Abstract
The first component (E1o) of the Escherichia coli 2-oxoglutarate dehydrogenase complex (OGDHc) was engineered to accept substrates lacking the 5-carboxylate group by subjecting H260 and H298 to saturation mutagenesis. Apparently, H260 is required for substrate recognition, but H298 could be replaced by hydrophobic residues of similar molecular volume. To interrogate whether the second component would enable synthesis of acyl-coenzymeA derivatives, hybrid complexes consisting of recombinant components of OGDHc (o) and pyruvate dehydrogenase (p) enzymes were constructed, suggesting that a different component is the ‘gatekeeper’ for specificity for these two multienzyme complexes in bacteria, E1p for pyruvate, but E2o for 2-oxoglutarate.
In this work, we are interested in elucidating the factors that govern specificity of 2-oxoglutarate dehydrogenase multienzyme complex (OGDHc) towards its 5-carboxyl substituent, with the goal to synthesize acylcoenzymeA analogs, comprising a large class of metabolically relevant compounds participating in many metabolic pathways. The OGDHc catalyzes the rate-limiting step in the citric acid cycle (1,2), which is the common pathway for the oxidation of fuel molecules, including carbohydrates, fatty acids and amino acids, and catalyzes the formation of succinyl coenzyme A (succinyl-CoA) according to equation 1.
| (1) |
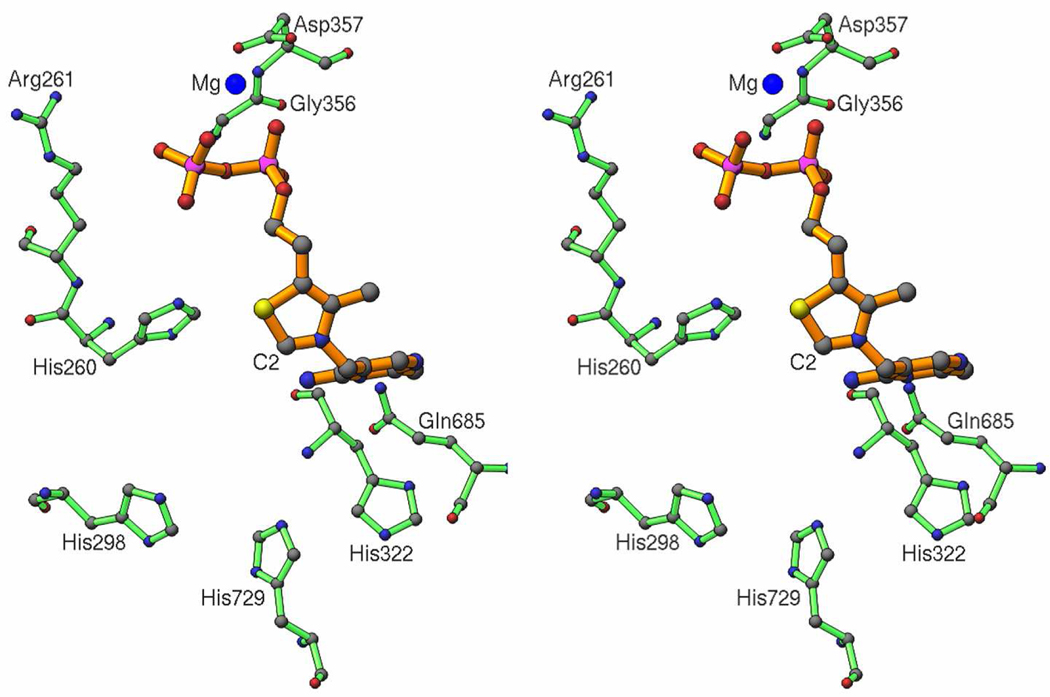
The OGDHc is composed of multiple copies of three components (3–6): (1) thiamin diphosphate (ThDP) dependent 2-oxoglutarate dehydrogenase (E1o, EC 1.2.4.2). (2) dihydrolipoylsuccinyl transferase (E2o, EC 2.3.1.6) and (3) dihydrolipoyl dehydrogenase (E3, EC 1.8.1.4). The first two components carry out the principal reactions for succinyl-CoA formation while the third one reoxidizes dihydrolipoamideE2 to lipoamideE2. This mechanism is similar to other 2-oxoacid dehydrogenase complexes, including pyruvate dehydrogenase (PDHc) and branched-chain 2-oxoacid dehydrogenase. According to the x-ray structure of E1o, there were three His residues (H260, H298 and H729) positioned near the thiazolium ring of ThDP (3) (Figure 1), and substitution of H260 and H298 to Ala drastically reduced the activity. The results suggested that the histidine side chains interacted with the distal carboxylate of 2-oxoglutarate (2-OG) (3).
Figure 1.
Stereo view showing histidines and a few other residues near the active site of the E. coli 2-oxoglutarate dehydrogenase multienzyme complex E1 component, showing their proximity to the reactive center C2 atom on thiamin diphosphate (ThDP). Coordinates for the protein atoms were obtained from the PDB entry 2JGD, which is described in ref. 3, but there were no coordinates for ThDP. The ThDP coordinates added were obtained by Least-Squares superposition of the active site area in the E. coli pyruvate dehydrogenase multienzyme complex E1 component (PDB code 2IEA), to that of the same area in the reported apo-structure 2JGD. The resulting figure here shows is in roughly the same orientation as in Figure 3a of ref.3, with the added ThDP nearly identical in conformation/position to that shown in ref. 3.
We constructed saturation mutagenesis libraries of H260, H298, and H260/H298 and screened for activity towards 2-OG, and an unnatural substrate, 2-oxovalerate (2-OV), in which a nonpolar methyl group replaces the charged carboxylate. Several E1o variants were isolated, some with the ability to decarboxylate 2-OV. The E1o variants created by the H260 and H298 substitutions were shown to be functionally competent according to their ability to produce a ThDP-bound pre-decarboxylation intermediate (7). Next, we wished to determine whether the second E2o component would allow synthesisof acyl-coenzymeA derivatives. Hybrid complexes consisting of recombinant components of the E. coli 2-oxoglutarate (o) and pyruvate dehydrogenase (p) enzymes were constructed (the E3 component is common to both) and it was demonstrated that E1p imparts specificity for acetyl-CoA formation from pyruvate, but E2o controls specificity for succinyl-CoA formation by OGDHc in Gram-negative bacteria.
Results of screening revealed that of the 352 colonies screened for H260 substitutions, 7 were found positive with 2-OV and 61 with 2-OG. Of the 7 colonies found positive for 2-OV, DNA sequencing revealed that all were wild type E1o. Of the 8 colonies screened positive with 2-OG, 7 were identified as wild type E1o and one was H260E. This immediately suggests that H260 is crucial for substrate binding. At position H298, 440 colonies were screened for 2-OG activity and 350 for 2-OV activity. DNA sequencing identified the H298T and H298L substitutions active with 2-OG, and the H298D and H298V substitutions active with 2-OV. Screening for dual H260/H298 substitutions with 2-OG (1232 colonies) and with 2-OV (1672 colonies) revealed several active variants: H260/H298T (with 2-OG), H260/H298D, H260/H298T, and H260E/H298N (with 2-OV). The E1-specific activity was unaffected when E1o was reconstituted into E1o-E2o-E3 or E1o-E2p-E3 complexes. Similar E1-specific activity was found with 2-OG, pyruvate or 2-OV using E1o by itself or assembled in the OGDHc or hybrid (E1o-E2p-E3) complex (Table 1-top). The activity of E1o was 24% towards pyruvate and 19% towards 2-OV compared to 2-OG. The E1-specific activity in complex reconstituted from E1o and the E2o and E3 components (36% with pyruvate and 21% with 2-OV) remained similar to that with E1o by itself. Reconstitution of E1o in the hybrid complex with E2p and E3, led to an E1-specific activity of 34% with pyruvate and 23% with 2-OV. This indicated that assembly into the OGDHc or hybrid complex does not affect significantly the E1-specific rates (Table 1, top). These results gave important evidence that pyruvate and 2-OV were substrates for E1o, as the DCPIP reduction assay clearly indicates that decarboxylation has taken place. No overall activity was detected with pyruvate or 2-OV for either the OGDHc or the hybrid E1o-E2p-E3 complex. The E1o-E2p-E3 hybrid complex exhibited detectable activity (2.2%) with 2-OG. In contrast to E1o, E1p displayed activity only with pyruvate. In the DCPIP assay, E1p by itself showed no activity towards 2-OG or 2-OV. Furthermore, there was no activity for 2-OG or 2-OV for E1p reconstituted with either E2p+E3 or E2o+E3. Similar results were obtained in the overall activity assay (Table 1, bottom panel).
Table 1.
E1-specific and complex activity for E1o (top) and E1p (bottom).
| Substrate | DCPIP activity (µmol.min−1.mg−1) | Overall activity (µmol.min−1.mg−1 E1o) |
|||
|---|---|---|---|---|---|
| E1o | E1o-E2o-E3 | E1o-E2p-E3 | E1o-E2o-E3 | E1o-E2p-E3 | |
| 2-OG (2 mM) |
0.34± 0.01 (100%) |
0.37 ± 0.02 (109%) |
0.40 ± 0.01 (119%) |
16.± 0.4 (100%) |
0.36±0.01 (2.2%)a |
| pyruvate (25 mM) |
0.08 ± 0.01 (24%) |
0.12 ± 0.01 (36%) |
0.12 ± 0.01 (34%) |
no activity | no activity |
| 2-OV (25 mM) |
0.06±0.001 (19%) |
0.07 ± 0.03 (21%) |
0.08 ± 0.01 (23%) |
no activity | no activity |
| Detected by FTMS | Not measured | succinyl CoA (2-OG) |
succinyl-CoAb (2-OG) |
||
| Substrate | DCPIP activity (µmol.min−1.mg−1) | Overall activity (µmol.min−1.mg−1 E1p) |
|||
|---|---|---|---|---|---|
| E1p | E1p-E2p-E3 | E1p-E2o-E3 | E1p-E2p-E3 | E1p-E2o-E3 | |
| pyruvate (2 mM) |
0.75± 0.01 (100%) |
0.64 ± 0.07 (85%) |
0.70 ± 0.02 (93%) |
28 ± 1.0 (100%) |
No activity |
| 2-OGc, 2-OVd | No activity | No activity | No activity | No activity | No activity |
The lowest activity detected was 0.068±0.004 mmol.min−1.mg−1 E1o.
Apparently, even 2.2% could be detected by FTMS, confirming a low percentage activity.
2 mM.
45 mM.
To provide further evidence that pyruvate is indeed a substrate for E1o, we carried out the following studies: (1) The carboligase side reactions commonly accompany ThDP-catalyzed decarboxylations. These reactions involve nucleophilic addition of the enamine (Scheme S1 Supporting Information) to the carbonyl carbon of reactant or product, resulting in the formation of acetoin-like or acetolactate-like ligated products. Observation of carboligase products provides strong confirmation that decarboxylation of substrate had taken place. On addition of pyruvate to E1o, a negative CD band developed at 300 nm, indicating formation of optically active (R)-acetolactate, (Figure S1 Supporting Information). The negative circular dichroism (CD) band at 300 nm was still present after removal of protein (not shown); both the sign of the band and its location are similar to those observed with the E636A variant of E1p (ref. 8, confirmed on that enzyme both by CD and NMR). (2) It was next demonstrated that the pyruvate decarboxylated by E1o could reductively acetylate lipoyl domain derived from E2p (LD-E2p), which could be detected by Fourier Transform Mass Spectrometry (FTMS). The rate constant for reductive acetylation of LD-E2p by (E1o+pyruvate) was 0.0056 ± 0.001 s−1, compared to 51.7 ± 5.4 s−1 for (E1p+pyruvate) under the same conditions. The reductive acetylation of the di-domain comprising lipoyl and subunit binding domains of the E2p was also demonstrated (not shown). Formation of succinyl-CoA by E1o-E2o-E3 and 2-OG was confirmed by both the isotopic pattern and m/z ratio (868.14) of a succinyl-CoA standard in FTMS (Figure S2 Supporting Information). Next, E1o-E2p-E3 was reacted with 2-OG to produce detectable amounts of succinyl-CoA (Table 1-top), displaying a similar isotopic distribution with the standard spectrum of succinyl-CoA (Figure S3 Supporting Information).
These additional experiments were carried out, in part, to also address the finding by Frey’s group that in the OGDHc isolated from E. coli, there is indeed found as much as 10% E1p (9), whose presence would confound our interpretation. We needed to demonstrate that the pyruvate-decarboxylating activity displayed by E1o was not an artifact of the presence of E1p. Our experiments above, and the fact that all of the components of OGDHc were His-tagged and independently overexpressed, rule out any significant contamination from intrinsic E1p components.
Next, the effect of H260 and H298 substitutions on E1o activity was examined. Saturation mutagenesis data revealed that H298 could tolerate substitution. The DCPIP activity for the E1o variants with 2-OG ranged from less than 1% (H298T) to 19% (H298L) (Table 2A). The Km for 2-OG increased for some E1o variants, while the catalytic efficiency (kcat/Km) of all variants with 2-OG was lowered. The catalytic efficiency for the best E1o variants decreased ~7-fold for H298L and ~17-fold for H260E/H298N, and was severely compromised for other variants (Table 2A). Remarkably, the H298D and H298V substitutions converted E1o to 2-oxovalerate dehydrogenase with activities comparable to that observed with 2-OG (Table 2B). The H298 substitution in E1o also affected the overall OGDHc activity (Table S1 Supporting Information). The relative activities according to the E1-specific and overall activity assays were approximately paralleled. Finally, OGDHc did not show any overall activity towards 2-OV, again implying discrimination at the E2o level.
Table 2.
Effect of H298 and H260/H298 substitutions on E1-specific activity of OGDHc.Top panel, 2-OG, bottom panel, 2-OV.
| Substitution | DCPIP activity (µmol.min−1.mg−1) |
kcat (s−1) |
Km (mM × 10−3) |
kcat/Km (s−1 mM−1) |
|---|---|---|---|---|
| none | 0.620 ± 0.03a | 2.15 ± 0.10 | 2.61 ± 0.376 | 824 |
| H298L | 0.120 ± 0.028 | 0.415 ± 0.098 | 3.41 ± 0.237 | 122 |
| H298T | 0.0018 ± 0.0001 | 0.0064 ± 0.0002 | 4.25 ± 0.160 | 1.5 |
| H298D | nd b | nd | nd | nd |
| H298V | 0.029 ± 0.003 | 0.099 ± 0.009 | 165 ± 6 | 0.60 |
| H260E/H298N | 0.039 ± 0.004 | 0.134 ± 0.013 | 2.73 ± 0.469 | 49.1 |
| H260E | 0.0091 ± 0.0007 | 0.032 ± 0.003 | 383 ± 35.5 | 0.0084 |
| Substitution | DCPIP activity (µmol.min−1.mg−1) |
kcat (s−1) |
Km (mM) |
kcat/Km (s−1 mM−1) |
|---|---|---|---|---|
| none | 0.022 ± 0.002 | 0.076 ± 0.007 | 16.3 ± 4.00 | 0.0047 |
| H298L | 0.027 ± 0.003 | 0.094 ± 0.011 | 6.30 ± 0.64 | 0.015 |
| H298T | 0.0086 ± 0.0009 | 0.030 ± 0.003 | 15.3 ± 1.00 | 0.0020 |
| H298D | 0.357 ± 0.018 | 1.24 ± 0.062 | 7.02 ± 0.023 | 0.18 |
| H298V | 0.160 ± 0.005 | 0.556 ± 0.018 | 8.96 ± 0.005 | 0.062 |
E1o was from a different preparation than in Table 1.
Not detectable above the background.
CD experiments revealed formation of a pre-decarboxylation intermediate analog between E1o and 2-oxophosphonate and 2-oxophosphinate analogs. The Rutgers group has published extensively on CD detection of ThDP-bound covalent intermediates on enzymes with substrate mimics derived from methyl acetylphosphonate (MAP) and acetylphosphinate (AcP−), which are analogs of pyruvate (Figure S4 Supporting Information) (7,10,11). It had been reported that succinyl phosphonate (SP2−) and its monomethyl phosphonate ester (SPME−), which are analogs of 2-OG, inhibit partially purified OGDHc complex from brain with S0.5, SP− = 0.12 mM, which is in the range of values of KM, 2-OG =0.1–0.2 mM reported for OGDHc from different sources (see Supporting Information for synthesis of the substrate analogs) (12). The inhibitory effect of SPME− was also demonstrated for OGDHc from E. coli and pigeon breast muscle (13) and very recently for MenD (14). First, E1o was titrated with AcP− because this analog was found to bind strongly to a number of ThDP enzymes (7). CD spectra of E1o titrated with AcP− revealed the generation of two CD bands: a positive one at 297 nm earlier assigned to the 1’,4’-iminopyrimidine tautomer (IP) of the first covalent intermediate (pre-decarboxylation in Scheme S1 Supporting Information) and a negative one at 330 nm assigned to a Michaelis complex (11). The calculated values of Kd for AcP− were 0.32 mM (at 297 nm) and 0.31 mM (at 330 nm) (Figure S5A, Table S2 Supporting Information) as compared with KM, 2-OG of 90 µM (Table S1 Supporting Information). Next, SPME−, SP2− and PP−, analogs for 2-OG and 2-OV, respectively, were evaluated in the CD experiment (Table S2, Figures S5B, S5C Supporting Information). These are the first CD experiments to demonstrate that on addition of substrate analogs, E1o forms a tetrahedral ThDP-bound pre-decarboxylation intermediate analog, resembling those formed from substrates. The Kd values determined are in the µM range (SPME− phosphonate monoester gives the best Kd =10 µM, while the diacid SP2− is approximately three times weaker) demonstrating that some of the tested compounds could be powerful inhibitors of the OGDHc E1o component (Table S2 Supporting Information). Similar CD experiments with the H298 E1o variants by phosphonate and phosphinate analogs of 2-oxoacids (Figures S6–S9 Supporting Information) indicated that this substitution is not favorable for binding of SPME−, excepting H298T. For the H260E/H298N variant, no CD band was detected at 300 nm. On the other hand, H298 substitutions and the double H260E/H298N substitution were favorable for binding of PP−, which is a substrate analog for 2-OV (the H298D variant has a Kd value of 9.6 µM as compared with 39 µM for E1o (Figure S10, Table S2 Supporting Information). In general, the Kd,PP− were smaller (binding was stronger) than that for E1o, and Kd,PP− ranged from 5-22 µM (Figures S11-S14 Supporting Information).
The following could be concluded about the roles of His260 and His298 in E1o. The H260E, H298T, H298V and H298L substitutions displayed activity with 2-OG. The H298D and H298V substitutions led to active enzyme with 2-OV, displaying improvement in kcat/Km in comparison to the E1o. While finding a positively charged or hydrophilic side chain in its place could be anticipated, the most active variant, H298L is unexpected, with a Km comparable to that of E1o. Being only slightly larger than His, this substitution to Leu may only fulfill a volume constraint (the van der Waals volumes for Leu and His are 124 Å3 and 118 Å3, respectively). A study on the active center residues of the ThDP enzyme benzoylformate decarboxylase led to a similar conclusion on residue His281: it could be replaced by Phe or Leu without significant activity loss (15). Randomization at His260 yielded only one E1o variant (H260E) with low activity toward 2-OG and much better activity towards 2-OV.
The active variants identified by E1-specific assay were shown to be functionally competent according to their ability to form pre-decarboxylation covalent intermediate analogs between the ThDP and phosphono- or phosphino- analogs of 2-OG and 2-OV as judged by CD. The values of Kd calculated from CD data are in the µM range as compared with reported values of Km,2-oxoglutarate=0.1-0.2 mM (12), and point to increasing binding potency with increasing chain length. For all H298 variants, PP− was more firmly bound than SPME−, suggesting conversion of function from 2-oxoglutarate dehydrogenase to 2-oxovalerate dehydrogenase, especially with the H298D and H298V variants, which display relatively high activity with 2-OV. The single low activity H260E E1o variant did not display measurable CD signal with PP− or SPME−, consistent with kinetic analysis. The randomization experiment, kinetic study and CD detection of covalent intermediate analogs provide strong evidence that H260 is crucial and indispensible for 2-OG recognition.
Surprisingly, the E1o could decarboxylate 2-OV and pyruvate, in addition to 2-OG according to the DCPIP assay, an assay adequate to confirm decarboxylation of the substrates. To interrogate whether the second component would enable synthesis of acyl-coenzymeA derivatives from substrates accepted by the engineered E1o, hybrid complexes consisting of recombinant components of the E. coli OGDHc (o) and PDHc (p) were constructed (the E3 component is common to both). On reconstitution of E1o with E2o and E3 in OGDHc, the overall activity with 2-OG was 17 µmol.min−1.mg E1o−1 and correlated well with recently published data on E1o (3). No NADH production was detected with pyruvate or 2-OV in the E1o-E2o-E3 and E1o-E2p-E3 hybrid complexes. Detection of succinyl-CoA formation by mass spectrometry, and of reductively acetylated and succinylated LD-E2p, and of activity of the reconstituted complex by NADH kinetic assay, allowed us to conclude that: (1) E1o-E2o-E3 and E1p-E2p-E3 produced the respective acyl-CoA products and NADH; (2) E1o-E2p-E3 could produce succinyl-CoA from 2-OG; (3) E1p-E2o-E3 could not produce acetyl-CoA from pyruvate; and (4) E1o could reductively acetylate, and reductively succinylate LD-E2p. Apparently, a different component is the ‘gatekeeper’ for specificity for acyl-CoA formation by these two important multienzyme complexes in Gram-negative bacteria, E1p for pyruvate, but E2o for 2-OG. The ability of E1o to reductively acetylate LD-E2p, and the ability of the hybrid E1o-E2p-E3 to produce succinyl-CoA provide strong confirmation of this statement.
The principal difference between this and earlier work is that here recombinant individual components were used, while earlier work used isolated complexes, PDHc and OGDHc, or their sub-complexes. Notably, Frey and associates demonstrated that 10% E1p co-purified with E. coli OGDHc, however, the overall activity was about 1% of that with PDHc, already suggesting that the E2o also conferred substrate specificity in terms of rates (9). deKok and coworkers used the OGDHc isolated from Azotobacter vinelandii and found modest overall activity with pyruvate, but no E1-specific activity was detected, in contrast to our findings (16). ‘Promiscuous’ substrate utilization has been identified in several ThDP enzymes: ‘engineering’ by single amino acid substitutions has been shown to lead to changing both substrate and reaction specificity from a decarboxylase/dehydrogenase activity to carboligase-like activity in yeast pyruvate decarboxylase (17), E1p (8), benzoylformate decarboxylase (18,19) among others; the specificity of acetohydroxyacid synthase toward pyruvate as donor has been attributed to hydrophobic residues (20).
Our results rule out an acid-base or hydrogen-bonding role for the residue H298, but confirm a hydrogen-bonding role for H260. Hence, to create complexes capable of accepting alternate 2-oxoacids, it will also be necessary to engineer the E2o active center.
Supplementary Material
Acknowledgement
E. coli E1o and E2o components from the National BioResource Project (NIG, Japan). The authors are grateful to Dr. K. Chandrasekhar at U. of Pittsburgh for kindly preparing Figure 1.
Abbreviations
- ThDP
thiamin diphosphate
- CoA
coenzyme A
- acetyl-CoA
acetyl coenzyme A
- succinyl-CoA
succinyl coenzyme A
- OGDHc
2-oxoglutarate dehydrogenase multienzyme complex from Escherichia coli
- E1o
wild type 2-oxoglutarate dehydrogenase component (EC 1.2.4.2)
- E2o
dihydrolipoylsuccinyl transferase component (EC 2.3.1.6)
- E3
dihydrolipoyl dehydrogenase component (EC 1.8.1.4)
- PDHc
pyruvate dehydrogenase complex from Escherichia coli
- E1p
pyruvate dehydrogenase component
- E2p
dihydrolipoylacetyl transferase component
- 2-OG
2-oxoglutarate
- 2-OV
2-oxovalerate
- CD
circular dichroism
- DCPIP
2,6-dichlorophenolindophenol
- AcP−
acetylphophinate
- MAP
methyl acetylphosphonate
- PP−
propionylphosphinate
- SP2−
succinylphosponate
- SPME−
succinylphosphonate methyl ester
- IP
1’, 4’-iminopyrimidine tautomer of ThDP
- LD-E2p
the lipoyl domain independently expressed from the 1-lipoyl domain E2p from Escherichia coli
Footnotes
Supporting information available: Materials and methods; Tables S1, S2, Scheme S1, Figures S1–S14. This material is available free of charge via the internet at http://pubs.acs.org.
Funding Sources Supported at Rutgers by NIH GM050380 (FJ) and at NJIT by NSF MCB0746078 (ETF)
References
- 1.Perham RN. Annu. Rev. Biochem. 2000;69:961–1004. doi: 10.1146/annurev.biochem.69.1.961. [DOI] [PubMed] [Google Scholar]
- 2.Hansford RG, Zorov D. Molecular and Cellular Biochemistry. 1998;184:359–369. [PubMed] [Google Scholar]
- 3.Frank RAW, Price AJ, Northrop FD, Perham RN, Luisi BF. J. Mol. Biol. 2007;368:639–651. doi: 10.1016/j.jmb.2007.01.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ricaud PM, Howard MJ, Roberts EL, Broadhurst RW, Perham RN. J. Mol. Biol. 1996;264:179–190. doi: 10.1006/jmbi.1996.0632. [DOI] [PubMed] [Google Scholar]
- 5.Knapp JE, Mitchell DT, Yazdi MA, Ernst SR, Reed LJ, Hackert ML. J. Mol. Biol. 1998;280:655–668. doi: 10.1006/jmbi.1998.1924. [DOI] [PubMed] [Google Scholar]
- 6.Robien MA, Clore GM, Omichinski JG, Perham RN, Appella E, Sakaguchi K, Gronenborn AM. Biochemistry. 1992;31:3463–3471. doi: 10.1021/bi00128a021. [DOI] [PubMed] [Google Scholar]
- 7.Nemeria N, Chakraborty S, Baykal A, Korotchkina LG, Patel MS, Jordan F. Proc. Natl. Acad. Sci. U. S. A. 2007;104:78–82. doi: 10.1073/pnas.0609973104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nemeria N, Tittmann K, Joseph E, Zhou L, Vazquez-Coll MB, Arjunan P, Hübner G, Furey W, Jordan F. J.Biol.Chem. 2005;280:21473–21482. doi: 10.1074/jbc.M502691200. [DOI] [PubMed] [Google Scholar]
- 9.Steginsky CA, Gruys KJ, Frey PA. J.Biol.Chem. 1985;260:13690–13693. [PubMed] [Google Scholar]
- 10.Nemeria NS, Korotchkina LG, Chakraborty S, Patel MS, Jordan F. Bioorg. Chem. 2006;34:362–379. doi: 10.1016/j.bioorg.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nemeria NS, Chakraborty S, Balakrishnan A, Jordan F. FEBS Journal. 2009;276:2432–2446. doi: 10.1111/j.1742-4658.2009.06964.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bunik VI, Denton TT, Xu H, Thompson CM, Cooper AJL, Gibson GE. Biochemistry. 2005;44:10552–10561. doi: 10.1021/bi0503100. [DOI] [PubMed] [Google Scholar]
- 13.Biryukov AI, Bunik VI, Zhukov YN, Khurs EN, Khomutov RM. FEBS Lett. 1996;382:167–170. doi: 10.1016/0014-5793(96)00166-4. [DOI] [PubMed] [Google Scholar]
- 14.Fang M, Toogood RD, Macova A, Ho K, Franzblau SG, McNeil MR, Sanders DA, Palmer DR. Biochemistry. 2010;49:2672–2679. doi: 10.1021/bi901432d. [DOI] [PubMed] [Google Scholar]
- 15.Yep A, Kenyon GL, McLeish MJ. Proc. Natl. Acad. Sci. U. S. A. 2008;105:5733–5738. doi: 10.1073/pnas.0709657105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bunik V, Westphal AH, deKok A. Eur. J. Biochem. 2000;267:3583–3591. doi: 10.1046/j.1432-1327.2000.01387.x. [DOI] [PubMed] [Google Scholar]
- 17.Sergienko EA, Jordan F. Biochemistry. 2001;40:7369–7381. doi: 10.1021/bi002856m. [DOI] [PubMed] [Google Scholar]
- 18.Müller M, Gocke D, Pohl M. FEBS J. 2009;276:2894–2904. doi: 10.1111/j.1742-4658.2009.07017.x. [DOI] [PubMed] [Google Scholar]
- 19.Yep A, McLeish MJ. Biochemistry. 2009;48:8387–8395. doi: 10.1021/bi9008402. [DOI] [PubMed] [Google Scholar]
- 20.Steinmetz A, Vyazmensky M, Meyer D, Barak Z, Golbik R, Chipman DM, Tittmann K. Biochemistry. 2010;49:5188–5199. doi: 10.1021/bi100555q. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.