Abstract
Resilin is an elastomeric protein found in specialized regions of the cuticle of most insects, providing outstanding material properties including high resilience and fatigue lifetime for insect flight and jumping needs. Two exons (1 and 3) from the resilin gene in Drosophila melanogaster were cloned and the encoded proteins expressed as soluble products in Escherichia coli. A heat and salt precipitation method was used for efficient purification of the recombinant proteins. The proteins were solution cast from water and formed into rubber-like biomaterials via horseradish peroxidase-mediated cross-linking. Comparative studies of the two proteins expressed from the two different exons were investigated by Fourier Transform Infrared Spectroscopy (FTIR) and Circular Dichrosim (CD) for structural features. Little structural organization was found, suggesting structural order was not induced by the enzyme-mediateed dityrosine cross-links. Atomic Force Microscopy (AFM) was used to study the elastomeric properties of the uncross-linked and cross-linked proteins. The protein from exon 1 exhibited 90% resilience in comparison to 63% for the protein from exon 3, and therefore may be the more critical domain for functional materials to mimic native resilin. Further, the cross-linking of the recombinant exon 1 via the citrate-modified photo-Fenton reaction was explored as an alternative dityrosine mediated polymerization method and resulted in both highly elastic and adhesive materials. The citrate-modified photo-Fenton system may be suitable for in-vivo applications of resilin biomaterials.
Introduction
Animal movement in nature often demands both high speed and power and has to overcome the constraints imposed by striated muscle [1–3]. Arthropods often overcome these limitations by slowly deforming elastic structures to maximize stored energy, and then deliver this stored energy by rapid recoil via elastomeric proteins [1–3]. An elastomeric protein resilin is found at many joints and tendons of arthropods where fast, repeated actions, or elastic energy storage are required, such as the flight systems of locusts and beetles [4, 5], the jumping mechanisms of fleas and froghoppers [6, 7], and the sound producing organ of cicadas [8].
In 2000 the complete Drosophila melanogaster genome became available, and the gene product CG15920 was considered to be a resilin precursor due to its amino acid composition and the presence of an N-terminal signal peptide sequence for secretion [9]. Further analysis showed that the protein was 620 amino acids long and divided into four distinct segments. The first 17 amino stand for a cuticlar secretion signal peptide. The middle segment contained 62 amino acids and showed consensus to a Rebers-Riddiford sequence [9, 10], which is exon 2 (Figure 1). This sequence is similar to a region conserved in a number of matrix proteins from insect cuticle. We have previously shown that Exon 2 is a chitin binding domain (ChBD) with high affinity to chitin, implying a role in the formation of the resilin-chitin composites in the cuticle [11, 12]. Resilin binds to cuticle chitin via the ChBD and is further polymerized through oxidation of the tyrosine residues, resulting in the formation of dityrosine bridges [11]. Within the Drosophila resilin gene sequence, two significant exons flanking exon 2 in the gene CG15920 were also identified, exon 1 and exon 3 (Figure 1). An N-terminal domain comprising 18 pentadecapeptide repeats (GGRPSDSYGAPGGGN) is found entirely on exon 1 and a C-terminal domain comprising 11 tridecapeptide repeats (GYSGGRPGGQDLG) dominates exon 3 [9]. Both of these exons have a high content of glycine and proline, and lack sulphur-containing amino acids or tryptophan (Suppl. Info. Table S1). The exon 1 repeats contain two proline residues and the exon 3 repeats contain one. The strong conservation of the positions of proline and glycine residues encoded in exon 1 and exon 3 in Drosophila resilin indicate that they are important for chain folding. This amino acid sequence led Ardell and Andersen [9] to predict that resilin forms irregular, extended beta-spiral structures with long-range elasticity, similar to elastin. Our recent finding [12] as well as those of others showed that resilin is mainly unstructured and might form β-turns as well as more extended poly-proline II (PPII) secondary structures, while no β-spiral structures were identified [13, 14].
Figure 1. Conceptual amino acid sequence of Drosophila gene product CG15920 after cleavage of the signal peptide.
The regions corresponding to exon 2 conserved in a number of matrix proteins from insect solid cuticle are indicated by bold type, and the remaining, exon 1 parts of the native resilin are indicated by italics; exon 3 parts of native resilin are indicated by single underlining.
Resilin protein biomaterials can be generated by genetic engineering of resilin-encoding genes [15]. In addition, a simple salt precipitation and heat purification method allowed rapid and efficient downstream processing of the soluble recombinant resilin [15]. Exon 1 (Figure 1), encoding the N-terminal domain in native resilin, was cloned and expressed as a ~30 kDa soluble protein in Escherichia coli [16]. Based on scanning probe microscopy and tensile testing, this sample had up to 92% resilience and could be stretched to over 300% of its original length before breaking [16]. Thus the physical properties of the rec1-resilin protein encoded by exon 1 of D. melanogaster recombinant resilin resembled that of natural resilin. More recent studies of rec1-resilin (exon 1) demonstrated assembly on different surfaces using atomic force microscopy (AFM) for imaging [17]. This approach is potentially useful in the design of hydrogel structures with controlled morphology from resilin proteins that could be exploited as a reservoir for drugs, nanoparticles, enzymes, catalysts and sensor applications [17]. The production of modular polypeptide materials, based on 15 exon 1 repeats in the native resilin, was also reported and these recombinant materials exhibited useful mechanical and cell adhesion behavior [18]. Exon 3 (Figure 1), encoding the C-terminal domain in native resilin, is a ~23 kDa protein. Compared with exon 1, little is known about the function of exon 3.
To date, studies of structural and functional properties from resilin are limited. The structural properties of a synthetic resilin construct, AN16 that was based upon 16 units of the consensus proresilin repeat from Anopheles gambiae, was investigated using NMR and Raman, and it was suggested that the resilin chains were mobile [13]. In our recent studies, we reported the expression of full length recombinant resilin protein (exon 1 + exon 2 + exon 3) from D. melanogaster [12]. Based on the secondary structure of this full length recombinant resilin, the majority of the resilin backbone exhibited a random coil conformation, indicating that the overall protein was dynamic and unstructured. A range of conformations were also present within the cross-linked protein [12]. Compared with 92% resilience of the partial resilin clone (exon 1), rec1-resilin [16], the full length resilin (exon 1 + exon 2 + exon 3) was 94% prior to crosslinking, and 96% when the protein was cross-linked. Thus the full length protein (exon 1 + exon 2 + exon 3) offered similar material functions to the partial clone (exon 1) [12, 16].
There is strong interest in biopolymer-based biomaterials in the field of regenerative medicine [19]. However, although generally biocompatible, most of these biomaterial scaffolds are insufficient for supporting load bearing tissues such as tendons, ligaments and the spine [20]. In addition, functionalizing scaffold–tissue attachments for adhesion is a critical feature enabling improved performance of scaffolds, a feature which remains a challenge [21]. Resilin, the most elastic biomaterial known, is a potential candidate for novel composite scaffolds for load bearing tissues [16]. The elastic properties of resilin are achieved by cross-links, formed by di-tyrosine bridges within the randomly coiled, isotropic resilin monomers [4, 9]. Several methods for in vitro cross-linking of recombinant resilin have been proposed [12, 16] and involve either the use of peroxidase enzymes (e.g., horseradish peroxidase (HRP)) or photosensitizers, such as [Ru(bpy)3]2+, which result in materials that are highly elastic with relatively low adhesiveness. Therefore, the development of new cross-linking methods for recombinant resilin to generate useful elastic and adhesive features would be beneficial as this could widen the potential use in biomaterials and tissue engineering applications.
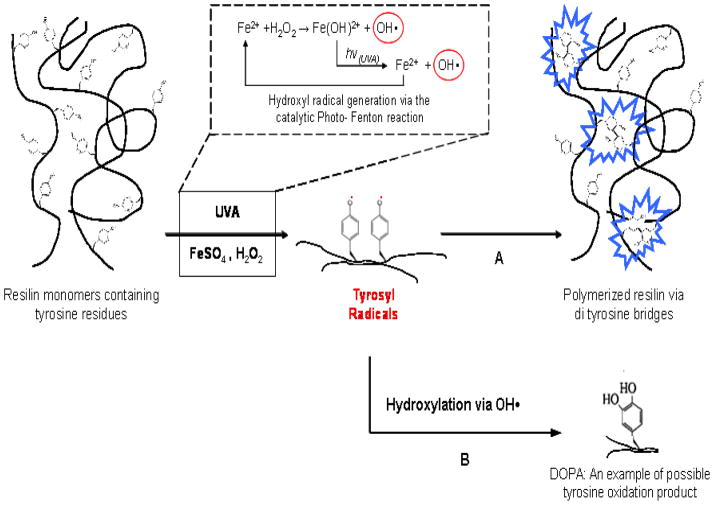
The peroxidase-hydrogen peroxide (H2O2) system was used for resilin cross-linking via the formation of tyrosyl radicals, which further yielded di-tyrosine cross-linked covalent bridges [22]. As previously suggested, the formation of tyrosyl radicals in the HRP-H2O2 system can be driven by two different mechanisms: The first involves a direct interaction of the enzyme active site with the hydroxyphenyl group. The second involves the generation of Reactive Oxygen Species (ROS) which can further interact with the hydroxyphenyl groups to generate the desired phydroxyphenyl radicals [23]. Hence, it is possible that ROS alone may lead to the desired resilin cross-linking. Moreover, it was shown that hydroxyphenyl residues can be further oxidized via ROS to yield 3,4-dihydroxy-DL-phenylalanine (DOPA), which is known for its outstanding surface attachment properties [24–26]. The proposed mechanism for the cross-linking and oxidation of resilin is presented in Scheme 1. We therefore explored this type of reaction in the present study using the citrate-modified photo-Fenton system.
Scheme 1. Possible mechanism for resilin crosslinking via the photo-Fenton reaction.
Hydrogen abstraction from tyrosine via strong oxidizing agent (•OH) can generate tyrosyl radical, resulting in di-tyrosine bridges (A) and other tyrosine oxidation products (B).
In an effort to further our understanding of the structural and functional properties of resilin, and to assess the importance of the two critical regions of the resilin encoding genes, exon 1 and exon 3, from the native resilin genes of the Drosophilla CG15920, the corresponding genes were cloned and expressed. The objective was to better define the respective roles of each exon in terms of resilin properties. This was carried out by enzymatic cross-linking of the two encoded proteins and assessment of structural and functional features. The structural and elastic properties of un-cross-linked and cross-linked resilin proteins were characterized by Fourier Transform Infrared Spectroscopy (FTIR), Circular Dichrosim (CD) and Atomic Force Microscopy (AFM). The data confirmed that the protein from exon 1 exhibited higher elasticity in comparison to the protein from exon 3, and therefore may be more critical towards material features that would mimic those of native resilin. In addition, the cross-linking of recombinant exon 1 resilin was studied via the photo-Fenton reaction, as a potential non-toxic method for recombinant resilin cross-linking towards biomaterial-related applications.
Materials and Methods
Gene designs and plasmid construction
To express native resilin gene fragments, total RNA was extracted from adult D. melanogaster using standard techniques [27]. RT-PCR was carried out following manufacturer’s protocols (Invitrogen Carlsbad, CA) as we have previously reported [12]. Exon 1 primers were 1PAF and 1PAR, with sequences: 1PAF: 5′–GGAATATGTTCAAGTTACTCGCGTTGACGCTGCTCATGGC–3′ and 1PAR: 5′–AATTCGTACTTGGCGGGCTCATCGTTATCGTAGTCAGCGC–3′. Exon 3 primers were 3PAF and 3PAR, with sequences: 3PAF: 5′–TGCCCGACGGAAGGAAGCAAATTGTGGAGTATGAAGCCGA–3′ and 3PAR: 5′–CTAGTACCGATAACCGCTGCCATCGTTGTCACCGTTCTGA–3′. These primers for polymerase chain reaction (PCR) were designed to amplify the two exons of the D. melanogaster resilin gene, CG 15920 [9]. Reagents were purchased from Invitrogen, and the following reaction mix was used for amplification of two exons: 5 μl Pfx Reaction Mix (22 U/ml Thermococcus species KOD thermostable polymerase complexed with anti-KOD antibody, 66 mM Tris-SO4 (pH 8.4), 30.8 mM (NH4)2SO4, 11 mM KCl, 1.1 mM MgSO4, 330 μM dNTPs, AccuPrime proteins, stabilizers), 0.5 μl (5 U/μl) AccuPrime platinum Pfx polymerase, 1 μl (20 mM) forward primer, 1 μl (20 mM) reverse primer, 37.5 μl distilled water, and 5 μl mixture of first strand cDNA synthesis reaction. The following thermocycler program was used: denature at 95°C for 2 minutes; 35 cycles of 95°C for 15 seconds, 55°C for 15 seconds, 68°C for 1 minute per kb to be amplified; and 68°C for 10 minutes. The PCR products were analyzed using agarose gel electrophoresis and purified by gel extraction. The PCR products (978 bp for exon 1 and 714 bp for exon 3) were inserted into the pCR-Blunt II-TOPO vector (Invitrogen Carlsbad, CA) and clones were selected for resistance to ampicillin (100 μg/ml). The DNA fragments of exon 1 and exon 3 were prepared for insertion into the bacterial T7-promoter expression vector pET22b (Novagen EMD Chemicals, Inc. CA) by partial digestion with NcoI and NotI. The NcoI/NotI fragments were inserted between the corresponding sites of pET22b. The recombinant expression plasmids, pET22b/exon1 and pET22b/exon3, were isolated from E. coli NEB5α cells (NEB, MA) with selection for ampicillin resistance (100 μg/ml), and the correct constructs were verified by DNA sequence analysis of the recombinant expression plasmids isolated from single transformants using standard procedures. By the similar strategy, the pET29 vector (Novagen EMD Chemicals, Inc. CA) was used to construct the expression plasmid pET29/6H-exon1 of 6H-exon 1 with a N-terminal His tag followed by a Tobacco Etch Virus (TEV) protease cleavage site enabling purification of the protein on Ni-NTA column and removal of the His tag when desired.
Expression and purification
The recombinant pET22b/exon1 and pET22b/exon3 plasmids were transformed into E. coli strain BL21 Star™ (DE3) (Invitrogen Carlsbad, CA) and selected on LB plates containing 100 μg/ml ampicillin. Authenticity of the clones was confirmed by DNA sequencing. The strains were grown overnight at 37 °C in LB medium containing antibiotics as previously described, and subsequently used to inoculate a 1-litre culture with similarly-supplemented LB medium. A final concentration of 1 mM IPTG (Sigma, St. Louis, MO) was added into the cultures to induce expression for a further 6 h. Cells were collected by centrifugation (10,000 g for 20 min at 4°C), and the cell pellets were frozen at −80°C.
Proteins were purified as previously described with some modification [12]. The cell pellets were thawed and resuspended in lysis buffer containing 1 X BugBuster Protein Extraction Reagent (Novagen EMD Chemicals, Inc. CA), Lysonase™ Bioprocessing Reagent (Novagen EMD Chemicals, Inc. CA) and 1 X phosphate-buffered saline (PBS, Invitrogen Carlsbad, CA). The cell suspension was lysed with shaking for 1 h. The soluble protein fraction was collected by centrifugation at 10,000 g for 1 h at 4°C. The resulting supernatant was heated for 30 min at 90°C and denatured proteins were removed by centrifugation at 10,000 g for 30 min at 20°C. Subsequently 30% solid ammonium sulfate was added to remove bacterial proteins, and 40% salt was then used to collect the proteins. Precipitating proteins were retained following centrifugation, resuspended in sterile phosphate-buffered saline (PBS), and dialyzed overnight in excess PBS. SDS-PAGE on 4–12% Bis-Tris precast gels (Invitrogen Carlsbad, CA) was used to assess recovery and purity. N-terminal amino acid sequence analysis of proteins was done in Tufts University Core Facilities (Boston, MA).
Full details of 6H-exon 1 with the His tag are provided in the Supplementary Information including the expression, purification and histidine-peptide cleavage of 6H-exon 1 and the purification of cleaved 6H-exon 1. The resulted recombinant exon 1 was generated and used for the photo-Fenton cross-linking reactions.
Cross-linking
Horseradish peroxidase was used to cross-link the two resilin proteins from exon 1 and 3, respectively [28]. To cross-link the tyrosine residues, 5 mg of lyophilized recombinant sample was dissolved in 100 μl 0.25 M sodium borate/boric acid buffer, pH 8.4, forming a solution with 50 mg/ml of the resilin protein. The sample solution was incubated at 37°C for 1 hour, and then 250 μl of a 1 mg/ml solution of peroxidase (HRP, Sigma, St. Louis, MO) was added and followed immediately with 10 mM hydrogen peroxide to initiate the reaction. The solution was allowed to incubate at 37°C for an hour. Successful cross-linking was checked by gel electrophoresis and fluorescence detection with a microplate spectrofluorometer (Molecular Devices, CA). Exon 1 polymerization was also performed via the citrate-modified Photo-Fenton reaction. All cross-linked polymers in this method were originated from 200 mg/ml resilin solutions (containing 140 mM tyrosine residues). In addition, all polymerization trials were conducted in citrate:Fe:H2O2 mole ratio of 5:1:20/10, respectively at pH 5. A concentrated resilin solution containing different concentrations (3–140 mM) of FeSO4 was vigorously mixed with H2O2, following 10 to 15 second vortex. The resulted mixtures were transferred into a 50 μl Teflon molds followed by illumination via UV LEDs (370 nm, 10 mW/cm2, 10 to 15 min). The polymers were detached from the Teflon molds and qualitatively evaluated for curing time, homogeneity and elasticity. A Nikon microscope 80i with a 2A filter cube (Excitation: 330–380 nm, Emission >420 nm) was used for polymerized resilin photomicrography. The polymers were washed in the citrate buffer prior to photomicrography under white or ultraviolet light.
Amino acid analysis of polymerized exon 1 resilins
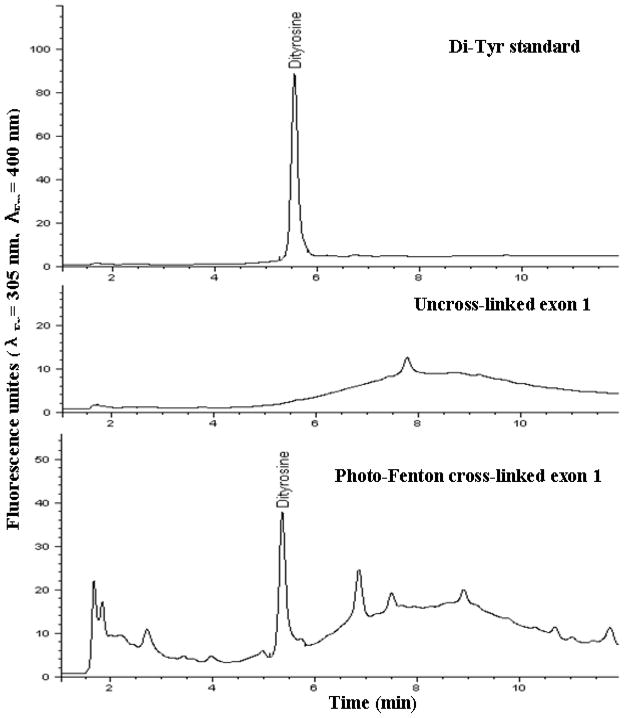
Five mg polymerized and non-polymerized resilin samples were hydrolyzed in 6 N HCl containing 0.1% phenol (145 °C, 4 hr), following filtration though 0.22 μm syringe membrane. Hydrolysis products were analyzed in an Agilent 1200 HPLC system with Diode Array and fluorescence detectors (Santa Clara, CA, USA). UV absorbance and fluorescence were measured at 280 nm, and at typical excitation/emission spectra for di-tyrosine (λEx 305 nm: λEm 400 nm) and DOPA (λEx 280 nm: λEm 320 nm) detection. The chromatography system was used with a LiChrospher 100 RP-18 column (5 μm). Hydrolysis products were eluted using increasing acetonitrile (containing 0.1% TFA) gradient. Additional analysis of the hydrolysis products was carried out in a Thermo Ltq-Orbitrap MS and an Accela High Speed HPLC (LC-MS) with an Agilent Zorbax Exlipse XDB-C18 column (2.1×100 mm, particle size 1.8 μm).
Di-tyrosine standard
A 20 μg/ml HRP solution was added to 5 mM L-tyrosine in borate buffer (50 mM, pH 9), following the immediate addition of 3 mM H2O2. After incubation for 30 minutes at 37 °C, the reaction mixture was centrifuged through a Centricon™ tube (molecular weight cut-off of 10 kDa and 3 kDa) for HRP removal. The filtrate was stored (−18°C) until use. Di-tyrosine was verified in the filtrate using LC-MS. The di-tyrosine/tyrosine mixture was used as a standard for di-tyrosine and tyrosine retention times and UV spectra.
Biomaterial properties
Structural Assessments
Resilin samples were lyophilized and the structural characteristics were observed using Fourier transform infrared spectroscopy (FTIR) as we have previously reported [29, 30]. The fractions of secondary structural components including random coil, alpha-helices, beta-strands and turns were evaluated using Fourier self-deconvolution (FSD) of the infrared absorbance spectra. FSD of the infrared spectra covering the amide I region (1595–1705 cm−1) was performed by Opus 5.0 software. The second derivative was first applied to the original spectra in the amide I region with a nine-point Savitsky-Golay smoothing filter. Deconvolution was performed using Lorentzian line shape with a half-bandwidth of 25 cm−1 and a noise reduction factor of 0.3 [29, 30]. Circular Dichroism (CD) spectra were also studied for the secondary structure of resilin samples as previously described [31]. All spectra were recorded at room temperature (25 °C) using a 1 mm path-length quartz cell. Resilin samples were measured in 1 X PBS with concentrations in the range of 1.0–3.0 mg/ml. CD data were analyzed using a DICHROWEB program (http://dichroweb.cryst.bbk.ac.uk) [32, 33].
Mechanical Assessments
The elastic properties of uncross-linked and cross-linked proteins were performed using Atomic Force Microscopy (AFM) as previously described [12, 16, 34]. A force-distance curve was obtained for evaluation of material properties. Resilin samples were dried on the surface of mica. Measurements on uncross-linked and cross-linked were conducted in force mode with 20 independent trials and mica was used as a control to calibrate the instrument. Resilience was calculated as the ratio of the area under the penetration and retraction curves in the force-distance curves [12, 16, 34].
Results
Gene construction, protein purification and cross-linking
The exons plasmids were constructed successfully by PCR amplification with exon 1 and exon 3 genes, 978 bp and 714 bp, respectively (Figure 2A). After the plasmids were transformed into E. coli strain BL21 Star™ (DE3), expression was carried out for 6 h at 30 °C. A heat and salting-out method was used to purify the resilin proteins, with approximately 30 kDa for exon 1 and 23 kDa for exon 3 detected by SDS-PAGE (Figure 2B). The final purified yield of the two proteins was about 25 mg per liter of culture for both exon 1 and exon 3 encoded proteins. N-terminal amino acid sequence analysis was performed on the purified resilins. MVRPEPPVNSYLPPSDSYGA for exon 1 and MGIVEYEADQQGYRPQIRYE for exon 3 were found, confirming the expected sequence based on the DNA sequence analysis. Heat stability is an important characteristic for elastic proteins. As with full length resilin [12], both proteins encoded by exon 1 and exon 3 exhibited heat stability up to 100 °C for 30 min during purification.
Figure 2. Gene construction, protein purification and cross-linking of soluble resilin.
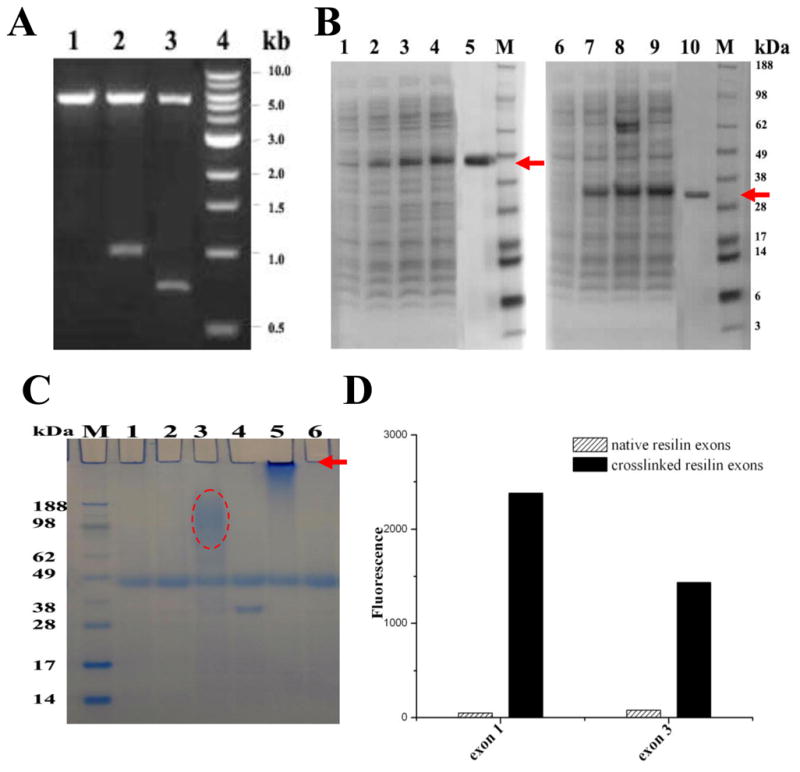
A. Gene construction.
Lane 1, pET22b vector digested by NcoI and NotI; Lanes 2, pET22b/exon1 plasmid digested by NcoI and NotI; Lanes 3, pET22b/exon3 plasmid digested by NcoI and NotI. Expected size of digestion product is 978 bp for exon 1 and 714 bp for exon 3. Size marker is Quick-load™ 1 kb DNA ladder (NEB, MA) in lane 4.
B. 4–12% SDS-PAGE of samples from purification.
Lane 1–4, cleared supernatant from lysed cells of exon 1 expressed for 0, 2, 4, 6 h; Lane 5, soluble native exon 1 purified by a heat and salting-out method (arrow); Lane 6–9, cleared supernatant from lysed cells of exon 3 expressed for 0, 2, 4, 6 h; Lane 10, soluble native exon 3 purified by a heat and salting-out method (arrow). Seeblue plus2 pre-stained standard (Invitrogen Carlsbad, CA) was used as size markers.
C. HRP cross-linking.
The solution containing 100 μl 50 mg/ml soluble resilin exons, 250 μl 1 mg/ml HRP and 10 mM hydrogen peroxide produces polymerized resilin exons which barely enter the gel (arrow). Lane 1, HRP solution with H2O2; Lane 2, HRP solution without H2O2; Lane 3, mixture of exon 3 and HRP solution with H2O2; Lane 4, mixture of exon 3 and HRP solution without H2O2; Lane 5, mixture of exon 1 and HRP solution with H2O2; Lane 6, mixture of exon 1 and HRP solution without H2O2.
D. Fluorescence of resilin.
The absorption and emission maxima were 320 and 400 nm, respectively, for native (uncross-linked) and cross-linked resilin exons.
Horseradish peroxidase was used to cross-link the proteins. Cross-linked exon 1 protein samples showed high molecular weight which barely entered 4–12% SDS-PAGE gels, indicating significant cross-linking. Similar smears were observed for cross-linked protein from exon 3, based on SDS-PAGE gel (Figure 2C). Fluorometric analysis showed the cross-links, with absorption and emission maxima at 320 and 400 nm, respectively. The fluorescence of both cross-linked proteins was higher than that of the uncross-linked proteins, and cross-linked protein from exon 1 showed higher fluorescence and more cross-links than that from the cross-linked protein encoded by exon 3 (Figure 2D). These results confirmed those from SDS-PAGE analysis. The 6H-exon 1 (a His tag on the N-terminal of exon 1) were also purified by Ni-NTA chromatography and observed as a ~35 kDa band in the SDS-PAGE gel (Suppl. Info. Figure S1), subsequently the histidine tag (6H) was cleaved by recombinant TEV protease (rTEV) followed by second Ni-NTA purification where the purified protein was collected from the column flow through (Suppl. Info. Figure S2). Recovered proteins were pooled, dialyzed against DDW and lyophilized for stable long-term storage (4 °C).
Structure and properties of resilin exons
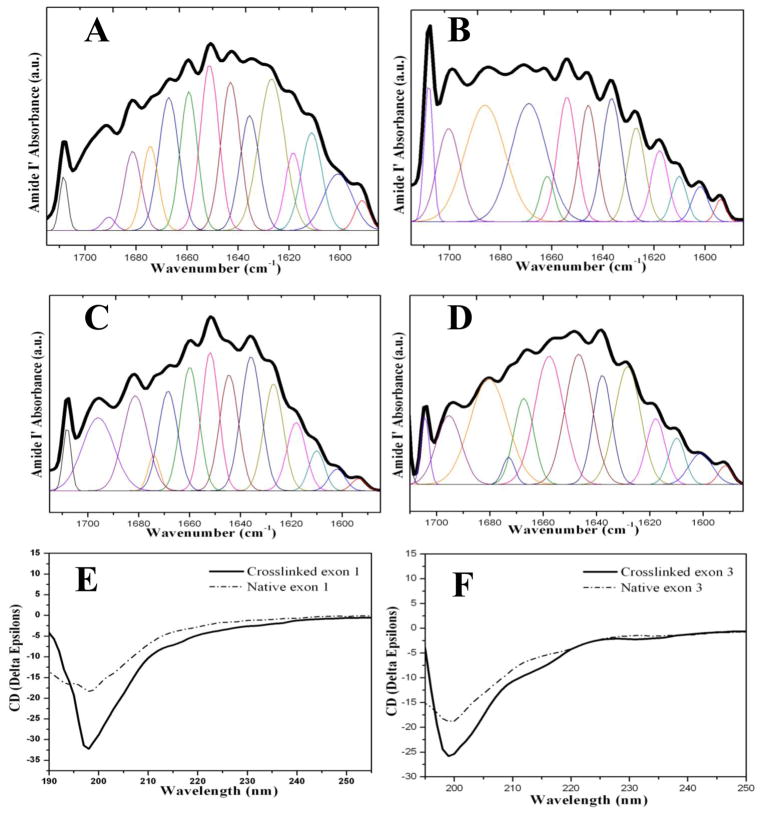
Based on the FTIR spectra of native (uncross-linked) and cross-linked exons, an expansion of the Amide I region was carried out for secondary structure (Figure 3). A range of heterogeneous conformations was indicated from the broad nature of the Amide I bands (centered around 1650 cm−1), and potential contributions from all known secondary structures were suggested by peak deconvolution (Table 1). The band between 1635 and 1655 cm−1 (centered around 1650 cm−1) from the FTIR spectra of all samples exhibited the characteristics of random coil (more than 40%). This high degree of disorder was consistent with the CD spectra for native and cross-linked samples (Figure 3). Similar FTIR and CD spectra were found for both the uncross-linked and cross-linked proteins, thus they had similar secondary structure distributions (Table 1). More or the same random coil conformation was observed in the cross-linked proteins, especially in cross-linked protein from exon 1. These data suggest that the chains of recombinant resilins were mobile and sample a wide range of conformations, which is the case for both the uncross-linked and cross-linked proteins.
Figure 3. Secondary structure analysis of native (uncross-linked) and cross-linked resilin.
A&B. Selected FTIR spectra of native (uncross-linked) exon 1 (A) and cross-linked exon 1 (B) in the Amide I regions after Fourier self-deconvolution.
C&D. Selected FTIR spectra of native (uncross-linked) exon 3 (C) and cross-linked exon 3 (D) in the Amide I regions after Fourier self-deconvolution.
The heavy line represents the deduced absorbance band. The light lines represent the contributions to the amide I′ band.
E&F. Far-UV CD spectra of native (uncross-linked) and cross-linked resilin exons.
All spectra were recorded at room temperature (25 °C) using a 1 mm path-length quartz cell with resilin concentrations in the range of 1.0–3.0 mg/ml.
Table 1.
FTIR and CD secondary structure analysis of native (uncross-linked) and cross-linked resilin generated from exons 1 and 3.
| Helices (%) | Strands (%) | Turns (%) | Random coils (%) | ||
|---|---|---|---|---|---|
| Exon 1 | FTIR | 11.9 | 19.9 | 27.1 | 41.0 |
| CD | 4.9 | 23.2 | 18.4 | 42.0 | |
| Cross-linked Exon 1 | FTIR | 4.4 | 19.8 | 28.9 | 46.8 |
| CD | 4.3 | 24.2 | 19.6 | 42.1 | |
| Exon 3 | FTIR | 12.9 | 19.2 | 26.4 | 41.4 |
| CD | 4.9 | 23.3 | 19.2 | 40.8 | |
| Cross-linked Exon 3 | FTIR | 8.1 | 20.2 | 28.3 | 43.4 |
| CD | 4.4 | 24.2 | 18.4 | 40.9 |
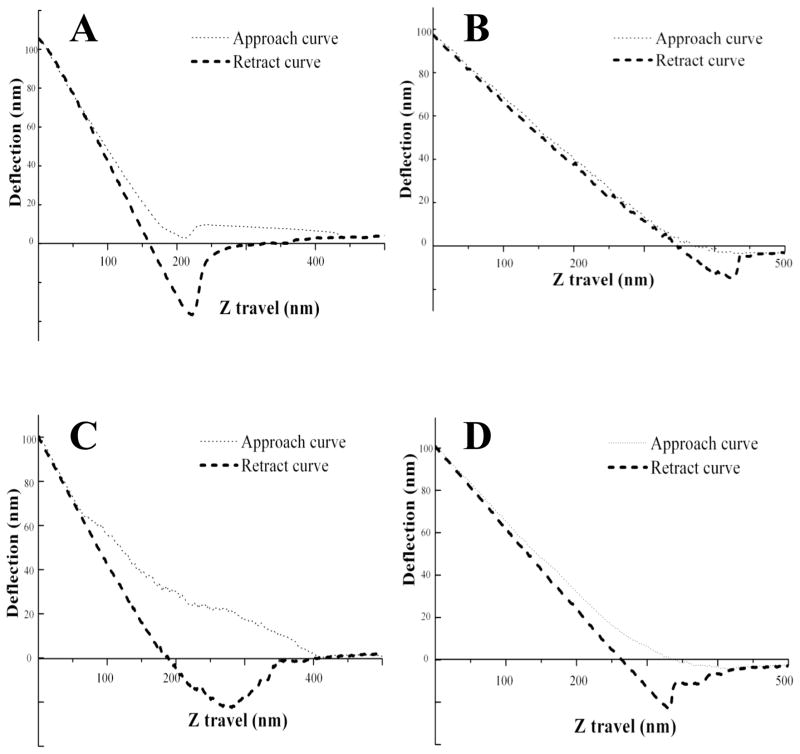
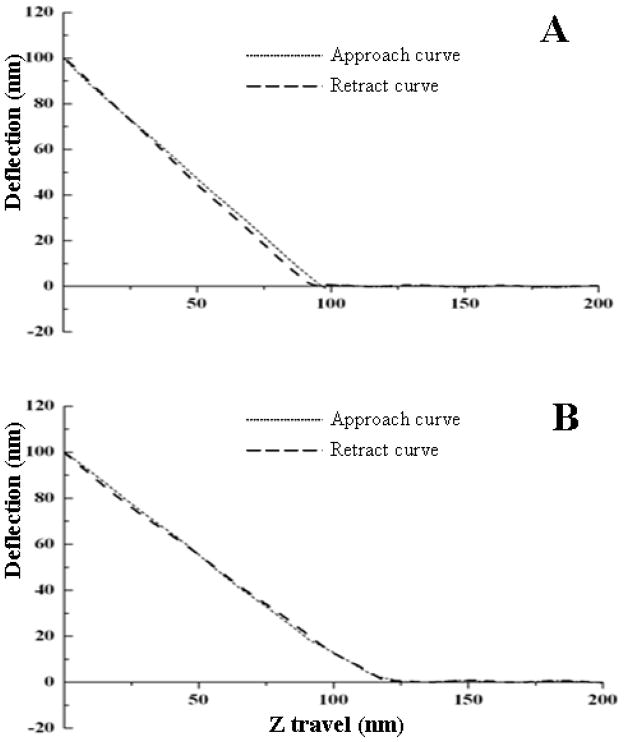
AFM has been used to measure the modulus or stiffness of materials, similar to conventional compression tests [16, 34]. Based on force-distance curves shown in Figure 4, 90±1% resilience for the uncross-linked protein from exon 1, and 93±2% for the HRP cross-linked protein from exon 1 was found. For protein from exon 3, 63±2% and 86±2%, were the corresponding values, respectively. Thus protein encoded by exon 1 exhibited more resilience than that encoded by exon 3. The difference in resilience as result of the crosslinking was minor in exon 1 compared to exon 3. The results of mechanical properties paralleled the results from the enzymatic cross-linking.
Figure 4. Elasticity of native (uncross-linked) and HRP-H2O2 cross-linked resilin exons by AFM.
Force-distance curves were recorded for native (un-cross-linked) exon 1 (A), cross-linked exon 1 (B), native (un-cross-linked) exon 3 (C) and cross-linked exon 3 (D).
Resilin photo-Fenton polymerization and mechanical properties
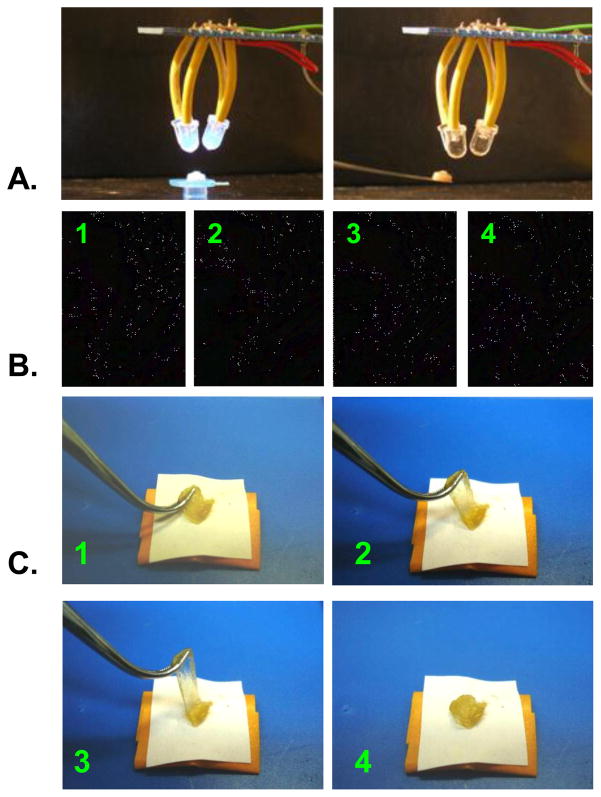
Different concentrations and ratios of the Fenton components (FeSO4 and H2O2) were evaluated to explore optimal conditions for exon 1 resilin polymerization, while maintaining a constant protein concentration (see experimental section). Resilin polymerization was obtained under all conditions evaluated. When high Fe concentrations were applied (tyrosine:Fe molar ratio of 1:1- 0.4), immediate polymerization was observed and inhomogeneous polymers formed prior to UV illumination. These reactions were hard to control while the polymers visually showed good strength and elasticity when pressed by tweezers. Consequently lower Fe concentrations were used and with tyrosine:Fe mol ratio of 1:0.2–0.03, reactions resulted in a more controllable polymerization leading to elastic polymers. These conditions enabled convenient handling of the resilin followed by UV illumination for final polymerization. In addition, polymers resulting from 30 mM and 3 mM FeSO4 were brown-yellow color and exhibited high elasticity and rubber-like properties when compression and extension forces were applied by tweezers (Figure 5). The polymer color varied dependant on Fe content; high Fe resulted in brownish polymers (Figure 5B) while low Fe resulted in yellowish polymers (Figure 5C). Moreover, the polymers displayed adhesive features when placed on matrixes such as stainless steel plastics and paper.
Figure 5. Photo-Fenton cross-linked exon 1 resilins (A) are squashed (B) and extended (C) over 50 cycles via tweezers, without any plastic deformation, showing high resilience.
The polymers color is Fe content dependant.
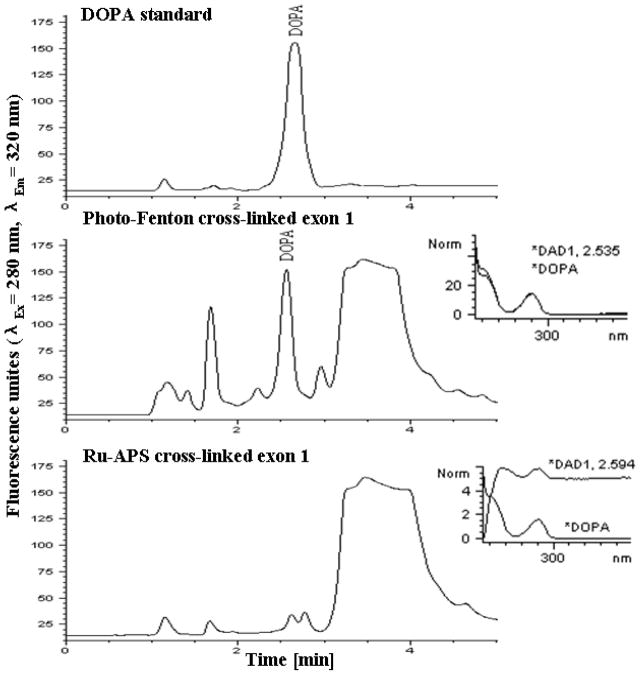
Fluorescence microscopy at di-tyrosine excitation/emission resulted in vivid blue fluorescence (Figure 6) as shown in natural resilin samples due to the presence of di-tyrosine bridges [35]. Amino acid analysis of acid hydrolyzed resilin polymers by LCMS and HPLC revealed di-tyrosine formation during the photo-Fenton polymerization (Figure 7). The content of di-tyrosine formed in the photo-Fenton reaction was less than 5% of the available tyrosines, significantly lower compared to the Ru-APS resilin polymerization method previously reported [16]. In addition to di-tyrosine, other unidentified fluorescent peaks with the same excitation-emission spectra were also observed. Hence we assume that additional tyrosine oxidation products and complexes with other amino acids were generated during the photo-Fenton cross-linking. Using LCMS analysis, another tyrosine oxidation product, DOPA was identified, which is important related to ROS protein oxidation. Interestingly, the formation of DOPA was observed only in the photo-Fenton polymerization system and did not occur in Ru-APS polymerization (Figure 8). These results can also explain the adhesive properties of polymers resulting from the photo- Fenton system, as DOPA has been reported as an adhesive component in mussel proteins [26].
Figure 6. Fluorescence analysis of photo-Fenton cross-linked exon 1 sample.
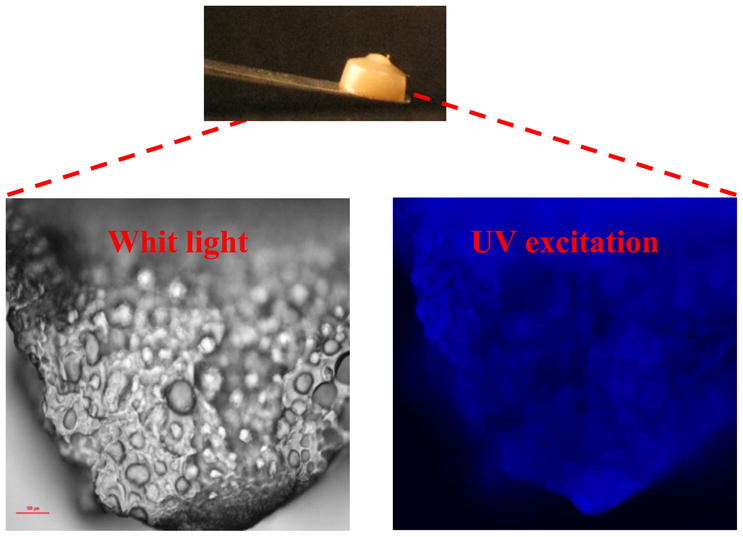
The pictures were taken by Nikon microscope with a 2A filter cube (Excitation: 330–380 nm, Emission > 420 nm).
Figure 7. Amino acid analysis by HPLC separation.
Acid hydrolysis products of resilin samples separated on a C–18 reverse phase column with fluorescence analyzer for di-tyrosine detection.
Figure 8. Amino acid analysis by HPLC separation.
Acid hydrolysis products of resilin samples separated on a C-18 reverse phase column with fluorescence analyzer for DOPA detection.
As in the HRP crosslinking, AFM was also used to determine the elastic modulus and elasticity of non-polymerized and polymerized exon 1 resilins prepared by the photo-Fenton system. Based on force-distance curves, 91±3% resilience for the non-polymerized resilin and 94.5±3% for the polymerized resilin were obtained (Figure 9), consistent with the results when HRP was used for the crosslinking. In addition, the elastic modulus of both cross-linked and uncross-linked resilin was calculated at 9.8±2% and 10.2±2% MPa, respectively. These results indicated that photo-Fenton reactions did not alter the intrinsic properties of the protein and successfully mimicked the elasticity of the native resilin. The resulting cross-linked resilin maintained its rubbery nature compared to previously reported cross-linking methods [12, 16]. However, the cross-linking method presented here also resulted in highly adhesive polymers, compared with resilin polymers that were obtained using either peroxidase or the photosensitizer [Ru(bpy)3]2+. Hence, the photo-Fenton reaction provides a new, approach to cross-linking for recombinant resilin to generate useful bioadhesive and elastic materials.
Figure 9. Elasticity of photo-Fenton cross-linked exon 1 by AFM.
Force-distance curves were recorded for the uncross-linked exon 1 resilin (A) and photo-Fenton cross-linked exon 1 resilin (B)
Discussion
In the 17th century, Robert Hooke defined elasticity as the measure of an object’s ability to deform in proportion to the amount of an applied force and then return to its original state when the force is removed [36]. By this definition, many elastomeric proteins are found in a diverse range of animal species and tissues and possess rubber-like elasticity, undergoing high deformation without rupture, storing the energy involved in deformation, and then returning to their original state when the stress is removed [37]. Protein elasticity remains to be fully described due to the large size and complexity of these proteins which has led to difficulties in isolation and purification [36]. Only a few elastomeric proteins, especially elastin, abductin, resilin and some spider silks, have been studied for mechanical and biochemical properties, and their potential as biomaterials for industrial and biomedical applications has been documented [16, 18, 38, 39]. In nature resilin and elastin have achieved near perfection and these two “elastomer” proteins can be stretched more than twice their original length and recover more than 90% of the deformation energy once the stretching (compression) force is removed [36]. The latter property is called resilience, thus the name resilin. While resilin and elastin have similar properties, resilin has not been studied extensively due to difficulties in obtaining large amounts of pure resilin from natural sources until recently [36].
Recently, Ardell et al. identified the gene product CG15920 as a tentative D. melanogaster resilin precursor, which had the three exons: N-terminal region (exon 1, containing signal peptide and elastic repeats), chitin-binding domain (exon 2, ChBD), and C-terminal region (exon 3, with other types of elastic repeats) [9]. Exon 2 identified as ChBD type R&R-2 in D. melanogaster resilin has been studied experimentally for chitin binding to resilin, implying its role in the formation of the cuticle composite structures [12]. Exon 1 and exon 3 domains contain regions of highly repetitive segments (elastic repeats), which are rich in proline and glycine similar to most proteins that have long-range elasticity. Based on the first exon of CG15920, a recombinant resilin-like protein (rec1) was successfully expressed, purified by immobilized metal affinity chromatography (IMAC), and photo-chemically cross-linked to form an elastic biomaterial (rec1-resilin) with exceptional resilience [16]. Surface induced assembly of rec1-resilin on different surfaces was investigated through direct imaging using AFM [17], and the recombinant materials from exon 1 exhibited useful mechanical and cell adhesion behavior [18]. The full length resilin containing all three exons in D. melanogaster CG15920 was also expressed, purified without affinity tags, and cross-linked by HRP for structural and functional studies. A high degree of disorder and high resilience were important features of the full length resilin (exon 1 + exon 2 + exon 3) [12]. These studies have demonstrated the possibility to generate a rubbery biomaterial by genetic engineering. But to date, there is limited experimental evidence on the function of the C-terminal domain coded by exon 3 as well as experiments comparing the function of the latter with the N-terminal domain (exon 1).
In the present study, the first and third exons from the Drosophilla CG15920 gene, (approximately 30 kDa for exon 1 and 23 kDa for exon 3), were cloned and expressed. Both proteins were expressed in soluble form in bacteria. As with the full length resilin (approximately 60 kDa for (exon 1 + exon 2 + exon 3)), the proteins encoded by exon 1 and exon 3 were also stable to 100 °C, which facilitated purification. Compared with full length resilin that precipitated at 30% ammonium sulfate, the recombinant proteins encoded by the specific exons precipitated at 40% ammonium sulfate. Taking advantage of these features, both full length and smaller protein variants, are resistant to heat and can be selectively salted out at low concentrations of ammonium sulfate [15, 40].
Based on the amino acid composition of D. melanogaster resilin, it was suggested that β-turns would form and result in a β-spiral [9]. A β-spiral is composed of several repetitive β-turns which act as spacers between the turns of the spiral, suspending chain segments in a conformationally free state. Like the full length resilin (exon 1 + exon 2 + exon 3), the resilins generated from the two exons (exon 1 and exon 3) in the present work were also found to be amorphous random network polymers based on the FTIR and CD analysis. For cross-linked resilins, the structural characteristics were similar to the uncross-linked versions, which implied that a range of conformations were present within the cross-linked proteins. Similar structural properties were reported in studies with full length resilin. Furthermore, the secondary structure of a synthetic proresilin (AN16) based upon 16 repeats from Anopheles gambiae (AQTPSSQYGAP), was investigated by Raman and NMR spectroscopy and showed that this protein was intrinsically unstructured with no apparent α–helical or β–sheet features [13]. All of these results could help to confirm either the random-network elastomer model or the sliding β-turn model [13, 41, 42].
Based on force-distance curves in AFM, 90% resilience of resilin encoded by exon 1 was obtained, and 93% was for the cross-linked exon 1, while native exon 3 has 63% resilience, and 86% for cross-linked exon 3. Exon 1 had more resilience than exon 3 and contributes more elasticity for the native resilin [12]. Compared with the 92% resilience of rec1-resilin (cross-linked partial resilin clone from exon 1) and the 93% of cross-linked protein encoded by exon 1 in this paper, the cross-linked full-length resilin had 96% resilience [12, 16]. Moreover the elastic modulus that was measured was 9.8±2% and 10.2±2% MPa for monomeric and crosslinked resilin, respectively.
Furthermore the elastic modulus values measured were somewhat higher than previously reported measurements. Elvin et al, 2005 tested fully saturated recombinant resilin samples and reached the value of 2.5 kPa. Measurements that were performed by others on natural resilin samples resulted in values of 0.3 to 2 MPa, which are 2 to 3 orders of magnitude higher [16, 43, 44]. Our tests were performed by AFM on dry samples which would explain the higher modulus. These results can be explained by the fact that AFM assessments assess lower length scales or macromolecular interactions, and not necessarily macroscopic properties. Therefore the difference in the resilience resulting either from the crosslinking or from the dry conditions of the sample was not reflected in the evaluations due to the high intrinsic resilience of resilin protein chains. Macroscopic materials testing, such as via Instron mechanical tests, would be required, however, the limited yields of recombinant DNA derived proteins in the study were prohibitive of such evaluations.
Moreover we have previously performed compression tests on photo-Fenton polymerized recombinant resilin samples in a tensile tester which resulted in values of 22 kPa (data not shown) in accordance to Elvin et al, 2005, which demonstrate the different properties of the material at the macromolecular vs. macroscopic level [16]. Thus the partial protein generated from exon 1 offered similar material functions to the full length protein with all three exons. This resilience is superior to that of a known low-resilience rubbers, such as chlorobutyl rubber (56%), and even to high-resilience polybutadiene rubbers (80%) [16]. AFM resilience measurements previously performed on full length resilin [12] as well as the current study on the N-terminal domain show a high degree of resilience at the monomeric state. Therefore only slight increases in the resilience and modulus was observed post crosslinking, independent to the crosslinking method. These results indicate the contribution of the basic protein structure to the highly elastic and resilient polymer. Natural crosslinking is required to form a macromolecular network in an elastic polymer which could not be measured by the AFM. Further, the crosslinking may be more relevant to the stabilization of structures in the exoskeleton or to avoid resolubilization of the resilin, versus adding to the mechanical function. Taking these data together, it could be suggested that exon 1 of resilin plays an important role in the functional properties of natural resilin from insects.
Elastic proteins contain repeat sequences forming elastomeric domains, and additional domains that form intermolecular cross-links [45, 46]. Elasticity depends on the length of elastic sequence and the extent of cross-linking. The presence of a network of cross-links (covalent or noncovalent) is a common feature to most of these proteins [14, 41]. Cross-linking of the proteins from the two exons was achieved to produce high molecular weight cross-linked polymeric material using horseradish peroxidase, an enzyme known to catalyze dityrosine formation and present in extracts of resilin from the adult desert locust (Schistocerca gregaria) [4, 47]. Physical properties including structure and elasticity were measured for these recombinant resilin proteins in both uncross-linked and cross-linked versions. The elasticity of cross-linked exon 1, exon 3 and full length resilin proteins was 93%, 86% and 96%, respectively, confirming that both the full length resilin and exon-encoded resilins had higher resilience by cross-linking. The amino acid cross-links are di- and trityrosine [45, 46]. Typically exon 1 encodes for 21 tyrosines of the 326 amino acids (6.46% composition) and exon 3 encodes for 15 tyrosines of the 237 amino acids (6.38% composition). The cross-links give rise to fluorescence with a bluish color with a maximum emission at 420 nm under ultraviolet light. The excitation maximum varies from 285 nm to 315 nm as the pH is changed from acidic to alkaline. Both proteins encoded by exons 1 and 3 were cross-linked by HRP and fluoresce, with the absorption and emission maxima at 320 and 400 nm, respectively, as we have reported previously for full length resilin produced by recombinant DNA technology [12].
Natural resilin is cross-linked in insect cuticle via di-tyrosine formation via enzymes, resulting in an almost perfect 3D elastomer. Both enzymatic and Ru-based methods have been reported for resilin polymerization [12, 16]. In nature, oxidative stress caused by ROS exposure leads to protein cross-linking [24]. However, environmental conditions can alter protein oxidation products. Protein fragments along with carbonyl group formation may occur in oxygen-rich environments. Hydrogen abstraction from tyrosine via •OH generates tyrosyl radicals in hypoxic environments, leading to di-tyrosine cross-links. Hence, we hypothesized that recombinant resilin could be cross-linked by •OH generating systems.
Strong ROS, such as hydroxyl radicals (•OH), are known to be generated either by water hydrolysis, or UVC irradiation of aqua media [48, 49]. However, the necessity of costly radiation sources and relatively high radiation doses makes these approaches problematic. Another approach for •OH production is the photo-homolysis of H2O2 (Scheme 2). This approach was utilized for hydrogel formation and cross-linking of several vinyl-based polymers [50].
| (2) |
The Fenton system, based on reduction of H2O2 in the presence of bivalent cation, such as Fe2+, can rapidly generate strong ROS and iron complexes (Scheme 3) following a series of propagation reactions [51].
| (3) |
In addition, it is known that UVA (up to 410 nm) photolysis of the Fe(OH)2+ complex can regenerate Fe2+ and provide new source of •OH (Scheme 4) [52].
| (4) |
The photo-Fenton system can act as a catalyst, supplying strong ROS by low Fe2+ concentration and low radiation energy. Biomaterial-based hydrogels can serve as a versatile tool in the field of medicine. Medical hydrogels, generated via Fenton reactions, have been previously reported [53–57]. However, a disadvantage of iron precipitates (iron sludge) production, when working at pH>3 is an issue. Nevertheless, this disadvantage can be overcome when working with irons chelates. Citrate, a non toxic iron chelate, can efficiently bind ferrous and ferric ions and prevent iron precipitation, therefore utilizing the Fenton reaction over wide pH ranges can be used to address this issue [51, 52]. Moreover, citrate-iron complexes can limit the amount of available Fe2+ for the H2O2 reaction, hence controlling the rate of •OH production. In addition, toxicity tests for Fenton- based hydrogels previously performed by others revealed no toxic effect, suggesting the Fenton system may serve as a biocompatible method for in vivo polymerization [53–57].
The recombinant resilin was polymerized at near neutral conditions via the citrate-modified photo-Fenton catalytic reaction. Elastic resilin polymers were successfully generated from a wide range of Fe:H2O2 concentrations and the highly polymerized polymers were achieved when FeSO4 was used at lower concentrations up to 30 mM and 3 mM, while maintaining Fe:citrate:H2O2 molar ratios of 1:5:10 and 1:3:10, respectively. Homogeneous polymers were generated without macroscopic gas bubbles and the crosslinked resilin proteins were fluorescent in the typical excitation/emission range of di-tyrosine. Moreover, DOPA was found during the photo-Fenton reaction by LC-MS analysis, resulting in the adhesion of resilin polymers produced by the photo-Fenton system [24, 25]. In contrast, the Ru-APS system, which is known to be highly specific toward di-tyrosine formation [58, 59], did not result in adhesive polymers and DOPA was not detected in Ru-APS-polymerized resilin (Figure 8). Our polymerization studies were conducted with concentrated resilin solutions (200 mg/ml). However, solid hydrogels could be formed even from 85 mg/ml solutions, which was not the case when recombinant resilin was polymerized via the Ru-APS system [16]. Based on these data the photo-Fenton reaction is a useful method for polymerizing resilin, albeit not specific for di-tyrosine formation.
The fundamental understanding of resilin drives the formation of novel resilin-like biomaterials. In particular, because of the outstanding material properties of high resilience and high fatigue lifetime, resilin-like biomaterials offer potential to be used in the medical device field. The use of resilin as a biomaterial scaffold may be of particular interest for engineering tissues which need to be subjected to elastic movements to induce differentiation of cells seeded on these matrices, such as the pulsatile properties of vascular tissues or skin [60]. The stability and biocompatibility of resilin in vivo remain to be fully elucidated. Other well-studied elastic proteins, such as elastin and some spider silks seem to rely on different structural motifs for their particular combination of elasticity and strength [36]. The research findings presented provide several opportunities for future studies. Genes encoding the elastomeric repeats found in resilin and other fibrous proteins can be exploited for mechanical properties by generating modified genes or genetic composites [61]. For example, hydrophilic elastomeric biomaterials were reported, based on the highly resilience protein resilin, which was equipped with multiple biologically active domains. These recombinant materials exhibited useful mechanical and cell adhesion behavior [18].
Recently •OH generating systems were utilized for biopolymer-based hydrogel formation [22, 50, 53–57]. Moreover, poly (vinyl alcohol) (PVA), poly(N-vinyl-2-pyrrolidone) (PVP) and poly(ethyleneimine) (PEI) Fenton-based hydrogels were prepared for medical applications and found to have low cytotoxicity. Thus, we suggest that the photo-Fenton resilin polymerization method may also be applicable for in vivo applications. Further, it is anticipated that future characterization of these properties will provide insight into the ordering and cross-linking of resilin.
Conclusions
Recombinant exon 1 and exon 3 encoded resilins were generated and characterized for structure and function. Cross-linking between tyrosines in the resilin exons was conducted via an enzymatic free radical process and resilience of native exons was characterized for further biomedical applications. The protein from exon 1 exhibited resilience closer to native resilin, suggesting that this region was critical to the functional features of this unusual elastomer. Recombinant exon 1 encoded resilin was also polymerized via a new cross-linking strategy, which is the citrate-modified photo-Fenton reaction. Using recombinant exon 1 resilin, elastic and adhesive resilin polymers were produced using photo-Fenton system that may serve as an in vivo method for resilin polymerization.
Supplementary Material
Acknowledgments
Support from the NIH P41 Tissue Engineering Resource Center (NIBIB, P41 EB002520) is gratefully acknowledged, as is support from DARPA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Vogel S. Living in a physical world III. Getting up to speed. J Biosci. 2005;30:303–312. doi: 10.1007/BF02703667. [DOI] [PubMed] [Google Scholar]
- 2.Alexander RM, Bennet-Clark HC. Storage of elastic strain energy in muscle and other tissues. Nature. 1977;265:114–117. doi: 10.1038/265114a0. [DOI] [PubMed] [Google Scholar]
- 3.Gronenberg Fast actions in small animals: springs and click mechanisms. J Comp Physiol [A] 1996;178:727–734. [Google Scholar]
- 4.Weis-Fogh T. A rubber-like protein in insect cuticle. J Exp Biol. 1960;37:887–907. [Google Scholar]
- 5.Haas F, Gorb S, Blickhan R. The function of resilin in beetle wings. Proc Biol Sci. 2000;267:1375–1381. doi: 10.1098/rspb.2000.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bennet-Clark HC, Lucey ECA. The jump of the flea: a study of the energentics and a model of the mechanism. J Exp Biol. 1967;47:59–67. doi: 10.1242/jeb.47.1.59. [DOI] [PubMed] [Google Scholar]
- 7.Burrows M, Shaw SR, Sutton GP. Resilin and chitinous cuticle form a composite structure for energy storage in jumping by froghopper insects. BMC Biol. 2008;6:41. doi: 10.1186/1741-7007-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young D, Bennet-Clark HC. The role of the tymbal in cicada sound production. J Exp Biol. 1995;202:2937–2949. doi: 10.1242/jeb.198.4.1001. [DOI] [PubMed] [Google Scholar]
- 9.Ardell DH, Andersen SO. Tentative identification of a resilin gene in Drosophila melanogaster. Insect Biochem Mol Biol. 2001;31:965–970. doi: 10.1016/s0965-1748(01)00044-3. [DOI] [PubMed] [Google Scholar]
- 10.Lombardi EC, Kaplan DL. Preliminary characterization of resilin isolated from the cockroach, Periplaneta americana. Mater Res Soc Symp Proc. 1993;292:3–7. [Google Scholar]
- 11.Rebers JE, Riddiford LM. Structure and expression of a Manduca sexta larval cuticle gene homologous to Drosophila cuticle genes. J Mol Biol. 1988;203:411–423. doi: 10.1016/0022-2836(88)90009-5. [DOI] [PubMed] [Google Scholar]
- 12.Qin G, Lapidot S, Numata K, Hu X, Meirovitch S, Dekel M, et al. Expression, cross-linking and characterization of recombinant chitin binding resilin. Biomacromolecules. 2009;10:3227–3234. doi: 10.1021/bm900735g. [DOI] [PubMed] [Google Scholar]
- 13.Nairn KM, Lyons RE, Mulder RJ, Mudie ST, Cookson DJ, Lesieur E, et al. A synthetic resilin is largely unstructured. Biophys J. 2008;95:3358–3365. doi: 10.1529/biophysj.107.119107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bochicchio B, Pepe A, Tamburro AM. Investigating by CD the molecular mechanism of elasticity of elastomeric proteins. Chirality. 2008;20:985–994. doi: 10.1002/chir.20541. [DOI] [PubMed] [Google Scholar]
- 15.Kim M, Elvin C, Brownlee A, Lyons R. High yield expression of recombinant pro-resilin: lactose-induced fermentation in E. coli and facile purification. Protein Expr Purif. 2007;52:230–236. doi: 10.1016/j.pep.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 16.Elvin CM, Carr AG, Huson MG, Maxwell JM, Pearson RD, Vuocolo T, et al. Synthesis and properties of crosslinked recombinant pro-resilin. Nature. 2005;437:999–1002. doi: 10.1038/nature04085. [DOI] [PubMed] [Google Scholar]
- 17.Dutta NK, Choudhury NR, Truong MY, Kim M, Elvin CM. Physical approaches for fabrication of organized nanostructure of resilin-mimetic elastic protein rec1-resilin. Biomaterials. 2009;30:4868–4876. doi: 10.1016/j.biomaterials.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 18.Charati MB, Ifkovits JL, Burdick JA, Linhardt JG, Kiick KL. Hydrophilic elastomeric biomaterials based on resilin-like polypeptides. Soft Matter. 2009;5:3412–3416. doi: 10.1039/b910980c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Velema J, Kaplan D. Biopolymer-based biomaterials as scaffolds for tissue engineering. Tissue Engineering I: Scaffold Systems for Tissue Engineering. 2006;102:187–238. doi: 10.1007/10_013. [DOI] [PubMed] [Google Scholar]
- 20.Chen JM, Xu JK, Wang AL, Zheng MH. Scaffolds for tendon and ligament repair: review of the efficacy of commercial products. Expet Rev Med Dev. 2009;6:61–73. doi: 10.1586/17434440.6.1.61. [DOI] [PubMed] [Google Scholar]
- 21.Wang DA, Williams CG, Yang F, Elisseeff JH. Enhancing the tissue-biomaterial interface: tissue-initiated integration of biomaterials. Adv Funct Mater. 2004;14:1152–1159. [Google Scholar]
- 22.Bailey A. The chemistry of natural enzyme-induced cross-links of proteins. Amino Acids. 1991;1:293–306. doi: 10.1007/BF00813999. [DOI] [PubMed] [Google Scholar]
- 23.Calabro A, Richard AG, Aniq BD. 6982298. US Patent. 2006
- 24.Coles GS. Studies on resilin biosynthesis. J Insect Physiol. 1966;12:679–691. doi: 10.1016/0022-1910(66)90114-4. [DOI] [PubMed] [Google Scholar]
- 25.Xu D, Hong J, Sheng K, Dong L, Yao S. Preparation of polyethyleneimine nanogels via photo-Fenton reaction. Radiat Phys Chem. 2007;76:1606–1611. [Google Scholar]
- 26.Yu M, Hwang J, Deming TJ. Role of 1–3,4-dihydroxyphenylalanine in mussel adhesive proteins. J Am Chem Soc. 1999;121:5825–5826. [Google Scholar]
- 27.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 28.Malencik DA, Anderson SR. Dityrosine formation in calmodulin: cross-linking and polymerization catalyzed by Arthromyces peroxidase. Biochemistry. 1996;35:4375–4386. doi: 10.1021/bi9526037. [DOI] [PubMed] [Google Scholar]
- 29.Hu X, Kaplan D, Cebe P. Determining beta-sheet crystallinity in fibrous proteins by thermal analysis and infrared spectroscopy. Macromolecules. 2006;39:6161–6170. [Google Scholar]
- 30.Hu X, Kaplan D, Cebe P. Dynamic protein-water relationships during beta-sheet formation. Macromolecules. 2008;41:3939–3948. [Google Scholar]
- 31.Greenfield NJ. Using circular dichroism spectra to estimate protein secondary structure. Nature Protocols. 2006;1:2876–2890. doi: 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Whitmore L, Wallace BA. DICHROWEB, an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 2004;32:W668–673. doi: 10.1093/nar/gkh371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitmore L, Wallace BA. Protein secondary structure analyses from circular dichroism spectroscopy: methods and reference databases. Biopolymers. 2008;89:392–400. doi: 10.1002/bip.20853. [DOI] [PubMed] [Google Scholar]
- 34.Huson MG, Maxwell JM. The measurement of resilience with a scanning probe microscope. Polymer Testing. 2006;25:2–11. [Google Scholar]
- 35.Neff D, Frazier S, Quimby L, Wang R, Zill S. Identification of resilin in the leg of cockroach, Periplaneta americana: confirmation by a simple method using pH dependence of UV fluorescence. Arthropod Struct Dev. 2000;29:75–83. doi: 10.1016/s1467-8039(00)00014-1. [DOI] [PubMed] [Google Scholar]
- 36.Alper J. Stretching the limits. Science. 2002;297:329–330. doi: 10.1126/science.297.5580.329. [DOI] [PubMed] [Google Scholar]
- 37.Tatham AS, Shewry PR. Comparative structures and properties of elastic proteins. Phil Trans Roy Soc B. 2002;357:229–234. doi: 10.1098/rstb.2001.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoda RJ. Elastomers for biomedical applications. Biomater Sci Polym Ed. 1998;9:561–626. doi: 10.1163/156856298x00046. [DOI] [PubMed] [Google Scholar]
- 39.Martino M, Perri T, Tamburro AM. Biopolymers and biomaterials based on elastomeric proteins. Macromol Biosci. 2002;2:319–328. [Google Scholar]
- 40.Lyons RL, Lesieur E, Kim M, Wong DCC, Huson MG, Nairn KM, et al. Design and facile production of recombinant resilin-like polypeptides: gene construction and a rapid protein purification method. Protein Eng Des Sel. 2007;20:25–32. doi: 10.1093/protein/gzl050. [DOI] [PubMed] [Google Scholar]
- 41.Aaron BB, Gosline JM. Elastin as a random-network elastomer: a mechanical and optical analysis of single elastin fibers. Biopolymers. 1981;20:1247–1260. [Google Scholar]
- 42.Tamburro AM, Guantieri V, Pandolfo L, Scopa A. Synthetic fragments and analogues of elastin. II. Conformational studies. Biopolymers. 1990;29:855–870. doi: 10.1002/bip.360290419. [DOI] [PubMed] [Google Scholar]
- 43.Gosline J, Lillie M, Carrington E, Guerette P, Ortlepp C, Savage K. Elastic proteins: biological roles and mechnical properties. Philos Trans R Soc Lond B Biol Sci. 2002;357:121–132. doi: 10.1098/rstb.2001.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vincent JF, Wegst UG. Design and mechanical properties of insect cuticle. Arthropod Struct Dev. 2004;33:187–199. doi: 10.1016/j.asd.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 45.Andersen SO. Covalent cross-links in a structural protein, resilin. Acta Physiol Scand Suppl. 1966;263:1–81. [PubMed] [Google Scholar]
- 46.Andersen SO. Crosslinks in resilin identified as dityrosine and trityrosine. Biochim Biophys Acta. 1964;93:213–215. doi: 10.1016/0304-4165(64)90289-2. [DOI] [PubMed] [Google Scholar]
- 47.Giulivi C, Davies K. Mechanism of the formation and proteolytic release of H2O2-induced dityrosine and tyrosine oxidation products in hemoglobin and red blood cells. J Biol Chem. 2001;276:24129. doi: 10.1074/jbc.M010697200. [DOI] [PubMed] [Google Scholar]
- 48.Lop rgolo L, Lugao A, Catalani L. Direct UV photocrosslinking of poly (N-vinyl-2-pyrrolidone)(PVP) to produce hydrogels. Polymer. 2003;44:6217–6222. [Google Scholar]
- 49.Ferradini C, Jay-Gerin J. The effect of pH on water radiolysis: a still open question a minireview. Res Chem Intermed. 2000;26:549–565. [Google Scholar]
- 50.Fechine G, Barros J, Catalani L. Poly (N-vinyl-2-pyrrolidone) hydrogel production by ultraviolet radiation: new methodologies to accelerate crosslinking. Polymer. 2004;45:4705–4709. [Google Scholar]
- 51.Lewis S, Lynch A, Bachas L, Hampson S, Ormsbee L, Bhattacharyya D. Chelate-modified Fenton reaction for the degradation of trichloroethylene in aqueous and two-phase systems. Environ Eng Sci. 2009;26:849–859. doi: 10.1089/ees.2008.0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Katsumata H, Kaneco S, Suzuki T, Ohta K, Yobiko Y. Photo-Fenton degradation of alachlor in the presence of citrate solution. J Photochem Photobiol A: Chem. 2006;180:38–45. [Google Scholar]
- 53.Martens P, Grant M, Nilasaroya A, Whitelock J, Poole-Warren L. Characterisation of redox initiators for producing poly (vinyl alcohol) hydrogels. Macromol Symp. 2008;266:59–62. [Google Scholar]
- 54.Makuuchi K. Critical review of radiation processing of hydrogel and polysaccharide. Radiat Phys Chem. 2010;79:267–271. [Google Scholar]
- 55.Mawad D, Martens P, Odell R, Poole-Warren L. The effect of redox polymerisation on degradation and cell responses to poly (vinyl alcohol) hydrogels. Biomaterials. 2007;28:947–955. doi: 10.1016/j.biomaterials.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 56.Barros J, Fechine G, Alcantara M, Catalani L. Poly (N-vinyl-2-pyrrolidone) hydrogels produced by Fenton reaction. Polymer. 2006;47:8414–8419. [Google Scholar]
- 57.Giulivi C, Traaseth N, Davies K. Tyrosine oxidation products: analysis and biological relevance. Amino Acids. 2003;25:227–232. doi: 10.1007/s00726-003-0013-0. [DOI] [PubMed] [Google Scholar]
- 58.Fancy D, Kodadek T. Chemistry for the analysis of protein–protein interactions: rapid and efficient cross-linking triggered by long wavelength light. Proc Natl Acad Sci USA. 1999;96:6020. doi: 10.1073/pnas.96.11.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Elvin C, Vuocolo T, Brownlee A, Sando L, Huson M, Liyou N, et al. A highly elastic tissue sealant based on photopolymerised gelatin. Biomaterials. 2010;31:8323–8331. doi: 10.1016/j.biomaterials.2010.07.032. [DOI] [PubMed] [Google Scholar]
- 60.Lefebvre F, Droullet F, Savin de Larclasuse AM, Aprahamian M, Midy D, Bordenave L, et al. Repair of experimental arteriotomy in rabbit aorta using new resorbable elastin-fibrin biomaterial. J Biomed Mater Res. 1989;23:1423–1432. doi: 10.1002/jbm.820231205. [DOI] [PubMed] [Google Scholar]
- 61.Capello J, Crissman J, Dorman M, Mikolajczak M, Textor G, Marquet M, et al. Genetic engineering of structural protein polymers. Biotechnol Prog. 1990;6:198–202. doi: 10.1021/bp00003a006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.