNA binding, suggesting a lower degree of coordination of n = 1 or 2 (refs. 16,20–22). However, subunits may participate in the coordination even if they have lower affinity to ssDNA. The coordinated model formulated (Supplementary Discussion) is rather general and naturally takes this into account. Interestingly, it predicts that processivity sensitively depends on n as [dTTP] is increased in the presence of a fixed [ATP] – the larger the n, the more subunits participate in DNA binding, and the more steeply processivity increases with [dTTP]. Therefore we measured processivity with mixtures of ATP and dTTP (Fig. 3c). A global fit to the processivity data in Fig. 3b-c yielded n = 5.2 ± 0.4 (Methods Summary). In contrast, n = 2 does not agree with the measurements. These findings are further substantiated by experiments using UTP instead of ATP (Supplementary Fig. 10, n = 5.0 ± 0.3 ), experiments under a different unzipping force (Supplementary Fig. 11, n = 5.4 ± 0.3 ), and data on time between slips (Supplementary Fig. 12, n = 5.5 ± 0.4 ). Since n≤ 6 is expected for a hexamer, this finding indicates that nearly all subunits participate in the coordination ( n = 5 or 6) (Fig. 3d). Our findings suggest that only one subunit at a time can accept an incoming nucleotide, while the rest of the subunits are already nucleotide-bound and coordinate to prevent slippage and maintain high processivity.
The current work provides a quantitative description of nucleotide binding/hydrolysis and its coupling to DNA binding and translocation for T7 helicase. This was possible because unwinding and slippage events are clearly distinguished in single molecule traces. The slippage behavior is explained by a multiple-site coordinated model. For helicase to slip, all six subunits must simultaneously lose their grip on the DNA. This happens more often when helicase subunits are bound only to ribose nucleotides. Our data demonstrate that T7 helicase has a very weak DNA binding affinity in the presence of ATP but the addition of a small amount of dTTP to the ATP reaction increases the binding affinity of helicase to DNA. As a consequence, the presence of a single deoxythymidine-ligated subunit significantly decreases the chance of slippage so that helicase can still effectively unwind dsDNA with ATP. Thus T7 helicase, like most other helicases2, could still employ ATP as a main power source in vivo, under conditions such as those during phage infection of E. coli18 where ATP is most abundant. ATP could be used for rapid unwinding and dTTP for high processivity. Although the current work focuses on a comparison of dTTP with ATP for helicase unwinding, other deoxyribose nucleotides may also reduce the frequency of slippage (Supplementary Fig. 3). We speculate that slippage may also provide an evolutionary advantage for replication: when dNTP concentrations are low, slippage can slow down helicase to allow its synchronization with a slow-moving DNA polymerase.
METHODS SUMMARY
(1) Single-molecule assays were performed as described previously9 . Additional description of protein and DNA constructs, single molecule assays, data collection and analysis are given in Supplementary Information. (2) If dTTP and ATP compete for binding to helicase according to the kinetic pathway outlined in Fig. 2d, then the resulting unwinding rate is: , where for each type of nucleotide and Vmax = sk2 with s being the step size (in nt) (see Supplementary Discussion). (3) In the presence of dTTP and ATP, if n helicase subunits coordinate in their chemo-mechanical activities and DNA binding, then the resulting distance between slips (processivity) is: (Supplementary Discussion), with c being a proportionality constant. This expression was used to fit data in Fig. 3b-c with c and n as fit parameters.
Supplementary Material
Acknowledgments
We thank members of the Wang lab for critical reading of the manuscript. We also thank Dr. M. A. Hall for assistance with single molecule assays, data acquisition, and data analysis. We wish to acknowledge support from NIH grants (GM059849 to M.D.W.; GM55310 to S.S.P.), the NSF grant (MCB-0820293 to M.D.W.), and Cornell’s Molecular Biophysics Training Grant Traineeship (to D.S.J. and B.Y.S.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions B.S., D.S.J., S.S.P., and M.D.W. designed the experiments. D.S.J. discovered the helicase slippage with ATP. B.S. carried out all single molecule work, and together with B.Y.S., analyzed and interpreted single molecule data. G.P. performed all the ensemble experiments. M.P. and G.P. purified and analyzed the WT and mutant T7 gp4 proteins. M.D.W. formulated the theoretical models. B.S., D.S.J, B.Y.S., S.S.P., and M.D.W. wrote the manuscript.
Author information Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests. Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 2.Patel SS, Picha KM. Structure and function of hexameric helicases. Annu Rev Biochem. 2000;69:651–97. doi: 10.1146/annurev.biochem.69.1.651. [DOI] [PubMed] [Google Scholar]
- 3.Donmez I, Patel SS. Mechanisms of a ring shaped helicase. Nucleic Acids Res. 2006;34:4216–24. doi: 10.1093/nar/gkl508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matson SW, Tabor S, Richardson CC. The gene 4 protein of bacteriophage T7. Characterization of helicase activity. J Biol Chem. 1983;258:14017–24. [PubMed] [Google Scholar]
- 5.Matson SW, Richardson CC. DNA-dependent nucleoside 5′-triphosphatase activity of the gene 4 protein of bacteriophage T7. J Biol Chem. 1983;258:14009–16. [PubMed] [Google Scholar]
- 6.Hingorani MM, Patel SS. Cooperative interactions of nucleotide ligands are linked to oligomerization and DNA binding in bacteriophage T7 gene 4 helicases. Biochemistry. 1996;35:2218–28. doi: 10.1021/bi9521497. [DOI] [PubMed] [Google Scholar]
- 7.Lyubimov AY, Strycharska M, Berger JM. The nuts and bolts of ring-translocase structure and mechanism. Curr Opin Struct Biol. 2011;21:240–8. doi: 10.1016/j.sbi.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee SJ, Richardson CC. Molecular basis for recognition of nucleoside triphosphate by gene 4 helicase of bacteriophage T7. J Biol Chem. 2010;285:31462–71. doi: 10.1074/jbc.M110.156067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson DS, Bai L, Smith BY, Patel SS, Wang MD. Single-molecule studies reveal dynamics of DNA unwinding by the ring-shaped T7 helicase. Cell. 2007;129:1299–309. doi: 10.1016/j.cell.2007.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Myong S, Rasnik I, Joo C, Lohman TM, Ha T. Repetitive shuttling of a motor protein on DNA. Nature. 2005;437:1321–5. doi: 10.1038/nature04049. [DOI] [PubMed] [Google Scholar]
- 11.Myong S, Bruno MM, Pyle AM, Ha T. Spring-loaded mechanism of DNA unwinding by hepatitis C virus NS3 helicase. Science. 2007;317:513–6. doi: 10.1126/science.1144130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun B, et al. Impediment of E. coli UvrD by DNA-destabilizing force reveals a strained-inchworm mechanism of DNA unwinding. EMBO J. 2008;27:3279–87. doi: 10.1038/emboj.2008.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dessinges MN, Lionnet T, Xi XG, Bensimon D, Croquette V. Single-molecule assay reveals strand switching and enhanced processivity of UvrD. Proc Natl Acad Sci U S A. 2004;101:6439–44. doi: 10.1073/pnas.0306713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chemla YR, et al. Mechanism of force generation of a viral DNA packaging motor. Cell. 2005;122:683–92. doi: 10.1016/j.cell.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 15.Tsay JM, Sippy J, Feiss M, Smith DE. The Q motif of a viral packaging motor governs its force generation and communicates ATP recognition to DNA interaction. Proc Natl Acad Sci U S A. 2009;106:14355–60. doi: 10.1073/pnas.0904364106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singleton MR, Sawaya MR, Ellenberger T, Wigley DB. Crystal structure of T7 gene 4 ring helicase indicates a mechanism for sequential hydrolysis of nucleotides. Cell. 2000;101:589–600. doi: 10.1016/s0092-8674(00)80871-5. [DOI] [PubMed] [Google Scholar]
- 17.Patel SS, Rosenberg AH, Studier FW, Johnson KA. Large scale purification and biochemical characterization of T7 primase/helicase proteins. Evidence for homodimer and heterodimer formation. J Biol Chem. 1992;267:15013–21. [PubMed] [Google Scholar]
- 18.Mathews CK. Biochemistry of deoxyribonucleic acid-defective amber mutants of bacteriophage T4. 3. Nucleotide pools. J Biol Chem. 1972;247:7430–8. [PubMed] [Google Scholar]
- 19.Hingorani MM, Patel SS. Interactions of bacteriophage T7 DNA primase/helicase protein with single-stranded and double-stranded DNAs. Biochemistry. 1993;32:12478–87. doi: 10.1021/bi00097a028. [DOI] [PubMed] [Google Scholar]
- 20.Liao JC, Jeong YJ, Kim DE, Patel SS, Oster G. Mechanochemistry of T7 DNA helicase. J Mol Biol. 2005;350:452–75. doi: 10.1016/j.jmb.2005.04.051. [DOI] [PubMed] [Google Scholar]
- 21.Crampton DJ, Mukherjee S, Richardson CC. DNA-induced switch from independent to sequential dTTP hydrolysis in the bacteriophage T7 DNA helicase. Mol Cell. 2006;21:165–74. doi: 10.1016/j.molcel.2005.11.027. [DOI] [PubMed] [Google Scholar]
- 22.Yu X, Hingorani MM, Patel SS, Egelman EH. DNA is bound within the central hole to one or two of the six subunits of the T7 DNA helicase. Nat Struct Biol. 1996;3:740–3. doi: 10.1038/nsb0996-740. [DOI] [PubMed] [Google Scholar]
- 23.Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–5. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 24.Thomsen ND, Berger JM. Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell. 2009;139:523–34. doi: 10.1016/j.cell.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.