Bacterial replicases are complex, tripartite replicative machines. They contain a polymerase, Pol III, a β2 processivity factor and a DnaX complex ATPase that loads β2 onto DNA and chaperones Pol III onto the newly loaded β2. Many bacteria encode both a full length τ and a shorter γ form of DnaX by a variety of mechanisms. The polymerase catalytic subunit of Pol III, α, contains a PHP domain that not only binds to prototypical ε Mg++-dependent exonuclease, but also contains a second Zn++-dependent proofreading exonuclease, at least in some bacteria. Replication of the chromosomes of low GC Gram-positive bacteria require two Pol IIIs, one of which, DnaE, appears to extend RNA primers a only short distance before handing the product off to the major replicase, PolC. Other bacteria encode a second Pol III (ImuC) that apparently replaces Pol V, required for induced mutagenesis in E. coli. Approaches that permit simultaneous biochemical screening of all components of complex bacterial replicases promise inhibitors of specific protein targets and reaction stages.
Introduction
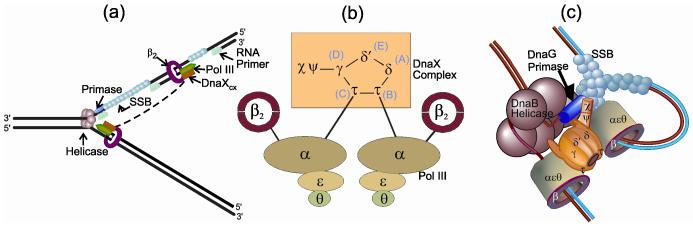
Cellular chromosomal replicases from all branches of life are tripartite. They contain a polymerase (Pol III in bacteria and Pol δ and ε in eukaryotes), a sliding clamp processivity factor (β2 in bacteria and PCNA in eukaryotes), and a clamp loader (DnaX complex (DnaXcx) in bacteria and RFC in eukaryotes). By themselves, replicative polymerases do not exhibit special properties that distinguish them from other polymerases, but together with the sliding clamp and clamp loader they become highly processive. β2 forms a ring that surrounds the DNA template and tethers the polymerase to it, enabling processive replication. Other features of bacterial replication that are conserved among all life forms are illustrated in Fig. 1.
Figure 1.
DNA Polymerase III Holoenzyme contacts at the replication fork. (a) A hexameric helicase translocates down the lagging strand template, splitting two strands apart in advance of the leading strand replicase, Pol III HE. Single-stranded regions of the lagging strand template are coated by SSB. Primase interacts with the helicase and synthesizes short RNA primers for Okazaki fragment synthesis that are extended by the Pol III HE until a signal is received to recycle to the next primer synthesized at the replication fork. For clarity, this view is drawn with a discrete DnaXcx on each Pol III; they are actually shared between the leading and lagging strand polymerase (dotted line). (b) Details of known subunit interactions within the Pol III HE. In addition, there is a transient interaction between δ and, perhaps, additional DnaXcx subunits with β2 during the clamp loading reaction. (c) A cartoon of the replication fork showing relevant protein-protein interactions, including dimerization of the leading and lagging strand polymerases through contact of domain V of τ with α. A contact between domain IV of two τs and two DnaB protomers anchors the replicase to the helicase, placing all replication fork components into one replisome.
Structure and function of α, the catalytic subunit
Like all polymerases, Pol III α contains palm, thumb and fingers domains, in the shape of a cupped right hand Fig 2. However, apo-enzyme structures of the full length Thermus aquaticus (Taq) and a truncated version of E. coli (Eco) α subunit revealed a big surprise: the palm domain has the basic fold of the X family of DNA polymerases, which includes the slow, non-processive Pol βs [1,2].
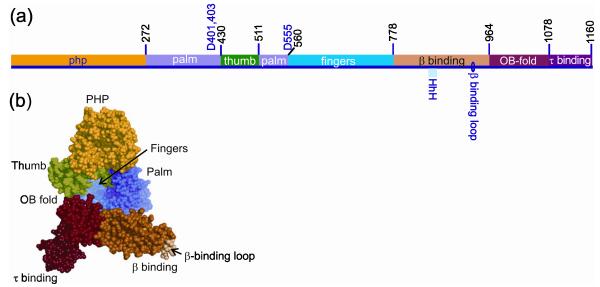
Figure 2.
Modular organization of Pol III α. (a) The residue numbers that define domain borders in E. coli α are shown above the bar in black. The positions of the three catalytic aspartates are indicated. The locations of the β binding loop and the dsDNA binding HhH motif are shown below the bar. (b) A space-filling representation of the Taq Pol III α structure [2], with the domains colored the same as in (a). The view is to the side where the dsDNA product emerges from the central channel.
A ternary complex of a dideoxy-terminated primer-template, incoming dNTP and full length Taq α provided significant insight into the function of Pol III α [3]. Among the template-primer induced conformational changes are movement of the thumb domain toward the DNA bound by the palm to make contacts with the sugar-phosphate backbone in the minor groove. The polymerase contacts the template from its terminus to a position 12 nucleotides behind the primer terminus. The fingers domain creates a wall at the end of the primer terminus that forces a sharp kink in the emerging template strand.
In the ternary complex structure of a Gram-positive Pol III, two novel elements, not present in Eco or Taq α, were identified [4]. The four-Cys Zn++ binding motif, shown earlier by Neal Brown and colleagues to be required for activity, serves an apparently structural function and is not part of the catalytic site. DNA binding through the thumb domain comes primarily from two β strands that interact with the minor groove [4].
PHP domain
A PHP domain (initially identified as having homology to phosphoesterases) is found in a wide variety of bacterial polymerases, including bacterial Pol βs (Fig. 2). Initially, it was proposed that this region might be involved in pyrophosphate hydrolysis, but such an activity has not been found [1]. This domain, at least in Taq Pol III, contains a second proofreading activity that is Zn++-dependent [5] and also binds the classical Mg++-based proofreading subunit, ε [6]. The structure of Taq α revealed a cluster of nine residues in the PHP domain that chelate three metal ions [2]. A structure of a Gram-positive PolC PHP domain also binds three metals, using the same nine ligands expected from the homologous Taq PHP structure. Kuriyan and colleagues, from the structure of Eco α, pointed out a channel between the polymerase active site and the proposed PHP active site [1].
β2 binding domain
A structure of Taq α revealed a well-organized β2-binding domain with a HhH motif that together with its flanking loops binds dsDNA [3] (Fig. 2). The β2 binding consensus sequence is presented in a loop that is oriented adjacent to dsDNA as it exits the polymerase, in the correct position to bind β2 as it surrounds DNA. The β2 binding domain rotates 20° and swings down into position as the enzyme binds DNA [3], a reorientation that is apparently driven energetically by the HhH motif binding to DNA and likely coupled to conformational changes of the thumb, palm, OB fold, and PHP domains.
OB-fold domain
The structure of the ternary complex of Taq α with primer-template and incoming dNTP reveals a striking conformational change that includes the OB-fold moving to a position near the single-strand template distal to the primer [3]. The path of the emerging template, which can be traced from electron density of the ribose-phosphate backbone, appears to come close to the OB-fold. The element of the OB-fold that comes closest to the ssDNA template, the β1-β2 loop, often contributes to ssDNA binding [7]. However, the β1-β2-β3 face that commonly interacts with ssDNA [7] appears to face away from the emerging template, toward the τ-binding domain.
A test of the importance of the OB-fold motif was made using a mutant in which three basic residues located in the β1-β2 loop were changed to serine [8]. The processivity of the mutant polymerase was decreased, an effect that was rescued by the presence of the τ-complex [8]. The latter observation would seem to suggest that although the OB-fold contributes to ssDNA affinity and processivity, it is not the processivity sensor, or at least that the residues mutated are not the key interactors.
In fact, the entire polymerase active site may function as the processivity switch [9]. Steitz and colleagues [3] have elegantly demonstrated a conformational change induced by substrate binding in which several elements move that include placement of the β2 binding domain in a position where it can productively interact with the β2 clamp on DNA. The geometry and spatial constraints around the active site when the exiting template is double-stranded might make insertion of the last nucleotide energetically unfavorable. Upon insertion, the product might lose affinity for the active site, triggering a reversal of the conformational changes that occurred upon primer-template and dNTP binding, switching the polymerase to a low-processivity mode.
τ-binding domain
The second half of the C-terminal domain in the Taq α structure revealed a domain containing an incompletely conserved sequence that binds weakly to β2, but is not required for processive replication in vitro or function in vivo. This domain is loosely packed against the OB fold, with many polar residues in the interface [2] (Fig. 2). Mutational studies support the importance of this subdomain in binding τ [10]. Possible sites of interaction of this extreme C-terminal domain with τ, derived from a genetic screen, have been reviewed recently [9].
Mechanism of clamp loading and initiation complex formation
Before DNA elongation begins, the Pol III holoenzyme (HE) forms an initiation complex in an ATP-dependent reaction. This reaction is often artificially divided into an ATP-dependent clamp loading reaction in which a β2 ring is loaded around DNA and a subsequent association of Pol III with the loaded clamp. Sliding clamps require ATP-powered clamp loaders for their assembly around DNA. The E. coli clamp loader is composed of seven subunits (Fig. 1). The ATP binding subunit that drives assembly is encoded by dnaX, which directs the synthesis of two proteins, τ and γ, by programmed ribosomal frameshifting. The shorter γ contains three domains; τ contains two additional domains (termed IV and V, respectively) that bind the replicative helicase DnaB6 and Pol III.
Pre-steady state kinetic studies of the β2 loading reaction have been performed by Bloom and colleagues. ATP serves as an allosteric effector, increasing the affinity of the DnaXcx for β2 and DNA, bringing all reaction participants together [11]. In a study investigating the rate of binding of β2 and primed DNA to γcx, it was found that β2 bound to γcx-ATP at a diffusion-controlled rate. In contrast, primed DNA bound slowly (limited by the slow ATP-induced conformational change within the γcx of 3.3 s−1) [11]. At replication forks, an ordered mechanism may be further enforced by the availability of primers for DNA replication that are only synthesized ca. every 2 s. Another investigation addressed the relative order of β2 release, primer-template release and ATP hydrolysis in the final stages of the γcx-catalyzed loading reaction [12]. Once a γcx-β2-ATP complex bound DNA, hydrolysis of three ATPs was triggered, followed by DNA release, then β2. It is not yet known when β2 becomes closed in this process.
The χ and ψ subunits of the DnaXcx perform several functions. The Bloom laboratory demonstrated important functional consequences that correlate nicely with a structure determined by Kuriyan and colleagues that reveals conformational changes induced by the interaction of an N-terminal peptide of ψ with γ3 in a complex with δδ’ [13,14]. It was concluded that ψ stabilizes an ATP-induced conformational state that binds DNA [13]. The structure also reveals how binding is restricted to a single copy of χψ: the N-terminus of ψ snakes through a collar of three DnaX domain IIIs in an asymmetric way with different contacts with each, but in a way that precludes further ψ association [14].
Binding of the ψ peptide stabilizes a conformation that binds the ATP analog, ADP-BeF3, in all three DnaX sites [14]. DNA binding favors a similar conformational change. This contrasts with structures in the absence of ψ where the DnaX subunit C (Fig. 1b) is in a conformation that cannot bind ATP. The DNA-ADP-BeF3-γ3δδ’-ψ peptide structure adopts a ‘notched screw cap’ conformation in which the ATPase subunits are symmetrically oriented in a spiral arrangement and contacts are made with the phosphates of the template strand through basic residues and α-helical dipoles. No contacts are made with the primer, other than a stack between δ Tyr316 and the terminal base of the primer. An interesting conjecture was made that this arrangement would allow for accommodating both RNA and DNA primers [14]. It will be interesting to see, in future studies, whether β2 is open when bound to this assembly and whether the ATP sites remain equivalent.
DnaX complex chaperones Pol III onto newly loaded β2
Recently, it has been demonstrated that τ-containing DnaX complexes serve another role in initiation complex formation: they chaperone the associated polymerase onto the β2 just loaded by the same complex [15]. Chaperoning significantly increases the rate of initiation complex formation and drops the Pol III concentration required for efficient assembly. Both of these features are likely critical for the cell to sustain a rate of initiation complex formation sufficient to support the rate of Okazaki synthesis required for chromosomal replication.
Initiation complex formation with τ-containing DnaXcx is stimulated by SSB, suggesting a role for SSB beyond that of protecting the DNA template. The only known SSB-interacting protein within Pol III HE is χ. The χ-SSB interaction has been shown to be important for allowing Pol III to replicate SSB-coated DNA and for stabilizing Pol III on DNA in strand displacement reactions [16]. Yet, the stimulation by SSB can occur in the absence of χ, revealing that an additional, undiscovered site for SSB interaction exists [15]. Polard and colleagues have detected a stable interaction between B. subtilis (Bsu) SSB and Bsu DnaE [17]. It would be interesting to identify the SSB binding site in Bsu DnaE and determine if a cryptic interaction occurs at the analogous site in E. coli.
Hydrolysis of an ATP by each DnaX protomer may not be required for initiation complex formation
Current models propose that the E. coli DnaXcx binds and hydrolyzes three ATPs in a synchronous wave during the clamp loading process [18]. Recently, both the γ and τ forms of DnaX with the critical Lys in the Walker A motif changed to a Glu have been purified [19]. Together with their wild-type counterparts, these mutants have been assembled into DnaX complexes and the ten possible resulting complexes purified and characterized. Surprisingly, complexes containing only one active ATP binding DnaX protomer function in initiation complex formation. This contradicts proposals regarding unique functions of individual DnaX protomers or the requirement for a synchronous wave of ATP hydrolysis to be absolutely required for β2 loading.
However, a very recent pre-steady-state kinetic study has confirmed that ATP hydrolysis at multiple sites is required for the fastest rates of initiation complex formation [20]. A DnaX complex containing only two active DnaX ATPases drives initiation complex formation 30-fold more slowly than a complex with three active ATPases. This same study showed that the benefit from chaperoning by τ-containing DnaX complexes is 100-fold. Thus, a complex containing τ and only two active ATPase subunits supports complex formation faster than a three-ATPase γ complex, the often used model assembly for β2 loading studies. It is likely that the presence of τ and chaperoning changes the kinetic pathway for initiation complex formation [20].
Possible new mechanism for recycling β2 at the replication fork
Until recently, it has been thought at Pol III*, when it cycles from a completed Okazaki fragment, leaves the old β2 behind and that a new β2 needs to be loaded on the next primer [21]. That may still be the most frequent event, but recent single-molecule experiments suggest that β2 might be recycled during Okazaki fragment synthesis [22]. Elongating complexes synthesize Okazaki fragments in the absence of added β2 in a flow system where unbound β2 was washed away. The authors acknowledge that an alternative interpretation could be that the lagging strand polymerase could function without β2 under the conditions they used [22]. Indeed, the presence of τ-containing DnaX complexes and interaction of χ with SSB coating the lagging strand stabilizes the replicase on replication forks [16]. The effective high local concentration of the lagging strand polymerase created by its association with the leading strand polymerase through a τ-τ bridge within the DnaX complex might support a β-less reaction. Thus, definitive testing for the presence of β2 with the lagging strand replicase under the conditions used should be pursued in future experiments.
DnaX complex composition and assembly within cells
The DnaX complex can be constructed, in vitro, to contain any combination of the τ and γ dnaX gene products [23]. A trimeric Pol III HE containing only τ was constructed in vitro and was shown to function in reconstituted rolling circle reactions [24]. A suggestion that it might represent the authentic replicase was made even though this form of the enzyme has never been isolated in preparations obtained from cells. All published preparations of Pol III HE obtained from cells contain both τ- and γ-subunits [9]. A recent report using Slimfield fluorescence microscopy, a technique that can visualize replication proteins tagged with YPet within foci of live cells, suggested three Pol IIIs and three τs are present per replisome [25]. The error was large enough to make this determination ambiguous (3.1 ±1.1 αs, for example). The interpretation was that the replicase contains three τs and three Pol IIIs, but the potential for other Pol III assemblies being in the vicinity performing mismatch or gap processing was not considered. Interestingly, mutation of dnaX to eliminate γ increased the τ stoichiometry 30% [25]— the amount expected with a Pol III HE dominated by a τ2γ stoichiometry. Other issues with interpretation and quantification are discussed elsewhere [9].
Replication of Gram-positive bacteria uses two DNA polymerase IIIs
In low-GC Gram-positive bacteria, two Pol IIIs exist, termed PolC and DnaE. They are homologous, but PolC has some of its domains rearranged, and it contains an endogenous proofreading activity. DnaE is more closely related to E. coli Pol III. A rolling circle replication system using 13 purified B. subtilis replication proteins has been reconstituted [26]. This system appears to accurately mimic the reaction at the replication fork of a Gram-positive bacterium, in terms of both its correspondence with genetic requirements and the replication fork rate in vivo. Leading strand replication requires 11 proteins, including the Pol III encoded by polC. In addition to these 11 proteins, lagging strand replication requires DnaE and primase [26]. This is consistent with proposals for a lagging strand role for DnaE [27]. However, the elongation rate of DnaE is too slow (~25 nt/s) to keep up with the replication fork. In contrast, PolC supports a physiologically relevant elongation rate (~500 nt/s). PolC discriminates against RNA primers; DnaE uses RNA primers efficiently [26]. These characteristics suggest a role for Bsu DnaE analogous to eukaryotic Pol α that extends RNA primers initially and then hands them off to a processive replicase.
Consistent with the eukaryotic Pol α role, model systems using RNA primed ssDNA show inefficient use by PolC, with a marked stimulation by low levels of DnaE to a level of synthesis greater than that achieved by DnaE alone [26]. Using a specific PolC inhibitor (HBEMAU) [28] that traps PolC in a dead end complex, drastic inhibition is observed when Pol C is added to DnaE RNA primer extension reactions, indicating the handoff to PolC occurs early in the reaction [26].
In E. coli, under appropriate conditions, Pol II, Pol IV and Pol V can invade elongating Pol III HE and gain access to the primer terminus [29,30]. Initially, a toolbelt model was proposed in which multiple polymerases might bind to multiple sites in an oligomeric sliding clamp processivity factor and switch out at the replication fork. However, interactions of polymerases with sliding clamps are weak when the polymerase cannot gain access to the primer terminus to acquire binding energy, and the off-rates are fast. Furthermore, Sutton and colleagues have provided evidence that would limit polymerase interaction to one set of adjacent sites on one half of the β2 sliding clamp [31]. So, if β2 is the initial contact with an exogenous polymerase, other interactions and contacts must be present to drive, and perhaps regulate, the exchange process. Goodman, Maki and colleagues have provided evidence that, in addition to the known E. coli Pol IV-β2 interaction, additional contacts between Pol IV and a component of Eco Pol III* (Pol III + τ-containing DnaXcx) triggers polymerase release [32]. Future research in this area requires an investigation of the role of the DnaXcx in the polymerase exchange mechanism.
Many bacteria use a special mutagenic Pol III in place of Pol V
In E. coli, a specialized class of PolY polymerases serves the role of induced mutagenesis and stress-induced adaptive modifications. With the sequencing of multiple bacterial genomes, it has become apparent that many bacteria have two E. coli-like dnaEs in their genomes (not including the PolC/DnaE combinations), with the second one apparently replacing Pol V, the major polymerase responsible for induced mutagenesis in E. coli [33-35].
In the most advanced studies in this area to date, it has been demonstrated that in Mycobacterium tuberculosis (Mtb), ImuB interacts with the replicative DnaE and ImuC (previously called DnaE2, see Table 1) and ImuA [36]. ImuB, in spite of being closely homologous to PolY error-prone polymerases, does not contain the triad of conserved Pol III catalytic acidic residues [36] and thus must be inactive as a polymerase. Mutation of ImuC’s predicted catalytic Asp residues ablates function (induced mutagenesis). Thus, ImuC is the error-prone polymerase in Mtb and presumably in other organisms that contain ImuA/B/C and lack Pol V homologs. Yet, ImuB interacts with β2, but ImuC does not. Thus, it appears that ImuB serves an important gatekeeper role, interacting with β2, the replicative DnaE, and the error prone polymerase ImuC.
Table I.
Three classes of DNA Polymerase IIIs
| Model Organism |
Name (or proposed name) |
Former Names |
Function | Accessory Factors |
|---|---|---|---|---|
| E. coli | α, DnaE | PolC |
|
β2, DnaXcx (τ2 γδδ’χΨ) |
| B. subtilis | PolC | DnaF |
|
β2, DnaXcx (τ3δδ’) |
| DnaE |
|
β2, DnaXcx | ||
| P. aeruginosa | DnaE |
|
β2, DnaXcx | |
| ImuC | DnaE2 |
|
β2, DnaXcx?, ImuA, ImuB |
There has been considerable confusion in the literature about the relationship between DnaE-like polymerases that coexist in PolC containing strains and those that coexist with replicative DnaEs. To provide criteria for distinguishing them, a comparison was made of the consensus sequences resulting from a comprehensive ImuC alignment with an alignment of DnaEs that exist alone or with PolCs. Both of the latter are very similar and readily distinguishable from ImuC. Key differences map to the active site and the end of the DNA binding channel [35]. If these changes relax substrate binding in a way that diminishes fidelity and permits bypass, this would appear to be consistent with the error-prone function attributed to ImuC.
Chemical Biology of DNA Replication
DNA replication is an essential process for the proliferation of all pathogens and offers a largely unexplored target for development of novel antibacterials. Most of the subunits of the bacterial DNA replication apparatus are essential, suggesting that their inhibition should lead to blockage of cell proliferation or death. This has been validated by a class of compounds, 6-anilinouracils, targeted to the polymerase subunit of the Gram-positive replicase, PolC. These compounds are not only potent biochemical inhibitors, but specifically block DNA replication in Gram-positive bacteria [37]. While screens targeting individual replicase subunits have been described, complete bacterial replicases have been explored only recently by chemical genetic approaches [38]. In a trial screen with a small (20,000-compound) library against full replication systems derived from model Gram-negative and Gram-positive organisms in parallel, it was possible to distinguish compounds that inhibited the replicase of a single species from compounds that exhibited broad spectrum potential. Counterscreens against non-orthologous enzymes with related activities revealed those compounds that are most likely to be target-specific.
Another source of useful in vitro inhibitors to support mechanistic studies derives from the discovery of bacteriophages that express peptides directed toward shutting down cellular processes, including DNA replication. For example, some Staphylococcus aureus phages produce peptides that bind to and inhibit the β2 sliding clamp and DnaI helicase loader [39,40]. And, coliphage N4 produces a peptide inhibitor of E. coli DnaXcx that functions through interaction with the δ subunit [41]. Crystal structures of complexes of these peptides and their targets should provide data that could support library design and development of small molecule inhibitors.
Conclusions
There have been several recent advances in our understanding of the structure and function of bacterial replicases. Structures of the apoenzyme and ternary complex forms of Pol III offer novel insights into the functions of a replicase and interaction with its processivity factor. These structures likely will provide a starting point for future work to understand the structural basis of interaction with DnaXcx, formation of initiation complexes upon the initiation of Okazaki fragment synthesis and regulated dissociation upon completion, and partitioning of the primer terminus to proofreading exonucleases.
New insight has emerged into the mechanism of initiation complex formation both in terms of the order of assembly and the discovery that the E. coli clamp loader also functions to chaperone the associated polymerase onto the newly loaded clamp. Future work will likely reveal if a reversal of this chaperoning reaction is responsible for dissociation of elongating complexes upon the completion of Okazaki fragment synthesis and polymerase exchange at the replication fork.
Reconstitution of a prototypical Gram-positive rolling circle replication system that mimics the enzymology of fork progression revealed the function of a second replicative polymerase: the extension of RNA primers before handoff to the principle replicase. Such a function was previously thought to be reserved to Pol α in eukaryotic systems. Future research will likely focus on the mechanism of the handoff between two polymerases, a reaction that may provide a general prototype for polymerase exchange during ongoing replication. Yet other bacteria contain two Pol IIIs for a different purpose: the second Pol III replaces a PolY polymerase for purposes of induced mutagenesis. The development of high throughput screens that include all proteins of complete replication systems promises specific small molecules that block these reactions at specific stages, providing new tools for biochemical and cell physiological investigations.
Highlights.
>Structures show Pol III active site has the fold like eukaryotic Pol β.
>The PHP domain of Pol III serves as a second proofreading exonuclease.
>ψ binds 3 DnaX protomers asymmetrically and stabilizes an active conformation.
>τ-containing DnaX complex chaperones Pol III onto newly loaded β.
>The second Gram-positive DnaE elongates RNA primers before handoff to PolC.
Acknowledgements
I thank Diane Hager for preparation of figures and Melissa Stauffer, PhD, of Scientific Editing Solutions, for editing the manuscript. Work from the author’s laboratory was supported by Grant RO1 GM060273 from the National Institute of General Medical Sciences and Grant MCB-0919961 from the National Science Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- •[1].Lamers MH, Georgescu RE, Lee SG, O’Donnell M, Kuriyan J. Crystal structure of the catalytic α subunit of E. coli replicative DNA polymerase III. Cell. 2006;126:881–892. doi: 10.1016/j.cell.2006.07.028. [DOI] [PubMed] [Google Scholar]
- ••[2].Bailey S, Wing RA, Steitz TA. The structure of T. aquaticus DNA polymerase III is distinct from eukaryotic replicative DNA polymerases. Cell. 2006;126:893–904. doi: 10.1016/j.cell.2006.07.027. The above two papers represent the first apoenzyme structures of Pol III α.
- ••[3].Wing RA, Bailey S, Steitz TA. Insights into the replisome from the structure of a ternary complex of the DNA polymerase III α-subunit. J Mol Biol. 2008;382:859–869. doi: 10.1016/j.jmb.2008.07.058. This work shows the structure of Pol III α in complex with a dNTP and primer-template. It reveals important conformational changes that occur to bring the β binding loop into position to productively interact with β during processive elongation.
- [4].Evans RJ, Davies DR, Bullard JM, Christensen J, Green LS, Guiles JW, Pata JD, Ribble WK, Janjic N, Jarvis TC. Structure of polC reveals unique DNA binding and fidelity determinants. Proc Natl Acad Sci U S A. 2008;105:20695–20700. doi: 10.1073/pnas.0809989106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •[5].Stano NM, Chen J, McHenry CS. A coproofreading Zn(2+)-dependent exonuclease within a bacterial replicase. Nat Struct Mol Biol. 2006;13:458–459. doi: 10.1038/nsmb1078. This work represents the first report of a novel Zn++-dependent second proofreading activity within Pol III.
- [6].Wieczorek A, McHenry CS. The NH(2)-terminal php domain of the α subunit of the E. coli replicase binds the ε proofreading subunit. J Biol Chem. 2006;281:12561–12567. doi: 10.1074/jbc.M513844200. [DOI] [PubMed] [Google Scholar]
- [7].Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annu Rev Biophys Biomol.Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Georgescu RE, Kurth I, Yao NY, Stewart J, Yurieva O, O’Donnell M. Mechanism of polymerase collision release from sliding clamps on the lagging strand. EMBO J. 2009;28:2981–2991. doi: 10.1038/emboj.2009.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- •[9].McHenry CS. DNA replicases from a bacterial perspective. Annu Rev Biochem. 2011;80:403–436. doi: 10.1146/annurev-biochem-061208-091655. This review includes models for a new assembly mechanism of the DnaXcx within cells and polymerase cycling upon completion of Okazaki fragment synthesis.
- [10].Dohrmann PR, McHenry CS. A bipartite polymerase-processivity factor interaction: Only the internal β binding site of the α subunit is required for processive replication by the DNA polymerase III holoenzyme. J Mol Biol. 2005;350:228–239. doi: 10.1016/j.jmb.2005.04.065. [DOI] [PubMed] [Google Scholar]
- • • [11].Thompson JA, Paschall CO, O’Donnell M, Bloom LB. A slow ATP-induced conformational change limits the rate of DNA binding but not the rate of β-clamp binding by the Escherichia coli γ complex clamp loader. J Biol Chem. 2009;284:32147–32157. doi: 10.1074/jbc.M109.045997. This expands on a series of elegant pre-steady state investigations of the mechanism of β loading by the DnaXcx. In this paper, the order of binding of participants to the DnaXcx is addressed.
- • [12].Anderson SG, Thompson JA, Paschall CO, O’Donnell M, Bloom LB. Temporal correlation of DNA binding, ATP hydrolysis, and clamp release in the clamp loading reaction catalyzed by the Escherichia coli gamma complex. Biochemistry. 2009;48:8516–8527. doi: 10.1021/bi900912a. The order of dissociation of components from the DnaXcx upon ATP hydrolysis during the β loading reaction is addressed.
- [13].Anderson SG, Williams CR, O’Donnell M, Bloom LB. A function for the ψ subunit in loading the Escherichia coli DNA polymerase sliding clamp. J Biol Chem. 2007;282:7035–7045. doi: 10.1074/jbc.M610136200. [DOI] [PubMed] [Google Scholar]
- • • [14].Simonetta KR, Kazmirski SL, Goedken ER, Cantor AJ, Kelch BA, McNally R, Seyedin SN, Makino DL, O’Donnell M, Kuriyan J. The mechanism of ATP-dependent primer-template recognition by a clamp loader complex. Cell. 2009;137:659–671. doi: 10.1016/j.cell.2009.03.044. This paper is the first report of an apparently active DnaXcx. It shows all three DnaX subunits in a conformation capable of binding and apparently hydrolyzing ATP. This work also reveals the mechanism for binding of one ψ subunit to the DnaXcx and reveals important conformational changes induced by this interaction.
- • • [15].Downey CD, McHenry CS. Chaperoning of a replicative polymerase onto a newly-assembled DNA-bound sliding clamp by the clamp loader. Mol Cell. 2010;37:481–491. doi: 10.1016/j.molcel.2010.01.013. This paper reveals that the DnaXcx clamp loader also chaperones the associated Pol III onto the newly loaded clamp. A new role for SSB by binding to a component other than χ in the clamp loading reaction is also demonstrated.
- [16].Yuan Q, McHenry CS. Strand displacement by DNA polymerase III occurs through a τ-ψ-χ link to SSB coating the lagging strand template. J Biol Chem. 2009;284:31672–31679. doi: 10.1074/jbc.M109.050740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Costes A, Lecointe F, McGovern S, Quevillon-Cheruel S, Polard P. The C-terminal domain of the bacterial SSB protein acts as a DNA maintenance hub at active chromosome replication forks. PLoS.Genet. 2010;6:e1001238. doi: 10.1371/journal.pgen.1001238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Johnson A, O’Donnell ME. Ordered ATP hydrolysis in the γ complex clamp loader AAA+ machine. J Biol Chem. 2003;278:14406–14413. doi: 10.1074/jbc.M212708200. [DOI] [PubMed] [Google Scholar]
- • [19].Wieczorek A, Downey CD, Dallmann HG, McHenry CS. Only one ATP-binding DnaX subunit is required for initiation complex formation by the E. coli DNA polymerase III holoenzyme. J Biol Chem. 2010;285:29049–29053. doi: 10.1074/jbc.C110.165076. This paper shows that DnaXcxs that contain fewer than three active ATPases can function in initiation complex formation.
- *[20].Downey CD, Crooke E, McHenry CS. Polymerase Chaperoning and Multiple ATPase Sites Enable the E. coli DNA Polymerase III Holoenzyme to Rapidly Form Initiation Complexes. J Mol Biol. 2011 doi: 10.1016/j.jmb.2011.07.051. In press.DOI: 10.1016/j.jmb.2011.07.051. This paper quantifies the kinetic advantage in initiation complex formation through chaperoning and multiple ATPases. It shows that the slow ATPγS supported initiation complex formation is accompanied by hydrolysis.
- [21].Stukenberg PT, Turner J, O’Donnell ME. An Explanation for Lagging Strand Replication: Polymerase Hopping among DNA Sliding Clamps. Cell. 1994;78:877–887. doi: 10.1016/s0092-8674(94)90662-9. [DOI] [PubMed] [Google Scholar]
- [22].Tanner NA, Tolun G, Loparo JJ, Jergic S, Griffith JD, Dixon NE, van Oijen AM. E. coli DNA replication in the absence of free beta clamps. EMBO J. 2011;30:1830–1840. doi: 10.1038/emboj.2011.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Pritchard AE, Dallmann HG, Glover BP, McHenry CS. A novel assembly mechanism for the DNA polymerase III holoenzyme DnaX complex: association of δδ’ with DnaX(4) forms DnaX(3)δδ’. EMBO J. 2000;19:6536–6545. doi: 10.1093/emboj/19.23.6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].McInerney P, Johnson A, Katz F, O’Donnell M. Characterization of a triple DNA polymerase replisome. Mol Cell. 2007;27:527–538. doi: 10.1016/j.molcel.2007.06.019. [DOI] [PubMed] [Google Scholar]
- [25].Reyes-Lamothe R, Sherratt DJ, Leake MC. Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science. 2010;328:498–501. doi: 10.1126/science.1185757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • • [26].Sanders GM, Dallmann HG, McHenry CS. Reconstitution of the B. subtilis replisome with 13 proteins including two distinct replicases. Mol Cell. 2010;37:273–281. doi: 10.1016/j.molcel.2009.12.025. This work represents the first reconstitution of an active replication fork in a Gram-positive organism using 13 purified proteins. This work also reveals a function for the second Pol III (DnaE) in replication. It elongates a nascent RNA primer and hands the product off to the PolC replicase.
- [27].Dervyn E, Suski C, Daniel R, Bruand C, Chapuis J, Errington J, Janniere L, Ehrlich SD. Two essential DNA polymerases at the bacterial replication fork. Science. 2001;294:1716–1719. doi: 10.1126/science.1066351. [DOI] [PubMed] [Google Scholar]
- [28].Tarantino PM, Zhi C, Gambino JJ, Wright GE, Brown NC. 6-Anilinouracil-based inhibitors of Bacillus subtilis DNA polymerase III: antipolymerase and antimicrobial structure-activity relationships based on substitution at uracil N3. J Med Chem. 1999;42:2035–2040. doi: 10.1021/jm980693i. [DOI] [PubMed] [Google Scholar]
- [29].Friedberg EC, Lehmann AR, Fuchs RP. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol Cell. 2005;18:499–505. doi: 10.1016/j.molcel.2005.03.032. [DOI] [PubMed] [Google Scholar]
- [30].Patel M, Jiang Q, Woodgate R, Cox MM, Goodman MF. A new model for SOS-induced mutagenesis: how RecA protein activates DNA polymerase V. Crit Rev Biochem Mol Biol. 2010;45:171–184. doi: 10.3109/10409238.2010.480968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Heltzel JM, Maul RW, Ponticelli SK Scouten, Sutton MD. A model for DNA polymerase switching involving a single cleft and the rim of the sliding clamp. Proc Natl Acad Sci U S A. 2009;106:12664–12669. doi: 10.1073/pnas.0903460106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Furukohri A, Goodman MF, Maki H. A dynamic polymerase exchange with Escherichia coli polymerase IV replacing polymerase III on the sliding clamp. J Biol Chem. 2008;283:11260–11269. doi: 10.1074/jbc.M709689200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Boshoff HI, Reed MB, Barry CE, Mizrahi V. DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell. 2003;113:183–193. doi: 10.1016/s0092-8674(03)00270-8. [DOI] [PubMed] [Google Scholar]
- [34].Galhardo RS, Rocha RP, Marques MV, Menck CF. An SOS-regulated operon involved in damage-inducible mutagenesis in Caulobacter crescentus. Nucleic Acids Res. 2005;33:2603–2614. doi: 10.1093/nar/gki551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- • [35].McHenry CS. Breaking the Rules: Multiple Replicases and DNA Polymerase IIIs in Bacteria. EMBO Rep. 2011;12:408–414. doi: 10.1038/embor.2011.51. This review presents a model for distinguishing Pol IIIs involved in mutatgenesis from Pol IIIs that serve a replicative function and suggests a nomenclature sytem for organisms containing more than one Pol III.
- • • [36].Warner DF, Ndwandwe DE, Abrahams GL, Kana BD, Machowski EE, Venclovas C, Mizrahi V. Essential roles for imuA’- and imuB-encoded accessory factors in DnaE2-dependent mutagenesis in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2010;107:13093–13098. doi: 10.1073/pnas.1002614107. This important work is the first to address molecular interactions that occur in ImuC (second DnaE-like polymerases) in induced mutagenesis.
- [37].Daly JS, Giehl TJ, Brown NC, Zhi C, Wright GE, Ellison RT., III In vitro antimicrobial activities of novel anilinouracils which selectively inhibit DNA polymerase III of Gram-positive bacteria. Antimicrob Agents Chemother. 2000;44:2217–2221. doi: 10.1128/aac.44.8.2217-2221.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Dallmann HG, Fackelmayer OJ, Tomer G, Chen J, Wiktor-Becker A, Ferrara T, Pope C, Oliveira MT, Burgers PM, Kaguni LS, McHenry CS. Parallel multiplicative target screening against divergent bacterial replicases: Identification of specific inhibitors with broad spectrum potential. Biochemistry. 2010;49:2551–2562. doi: 10.1021/bi9020764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Liu J, Dehbi M, Moeck G, Arhin F, Bauda P, Bergeron D, Callejo M, Ferretti V, Ha N, Kwan T, et al. Antimicrobial drug discovery through bacteriophage genomics. Nat Biotechnol. 2004;22:185–191. doi: 10.1038/nbt932. [DOI] [PubMed] [Google Scholar]
- [40].Belley A, Callejo M, Arhin F, Dehbi M, Fadhil I, Liu J, McKay G, Srikumar R, Bauda P, Ha N, et al. Competition of bacteriophage polypeptides with native replicase proteins for binding to the DNA sliding clamp reveals a novel mechanism for DNA replication arrest in Staphylococcus aureus. Mol Microbiol. 2006;62:1132–1143. doi: 10.1111/j.1365-2958.2006.05427.x. [DOI] [PubMed] [Google Scholar]
- • • [41].Yano ST, Rothman-Denes LB. A phage-encoded inhibitor of Escherichia coli DNA replication targets the DNA polymerase clamp loader. Mol Microbiol. 2011;79:1325–1338. doi: 10.1111/j.1365-2958.2010.07526.x. This work reveals a peptide encoded by a bacteriophage that shuts down host DNA replication by binding to the DnaXcx.