Abstract
Stochasticity in gene expression, protein or metabolite levels contributes to cell-cell variations, the analysis of which could lead to a better understanding of cellular processes and drug responses. Current technologies are limited in their throughput, resolution (in space, time, and tracking individual cells instead of population average) and the ability to control cellular environment. A few microfluidic tools have been developed to trap and image cells; however, in most designs available to date, there is a compromise among loading efficiency, speed, the ability to trap single cells, and density or number of trapped cells. To meet the needs of single-cell imaging studies, we developed a microfluidic platform for high-throughput capture and imaging of thousands of single cells. The optimized trapping mechanism enables 95% of the traps to be occupied with single cells, with a trap density of 860 traps / mm2. The dense array allows up to 800 cells to be imaged simultaneously with a 4× objective and a typical camera setup. Capture occurs with low shear and 94% viability after 24h. This platform is compatible with other upstream microfluidic components for complex cell stimulation patterns, and we show here the ability to measure heterogeneity in calcium oscillatory behavior in genetically identical cells and monitor kinetic cellular response to chemical stimuli.
Keywords: lab-on-a-chip, microfluidics, cell trap array, imaging, single-cell analysis, kinetics, calcium, lymphocytes
INTRODUCTION
Stochastic effects in gene expression and transcription events in mammalian cells lead to large variations in messenger RNA copy numbers, causing cell-to-cell variability in genetically identical cells1,2. A current view is that noise arising from stochastic fluctuations plays an essential role in key cellular activities3. For example, clonal populations of mouse multipotent progenitor cells4 or cancer cells5 have differential fate outcome in response to the same uniform stimulus because of heterogeneities in the dynamics of regulatory proteins5, in the expression level of basal signaling proteins4 or states of proteins regulating apoptosis6,7. Tracking single cell dynamic response is therefore necessary to monitor stochastic fluctuations among cell populations. At present, such experiments can be technically challenging if the cells of interests are non-adherent, if stimuli need to be delivered, or if studies on long time scales are desired. Flow cytometry is often the technique of choice to measure heterogeneity of suspension cell populations, as it is high-throughput and can distinguish subpopulations of cells. However, this technology is capable of neither monitoring temporal changes within the same cell, nor distinguishing population from noise due to temporal fluctuation within one cell. Quantitative time lapse microscopy is often required for these measurements, but it presents additional challenges, such as low throughput and movement of the target cells during imaging. It is particularly challenging to image suspension cells; one could use adhesion to a solid surface by use of an artificial membrane and receptor binding, but this may alter the biological behavior of the cells.
To overcome limitations of traditional real-time microscopy, microfluidics can be used to allow for increased throughput, control of cell location and extracellular conditions. Various microfluidic techniques have been developed to capture cells, retain them in a specific location, and control the environment surrounding them. Although very powerful, these methods have a limited throughput because the cell traps are spaced sparsely enough such that per view only a small number of cells are captured, and some are difficult to implement, or have side effects or other limitations. For example, active single-cell capture mechanisms use valves8,9 to control flow or dielectric forces with DEP10,11 or optical tweezers12 to control the location of cells in various environments. The use of dielectric forces on living cells limits cell viability due to buffer cytotoxicity and heat damage. Passive capturing mechanisms have also been proposed using gravity13–16 or fluid flow17 to direct cells into traps. Most microwell arrays rely on gravity to capture cells. Careful design of the microwells enables up to 70% single cell capture in densely packed wells, but once trapped, exposure to varying chemical solutions and manipulation of the cells are limited because the cells are not actively held in the wells13–16. Flow by diverting streamlines towards traps can also be used to transport and dock cells at specific locations17–21. Once a trap contains cells, fluid towards the trap is significantly reduced, and therefore incoming cells will be diverted to the next empty trap. Optimization of trap dimensions, location and spacing has been performed to increase capture efficiency or single cell trapping18,21,22. However, in most designs to date, there is a compromise between cell trap density per area and single cell capture efficiency.
To address the need of high-density and high-efficiency cell traps, we designed a microfluidic high-density single cell capture, stimulation, and imaging platform. The design principles of this chip were adapted from our previous work for high-density single embryo trapping23 to accommodate single cells, using hydrodynamic flow in conjunction with a careful disposition of the cell traps in an array formed by a serpentine channel. Our device is capable of passively trapping thousands of cells in less than a minute with a single-cell loading efficiency of 95%. Cells are captured sequentially and deterministically on chip with minimal shear. At low magnification, the trap array enables tracking of hundreds of cells simultaneously over time. At high magnification, spatial information can be resolved on a few precisely located single cells. Imaging can be performed on either live or immunostained cells. We show that various soluble stimuli can be delivered to the captured cells, and the trap arrays can be easily integrated with upstream microfluidic components capable of multiplexing several experiments on a single chip. Using this microfluidic platform, we studied the heterogeneity in calcium dynamics in resting and stimulated single Jurkat T cells.
EXPERIMENTAL METHOD AND MATERIALS
Fabrication of polydimethylsiloxane (PDMS) devices
The microfluidic devices were fabricated using soft lithography24. Negative molds were fabricated by UV photolithographic processes using a negative photoresist (SU8-2010, 14–16 µm, and SU8-2002, 1.5–3 µm in thickness) (Microchem, Newton, MA). Patterned wafers were then treated with tridecafluoro-1,1,2,2-tetrahydrooctyl-1-trichlorosilane vapor (United Chemical Technologies, Bristol, PA) in a vacuum desiccator to prevent adhesion of PDMS (Sylgard 184, Dow Corning, Midland, MI) before the molding process. PDMS mixture of A and B in 10:1 ratio was poured onto the mold to obtain a 5-mm thick layer and then fully cured at 70°C for 2 hours. The PDMS was peeled off the mold and individual devices were cut to size. Medical grade polyethylene (PE3) tubings (Scientific Commodities) were used for fluidic connections. Holes for fluidic connections were punched with 19 gauge needles. PDMS devices were plasma bonded onto either a cover glass or slide glass depending on applications.
Cell culture, stimulation and staining
Jurkat E6-1 human acute T cell lymphoma cells (ATCC) were cultured in RPMI 1640 medium with L-glutamine (Sigma-Aldrich, St. Louis, MO) with 10mM HEPES, 1mM sodium pyruvate, 1X MEM nonessential amino acids, and 100 units mL−1 penicillin streptomycin (Cellgro), supplemented with 10% certified heat inactivated fetal bovine serum (Sigma-Aldrich, St. Louis, MO), at 37 °C in a humidified 5% CO2 incubator. For nuclei visualisation, Jurkat cells were incubated with Hoechst 33258, at a final concentration of 1 µg mL−1, at 37 °C for 20 minutes. Cells were checked for viability using Live/Dead stain (Invitrogen) following manufacturer’s protocol. For high resolution microscopy, 106 cells were fixed in a 5% formalin solution (Sigma-Aldrich, St. Louis, MO) for 15 minutes at 37 °C, washed three times with cold PBS, and resuspended in 100 µL of ice cold 90% methanol. Immunostaining was performed on fixed cells using Hoechst 3342 for DNA staining, mouse α-calnexin (Abcam) for ER staining, and rabbit α-profilin-1 (Cell Signaling), as a cytoplasmic localized protein. Incubation with the primary antibody for one hour at room temperature was followed by three wash steps with a solution of 2% BSA in PBS and incubation for 40 minutes at room temperature with the following secondary antibodies, Alexa 488 α-mouse (Invitrogen) and goat α-rabbit TRITC (Southern Biotech). To monitor calcium signaling, Jurkat cells were incubated with 5 µM Fluo-3 (Invitrogen) for 20 minutes at 37 °C, washed with cold PBS and loaded into the cell traps. Trapped cells were stimulated with ionomycin (Sigma, St. Louis, MO) at various concentrations to release intracellular calcium.
Microfluidic system operation
Before each experiment, the microfluidic devices were primed using a solution of 2% BSA in PBS to remove any air bubbles and prevent undesirable cell adhesion to the wall. A pressure difference of 3.5 kPa (5.5 kPa for devices with upstream serial-dilution gradient generator) created by gravity was used to drive the flow, resulting to an average flow rate of ~ 2 µL h−1. Cell loading was obtained by pipetting 2 µL of 5×106 cell mL−1 of cell suspension on the chip positioned on the microscope stage. Further experiments (staining, stimulation) were performed by adding 5 µL of 4× chemicals in the inlet hole and flowing over the trapped cells for the desired time. All experiments were performed in a microcontrolled environment (temperature set at 37°C in a humidified 5% CO2 environment).
Quantification of the trapping efficiency
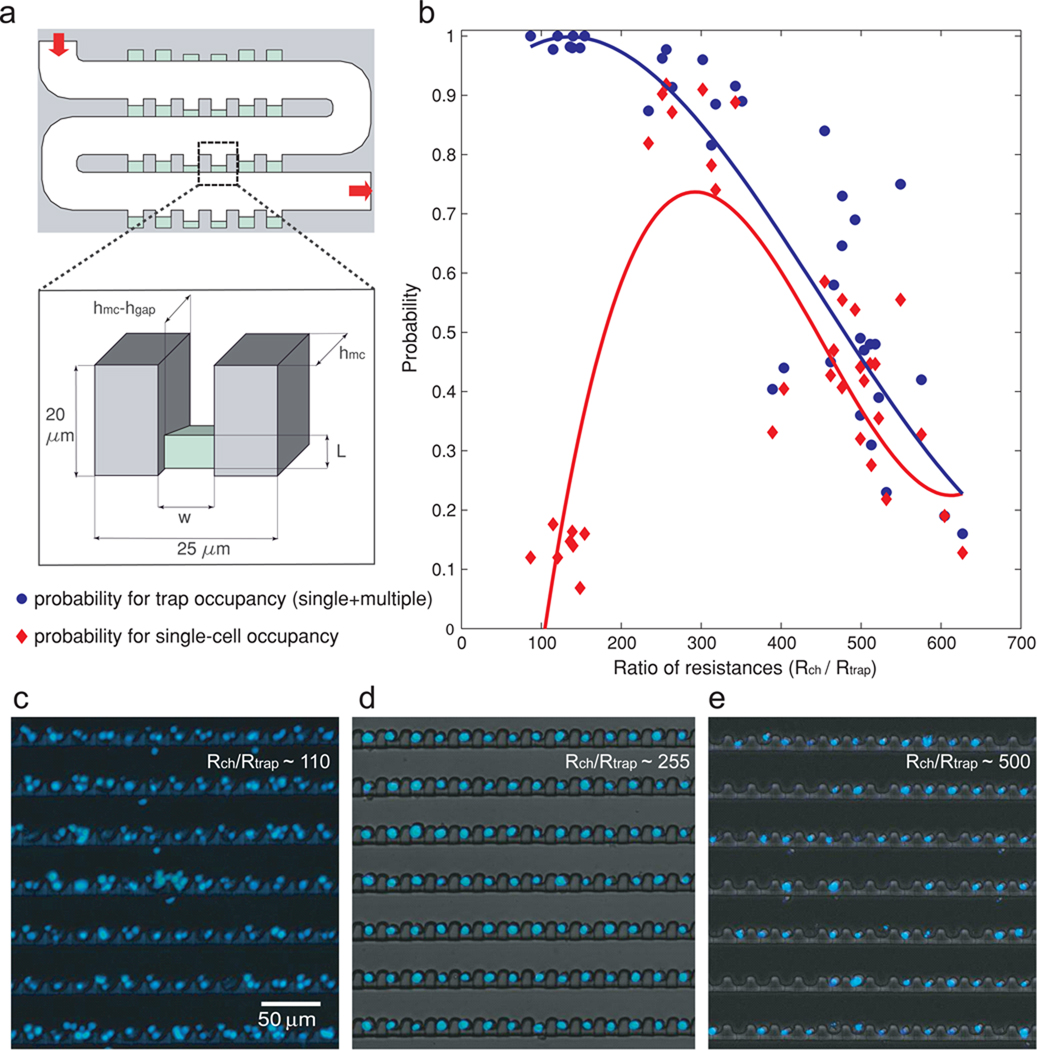
To determine trapping efficiency, devices with varying geometries were built (Fig. 2). The height of the main channel (hmc) was varied from 14 to 16 µm. Width of the traps (w) was varied from 8 to 15 µm. The length of the narrow microchannel (L) was varied from 3.3 to 8 µm. The height of the narrow microchannel (hgap) ranged from 1.5 to 3 µm. Conserved lengths are: width of the main channel (30 µm), total width of a trap (pocket and wall included: 25 µm) as well as total length of the trap (20 µm). Resistance of the main channel above a trap was estimated by and resistance of the trap .
Figure 2. Characterizing trapping efficiency of the devices with various geometric dimensions.
(a) A schematic representing the variables involved in loading efficiency optimization. (b) A plot showing probability of trap occupancy (blue circles) and probability of single-cell occupancy (red diamonds) in varying ratios of resistances (Rch / Rtrap). The trapping efficiency in a single device will be represented by a red and a blue point. The blue and red lines are guides for the eye. (c–e) Representative micrographs of cell trapping. (c) Rch / Rtrap ~ 110; (d) Rch / Rtrap ~ 255; (e) Rch / Rtrap~ 500.
Data collection and analysis
High resolution microscopy was performed on a 2-photon confocal microscope (Zeiss LSM 510 NLO). Time-lapse microscopy and device characterization experiments were performed on an epifluorescent (Nikon Eclipse Ti) microscope with an environment controlled chamber. Images from individual chambers were captured sequentially using an automated XYZ stage with a 0.7 second delay between each chamber. Custom Matlab® (MathWorks) scripts were written for semi-automated image processing. Briefly, images were cropped to contain the cell trapping area and a mask of the traps drawn from each picture by finding the areas of higher intensities. The ratio of the number of objects in the overlay of the mask on the original picture to the number of traps corresponds to the percentage of traps occupied. To discriminate traps occupied by a single versus multiple cells, several features were measured for each object, including area, convex area, eccentricity, solidity, perimeter, extent and orientation. A principal least square analysis (SIMCA-P, Umetrics) was run on a known dataset of objects to determine the two most informative predictors of the number of cells contained in an object. The perimeter and the extent (ratio of pixels in the object to pixels in the total bounding box) were found as being the most informative. To quantify single cell trapping efficiency, the distribution of objects in the perimeter-extent space was fitted to a 2-component Gaussian mixture model for each chamber trap array. The maximum likelihood parameters from each of the two subpopulations were retrieved and represented respectively the percentage of single cell objects and multiple cell objects.. Single cell response intensities were obtained by tracking the mean intensity of each object considered as a single cell in the overlaid mask and image over time.
RESULTS & DISCUSSION
Design of an efficient microfluidic single-cell trap array
To allow imaging of a large number of cells in a field of view, single cells need to be arranged with high efficiency and with uniform trapping conditions in an array of densely packed traps. To satisfy these requirements, we adapted the design principles that we previously developed for high-density embryo trapping23, and achieved capture of 4,000 single cells on 4.5 mm2 in 30 seconds, and with a loading efficiency over 95%.
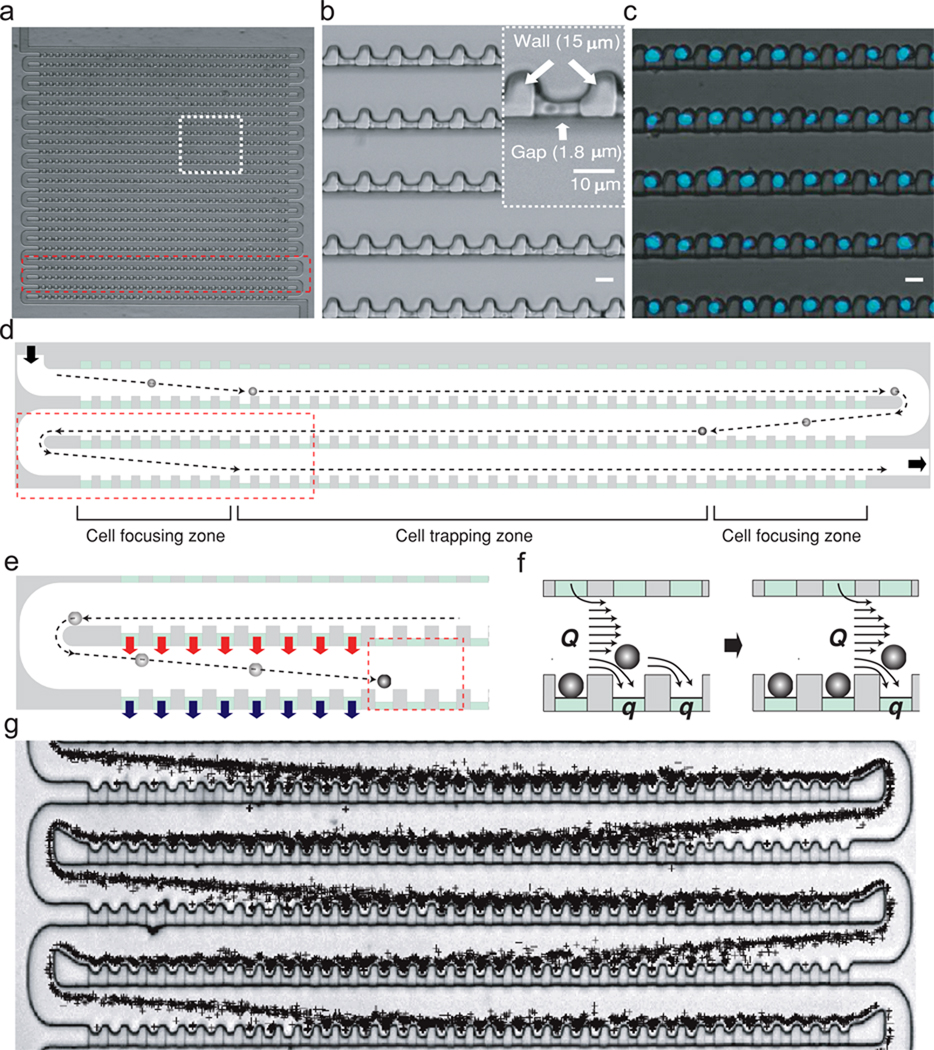
The microfluidic devices made from one layer of polydimethylsiloxane (PDMS) contain arrays of highly packed single cell traps (Fig. 1). Each array consists of a wide serpentine cell-delivery channel arranged in 26 column format and an array of cross-flow channels that connect each section of the serpentine channel (Fig. 1a–b). The width (~ 25 µm) and height (14 µm) of the cell-delivery channel are large enough to ensure cells easily moving without clogging. Each column includes 24 single cell traps (Fig. 1b–c) in the middle and 8 dummy traps at each end (Fig. 1d). The size of the cell trap is similar to that of cell of interest so that once a cell occupies a trap, it physically excludes the next cell and reduces the possibility of trapping of more than one cell. The cell traps are connected to the 1.8 µm deep shallow cross-flow channels (Fig. 1b inset). By minimizing space between neighboring traps (~ 8 µm in a column, ~ 33 µm between columns), we achieved a density of 860 traps / mm2, which is 2–3 orders of magnitude higher than what has been previously reported for deterministic single cell traps (175 ~ 700 traps/cm2)21.
Figure 1. Design and principle of single cell trapping array.
(a) Optical micrograph of the trap array fabricated using soft lithography. (b) Details of cell trap design (boxed region in a). The inset shows dimensions of a single trap. Scale bar, 10 µm. (c) Overlay of phase contrast and fluorescent images showing single cells trapped in the array. (d) Schematic drawing of three columns of the array showing trajectory of cells. Cell suspension enters the array from the top-left and exit to bottom-right. Dotted lines represent trajectory of cells. (e) Boxed region in d showing cell focusing mechanism. Converging flow (red arrow) and diverging flow (blue arrow) through the dummy traps focus cells toward the traps. (f) Boxed region in e describing two major streams that cells experience; main stream (Q) flowing along the delivery channel and a stream (q) directing the cell into the trap. (g) Overlay of a series of 1045 images showing cell trajectories during loading.
If flow through the traps has large variations throughout the array, the trap occupancy will be severely compromised. To make the trapping condition uniform, we engineered the geometry of the channels so that cells experience similar flow rates near each trap. Cells passing the focusing zones along the wide delivery channel are focused toward the traps by diverging (blue arrows in Fig. 1e) and converging flow (red arrow in Fig. 1e) through the dummy traps (Fig. 1d, e). The number of the dummy traps is optimized to make cells move closer to the trap after passing the focusing zone. This increases the frequency with which cells contact the traps and are loaded into them (Fig. 1g). After passing the focusing zone, cells close to a trap experience two streams; main stream (Q) flowing along the delivery channel and a stream (q) directing the cell into the trap (Fig. 1f). If the Q/q is in a proper range, as described previously19,21,23, cells can be guided into the trap and docked. Once all the traps in one row are occupied, extra cells pass another cell focusing zone without getting trapped and move to the next row. Despite the proximity of the dummy traps, cells are not captured because the size of the dummy traps is smaller than that of the cells. By optimizing various geometric parameters, such as the width and height of the channels and the number of traps, we were able to achieve over 95% single cell trapping efficiency throughout the device (Fig. 1c).
Optimization of single cell loading efficiency
Geometries of the docking sites were optimized in order to deterministically trap a single cell per trap. A cell close to a trap experiences forces in two directions due to the combined effect of the mainstream bulk flow (Q) and the cross-flow (q); large bulk flow moves the cell along the main channel and significant enough cross-flow guides the cell into the trap. However, with too large a cross-flow, additional cells can be forced to pile onto the already-occupied trap, reducing single-cell trapping efficiency. By optimizing the fluidic resistance of the cross-flow channel with respect to the resistance of the delivery channel, conditions for trapping a single cell in a single trap can be met.
In a previously published work19, spherical beads were captured hydrodynamically at the intersection of a bypass channel and a main channel. Although our trapping mechanism also relies on diverging flow from a main channel, the bypass channel is formed by a series of parallel traps so that traps can be incorporated at a higher density; additionally, the array format ensure identical flowrates throughout the entire chamber. Similar to our previous work23, if the cross-flow channel has significantly higher hydrodynamic resistance than that of the main delivery channel, Q/q stays relatively constant throughout the large array, ensuring Q/q at each traps being in a proper range for trapping single cells. By varying the length (L), width (w), and depth (hgap) of the trapping area and the overall depth of the main channel (hmc) (Fig. 2a), we were able to empirically determine the optimal geometry for trapping Jurkat cells (9 ±1 µm of diameter) (Fig. 2b). Fig. 2b presents the experimentally determined probability for a trap to be filled (blue circles) as well as the probability for single-cell occupancy (red diamonds). A low resistance ratio results in all traps being occupied, at the cost of having multiple cells per trap (Fig. 2c). For high resistance ratios, the flow going through the trap is not sufficient for optimal loading, resulting in very few traps being occupied but when occupied, only one cell is present (Fig. 2e). At the optimal ratio, 93±2% of the traps are occupied with 94±1% of the occupied trap with a single cell trapped (Fig. 2d and Supplementary Movies). Using the same optimized device, we were also able to trap efficiently various cell types (e.g. primary T cells and Mouse Embryonic Stem Cells size varying from 8 to 20 µm), suggesting that the optimal resistance ratio is conserved in this size range. In addition, loading efficiency is independent on the initial cell concentration; cell concentration only affects loading time with high concentration loading faster. Using 10,000 cells at 5×106 cells mL−1, full loading of a chamber takes less than a minute at a flow rate of 1 µl hr−1. At lower flow rates, loading time is longer and cells tend to settle in the inlet reservoir. For flow rates above 6 µl hr−1, cells will experience high shear stress and sometimes squeeze through the 1.8 µm deep shallow channel, but the time-saving is not significant, so in normal use of the device, we chose a flow rate of 1–2 µl hr−1. We also note that an additional benefit of this trap array design is the sequential capture of incoming cells (Supplementary Movies), preventing undesirable cell loss. Of a small number of cells (e.g. 100 cells) entering the cell trap chamber, all cells will be effectively captured. This could be especially useful for precious sample capturing where the tolerance of cell loss is very low.
On-chip microscopy and cell study
We showed that this microfluidic platform can successfully capture and immobilize both fixed and live cells (Figure 3). To ensure trapping and perfusion rate do not induce undesirable shear stress for long-term studies, we also performed a viability study on chip. Jurkat cells were loaded into the chip and perfused with medium for up to 24 hours in a microcontrolled chamber. After 24 hours, 94% of Jurkat cells were still viable by live-dead stain, comparable to conventional cell culture techniques in flasks. Viability was also observed to be uniform throughout the trap array chamber, suggesting the absence of high shear stress zones in the chamber and the compatibility of the chip with long-term dynamic studies.
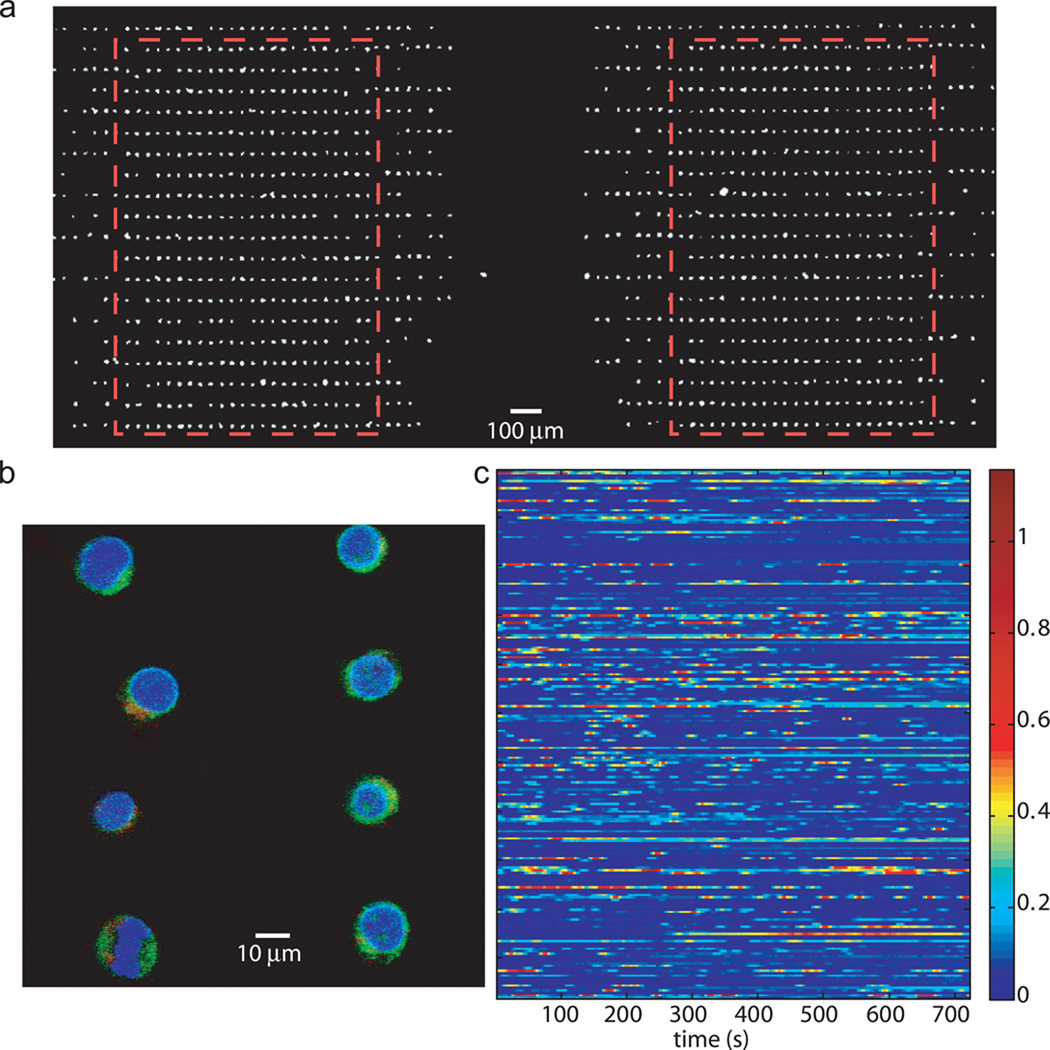
Figure 3. Using the array chip for high resolution imaging of cellular features and activities.
(a) Fluorescent microphotograph of two cell trap chambers (boxed areas). (b) Fluorescent image of immunostained Jurkat cells with confocal microscopy (100×). In blue, Hoechst for the nucleus, in green, calnexin, an ER-bound protein, and in red, profilin-1, a cytoplasmic protein. (c) Calcium dynamics in resting Jurkat cells: 216 cells are imaged every 3 seconds for 15 minutes. Each line corresponds to an individual cell in the array. The heat map indicates low (in blue) to high (in red) intracellular calcium concentration.
Another advantage of the device is that the high trap density allows for imaging large number of cells. For very bright signals, such as a DNA stain or calcium staining with Fluo3, low NA (low magnification) objectives can be used, and up to 800 single cells can be monitored in a field of view (Fig. 3a). In a typical flow cytometry experiment, 1,000 to 10,000 data points are collected. However, these 1,000 data points correspond to 1,000 different individual cells. With our chip and 1,000 cells, we are able to collect 1,000 data points per time point therefore increasing the experimental throughput and reaching similar statistical significance as flow cytometry.
The microfluidic chip is also compatible with immunostaining. Fixation, permeabilization, immunostaining and necessary wash steps can be performed on chip following standard protocol after cells are loaded into the traps. It is also possible to capture already immunostained cells, although chances of having multiple cells per trap increase due to the increased probability of adhesion of fixed cells to each other or to the device. Fig. 3b presents Jurkat cells immunostained off chip for calnexin (endoplasmic reticulum), profilin-1 (cytoplasmic cytoskeletal protein) and Hoechst (nucleus) and imaged by confocal microscopy. Cells trapped in microwells are not always compatible with high magnification imaging due to the depth of the substrate forming the wells.
In contrast, in our chip, cells are always located next to the coverslip, enabling high spatial resolution imaging of a few single cells at high magnification (Fig. 3b). Because the cells are also at known locations on the chip in an arrayed format, thousands of cells can be imaged in one single device repeatedly.
One advantage over flow cytometry is that our microfluidic chip coupled with real-time microscopy allows tracking of dynamic behavior of hundreds of cells and monitoring temporal changes within single cells, which cannot be measured by flow cytometry. As a proof of concept, we performed live cell imaging of intracellular calcium concentration in Jurkat cells (Fig. 3c). Two hundred trapped cells in a single chamber, loaded with a fluorescent intracellular dye specific for unbound calcium, were imaged every 3 seconds for duration of 15 minutes. The heat map presented in Fig. 3c highlights heterogeneity in behaviors of individual clonal cells. About 25% of these cells exhibited calcium oscillations under resting conditions, and removal of extracellular calcium abolished the oscillations in all of these cells. The oscillations are asynchronous and may arise from random fluctuations in ER calcium channels clustering25 rather than controlled periodic homeostatic behavior.
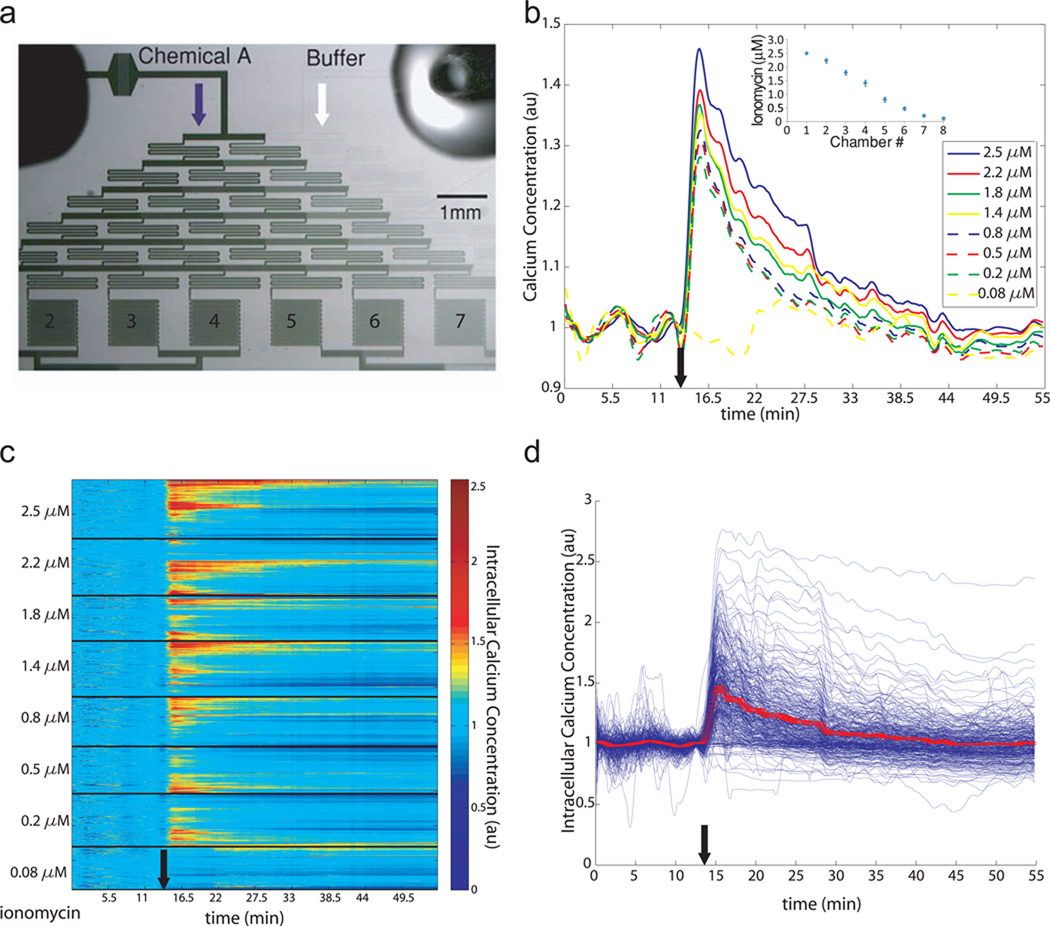
The trap arrays can also be easily integrated with upstream microfluidic components capable of multiplexing experiments, where one expose cells to different extracellular conditions on a single chip. To demonstrate this capability, we coupled the cell trap chambers with a linear serial dilution generator26 to expose each chamber to a different concentration of the stimulus. By fluorescence measurement, the gradient of concentration was observed to be linear and not disturbed by the high resistance of the loaded cell traps (Fig. 4b inset). After loading cells containing Fluo-3, more than 1,000 individual cells were monitored for an hour after the addition of different concentrations of the calcium ionophore ionomycin. Ionomycin increases intracellular calcium via mobilization of both extracellular and intracellular calcium stores27 in T cells. As expected, increased concentration of stimulus lead to increased average intracellular calcium concentration (Fig. 4b). Interestingly, when individual cells are monitored, it appears that only a fraction of the cell population are responding to the stimulus (Fig. 4c,d), and the fraction of responding cells increases linearly with increasing concentration of stimulus (R2=0.88) as shown by unsupervised clustering for each chamber in Figure 4c and Table 1; however, cell response was not correlated to position in the array nor to the presence of oscillations prior to stimulation. This observation of partial calcium response of a cell population to external stimulus is not unprecedented; as reported for clonal human embryonic kidney 293 cells when challenged with caffeine, only 40% respond with an elevation in intracellular calcium due to in endogenous protein expression levels28. Cellular response in terms of amplitude and duration of cytoplasmic calcium influx were not dependent on their respective position in the array. In addition, cells in the bottom rows receive the stimulus with minimal delay compared to cells in top rows of the array (Supplemental Figure 1), which suggests a homogenous stimulus delivery throughout the array.
Figure 4. Calcium dynamics in response to ionomycin stimulation of multiple cells tracked on chip.
(a) Microphotograph of cell trap arrays interfaced with a concentration gradient generator. The numbers represent the chamber number. (b) Average single cell calcium response for different concentrations of ionomycin. The inset show the linearity of the concentration gradient (n = 4). (c) Single cell response to ionomycin. Each line of the heat map corresponds to the dynamics of a single cell. The heat map is subdivided into 8 smaller heat maps, which correspond to decreasing ionomycin concentrations (cf table 1 for details about number of cells imaged and ionomycin concentration). In each subset, unsupervised clustering has been performed to cluster cells with similar responses. (d) Traces of single cell responses to 2.5 µM ionomycin. The red line corresponds to the average response ± SEM. The black arrow represents time of ionomycin addition.
Table 1.
Calcium response to various ionomycin concentrations
| Chamber No | Number of tracked cells |
Ionomycin conc. (µM) |
% Responding cells |
|---|---|---|---|
| 1 | 342 | 2.5 | 44% |
| 2 | 328 | 2.2 | 33.2% |
| 3 | 263 | 1.8 | 31.9% |
| 4 | 322 | 1.4 | 31.1% |
| 5 | 287 | 0.8 | 25.4% |
| 6 | 273 | 0.5 | 19.1% |
| 7 | 306 | 0.2 | 17% |
| 8 | 250 | 0.08 | 4% |
We performed a similar experiment using flow cytometry as a technique to measure calcium kinetics of Jurkat cell in response to 2.5 µM of ionomycin (Supplementary Figure 2). Average response of cells was comparable for both techniques, with a similar standard deviation due to population noise. It must be noted that to get 15 minutes of calcium dynamics, our chip only requires 300 cells as opposed 86,200 cells with a kinetic read with flow cytometry. Moreover, flow cytometry does not enable to monitor the early kinetics after addition of the stimulus (20 seconds), and because fluctuations within one cell cannot be quantified, flow cytometry cannot discriminate between oscillating and non-oscillating cells. In addition, discrimination between responding and non responding cells at different time points is facilitated by our chip.
CONCLUSION
We present here a microfluidic platform for single-cell capture, stimulation, and imaging. It is capable of passively trapping 4,000 single cells on a 4.5 mm2 footprint in 30 seconds, with a single-cell loading efficiency of 95%. The array format and optimized geometry allows for easy, robust and efficient single-cell loading, while maintaining captured cells in a low shear stress environment for long-term studies. Because cells are captured sequentially, this system is adequate for rare cell samples. Compared to previous designs, our higher cell trap density allows for imaging of increased cell numbers, therefore increasing throughput of single cell experiments, while being compatible with high resolution imaging at high magnification. Trapped cells can be exposed to various environmental conditions and chemical stimulus and their dynamic response can be monitored over time. The information gained from high-throughput, single-cell time lapsed imaging presents new opportunities in quantifying cellular responses, as averaged information by other measurement methods eliminates sub-population phenotypes. Because of the ease of use of this system, we envision this platform to be used for diverse applications, such as fundamental studies of stochastic behavior, diagnosis of patient samples or drug screens in cancer biology and stem cell biology.
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge the National Science Foundation (grant DBI-0649833 to HL), the Alfred P. Sloan Foundation (Research Fellowship to HL), the National Institutes of Health (grant R21NS058465 to HL, and grant R21CA134299 to HL and MLK) and the Georgia Tech Integrative Biosystems Institute (grant to MLK and HL).
REFERENCES
- 1.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Plos Biol. 2006;4:1707–1719. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elowitz MB, Levine AJ, Siggia ED, Swain PS. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- 3.Chang HH, Hemberg M, Barahona M, Ingber DE, Huang S. Nature. 2008;453 doi: 10.1038/nature06965. 544-U10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh DK, Ku CJ, Wichaidit C, Steininger RJ, Wu LF, Altschuler SJ. Mol Syst Biol. 2010;6 doi: 10.1038/msb.2010.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen AA, Geva-Zatorsky N, Eden E, Frenkel-Morgenstern M, Issaeva I, Sigal A, Milo R, Cohen-Saidon C, Liron Y, Kam Z, Cohen L, Danon T, Perzov N, Alon U. Science. 2008;322:1511–1516. doi: 10.1126/science.1160165. [DOI] [PubMed] [Google Scholar]
- 6.Spencer SL, Gaudet S, Albeck JG, Burke JM, Sorger PK. Nature. 2009;459 doi: 10.1038/nature08012. 428-U144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eldar A, Elowitz MB. Nature. 2010;467:167–173. doi: 10.1038/nature09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Irimia D, Toner M. Lab Chip. 2006;6:345–352. doi: 10.1039/b515983k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wheeler AR, Throndset WR, Whelan RJ, Leach AM, Zare RN, Liao YH, Farrell K, Manger ID, Daridon A. Anal Chem. 2003;75:3581–3586. doi: 10.1021/ac0340758. [DOI] [PubMed] [Google Scholar]
- 10.Taff BM, Voldman J. Anal Chem. 2005;77:7976–7983. doi: 10.1021/ac0513616. [DOI] [PubMed] [Google Scholar]
- 11.Voldman J, Gray ML, Toner M, Schmidt MA. Anal Chem. 2002;74:3984–3990. doi: 10.1021/ac0256235. [DOI] [PubMed] [Google Scholar]
- 12.Enger J, Goksor M, Ramser K, Hagberg P, Hanstorp D. Lab Chip. 2004;4:196–200. doi: 10.1039/b307960k. [DOI] [PubMed] [Google Scholar]
- 13.Rettig JR, Folch A. Anal Chem. 2005;77:5628–5634. doi: 10.1021/ac0505977. [DOI] [PubMed] [Google Scholar]
- 14.Yamamura S, Kishi H, Tokimitsu Y, Kondo S, Honda R, Rao SR, Omori M, Tamiya E, Muraguchi A. Anal Chem. 2005;77:8050–8056. doi: 10.1021/ac0515632. [DOI] [PubMed] [Google Scholar]
- 15.Figueroa XA, Cooksey GA, Votaw SV, Horowitz LF, Folch A. Lab Chip. 2010;10:1120–1127. doi: 10.1039/b920585c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park JY, Morgan M, Sachs AN, Samorezov J, Teller R, Shen Y, Pienta KJ, Takayama S. Microfluid Nanofluid. 2010;8:263–268. doi: 10.1007/s10404-009-0503-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Carlo D, Aghdam N, Lee LP. Anal Chem. 2006;78:4925–4930. doi: 10.1021/ac060541s. [DOI] [PubMed] [Google Scholar]
- 18.Skelley AM, Kirak O, Suh H, Jaenisch R, Voldman J. Nat Methods. 2009;6:147–152. doi: 10.1038/nmeth.1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan WH, Takeuchi S. P Natl Acad Sci USA. 2007;104:1146–1151. doi: 10.1073/pnas.0606625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Faley S, Seale K, Hughey J, Schaffer DK, VanCornpernolle S, McKinney B, Baudenbacher F, Unutmaz D, Wikswo JP. Lab Chip. 2008;8:1700–1712. doi: 10.1039/b719799c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobel S, Valero A, Latt J, Renaud P, Lutolf M. Lab Chip. 2010;10:857–863. doi: 10.1039/b918055a. [DOI] [PubMed] [Google Scholar]
- 22.Kim MC, Wang ZH, Lam RHW, Thorsen T. J Appl Phys. 2008;103 [Google Scholar]
- 23.Chung K, Kim Y, Kanodia JS, Gong E, Shvartsman SY, Lu H. Nat Methods. 2011;8 doi: 10.1038/nmeth.1548. 171-U103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDonald JC, Duffy DC, Anderson JR, Chiu DT, Wu HK, Schueller OJA, Whitesides GM. Electrophoresis. 2000;21:27–40. doi: 10.1002/(SICI)1522-2683(20000101)21:1<27::AID-ELPS27>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 25.Skupin A, Kettenmann H, Falcke M. Plos Comput Biol. 2010;6 doi: 10.1371/journal.pcbi.1000870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeon NL, Dertinger SKW, Chiu DT, Choi IS, Stroock AD, Whitesides GM. Langmuir. 2000;16:8311–8316. [Google Scholar]
- 27.Imboden JB, Weiss A. Biochem J. 1987;247:695–700. doi: 10.1042/bj2470695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakamura N, Yamazawa T, Okubo Y, Iino M. Mol Syst Biol. 2009;5 doi: 10.1038/msb.2009.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.