Abstract
The sodium-potassium (Na/K) pump is a P-type ATPase that generates Na+ and K+ concentration gradients across the cell membrane. For each ATP molecule, the pump extrudes three Na+ and imports two K+ by alternating between outward- and inward-facing conformations that preferentially bind K+ or Na+, respectively. Remarkably, the selective K+ and Na+ binding sites share several residues, and how the pump is able to achieve the selectivity required for the functional cycle is unclear. Here, free energy perturbation molecular dynamics (FEP/MD) simulations based on the crystal structures of the Na/K pump in a K+-loaded state (E2·Pi) reveal that protonation of the high-field acidic side-chains involved in the binding sites is critical to achieve the proper K+ selectivity. This prediction is tested with electrophysiological experiments showing that the selectivity of the E2P state for K+ over Na+ is affected by extracellular pH.
The Na/K pump is a large heterodimer membrane-bound protein comprising a catalytic α subunit (<105 kD) and a glycosylated β subunit (<55 kD)1. For each hydrolyzed ATP molecule, the Na/K pump transports three Na+ outward and two K+ inward by toggling between two major conformations, E1 and E2, according to the “alternating access” scheme2–4. Under physiological conditions, the outward-facing state E2P preferentially binds two K+ in the presence of a 30-fold greater external concentration of Na+ ions, while the inward-facing state E1 selects three Na+ in the presence of a 10-fold higher internal concentration of K+ ions. Results from several investigations indicate that there is a total of three distinct cation binding locations involved in the functional pumping cycle5. The same residues are implicated in two of the three sites (I and II), which select Na+ in the inward-facing conformation and K+ in the outward-facing conformation, while a third site (III) exclusively binds Na+ in the inward-facing conformation5. The microscopic mechanism underlying the alternating selection of Na+ and K+ by the shared sites I and II is not understood. In the present study, we used computations based on atomic models together with electrophysiological experiments to show that protonation of several acidic residues forming the cation binding sites is critical to achieve the K+ selectivity in the Na/K pump from shark rectal glands.
RESULTS
FEP and pKa calculations
The determination of high resolution crystal structures of the Na/K pump captured in the K+-loaded E2·Pi state6,7 gives a fresh impetus to computational studies aimed at elucidating the transport mechanism at the atomic level. To characterize the selectivity of the K+ binding sites in the E2·Pi state, atomic models were constructed on the basis of the two available crystal structures, 2ZXE7 and 3B8E6, and free energy perturbation molecular dynamics (FEP/MD) simulations were carried out (see Supplementary Table 1). Of particular importance are four titratable residues directly implicated in the two shared binding sites: Glu334, Glu786, Asp811 and Asp815 in the 2ZXE crystal structure (see Supplementary Table 2 for corresponding residue numbers in different pump sequences). The shortest distances between the carboxylate oxygen atoms of Glu786 and Asp811 are 2.8 Å in 2ZXE7 and 2.7 Å in 3B8E6. Such short distances between negatively charged moieties are unlikely, suggesting that they probably share a proton and are involved in a so-called strong di-acid hydrogen bond8. To clarify this issue, the pKa of the four acidic binding site residues was calculated using different computational methods to ascertain their protonation states (see METHODS). The pKa calculations and the results of the FEP/MD simulations on ion selectivity are summarized in Tables 1 and 2.
Table 1.
The pKa values of the binding site residues of the Na/K pump at the E2·P state.
| Na/K Pump | Ca2+ pump | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| PDB 3B8E | PDB 2ZXE | PDB 1WPG | |||||||
| Residues | MCCE | MCCE | PROPKA | Residues | MCCE | MCCE | PROPKA | Residues | MCCE11 |
| εp = 4 | εp = 8 | εp = 4 | εp = 8 | εp = 4 | |||||
| Asp811 | 2.6 | 4.2 | <0 (0.9) | Asp811 | 1.0 | 2.8 | <0 (3.7) | Asp800 | 7.1 |
| Asp815 | 13.1 | 8.3 | 3.6 (6.8) | Asp815 | 3.0 | 3.8 | 4.7 (5.8) | Glu908 | >14 |
| Glu334 | >14 | 12.3 | 8.5 (10.9) | Glu334 | 13.8 | 8.4 | 8.3 (8.3) | Glu309 | 8.4 |
| Glu786 | 12.9 | 9.1 | 7.5 (9.8) | Glu786 | >14 | >14 | 7.9 (10.7) | Glu771 | >14 |
The pKa calculations were based on the Poisson-Boltzmann equation with MCCE37 and the empirical method PROPKA version 2.0 (the number in parenthesis are from the version 3.0 of the program)38. The corresponding values for the Ca2+ pump at the E2·Pi state were taken from Ref. 11. Corresponding residue numbers for different Na/K pumps are given in Supplementary Table 2.
Table 2.
FEP/MD calculations for the cation binding sites I and II
| Simulations | Binding site residues | ΔΔGNa,K (kcal·mol−1) | ||||
|---|---|---|---|---|---|---|
| Site I | Site II | |||||
| A | E334- | E786- | D811- | D815- | −2.5 (−1.7) | −2.7 (−4.5) |
| B | E334 | E786 | D811- | D815 | +1.9 (+3.0) | +4.0 (+1.7) |
| C | E334 | E786 | D811- | D815- | −1.5 | +3.5 |
| D | E334- | E786 | D811- | D815 | −1.2 | −7.7 |
| E | E334 | E786- | D811- | D815 | −2.9 | −3.0 |
| F | E334 | E786 | D811- | D815N | +3.1 | +6.3 |
| G | E334Q | E786 | D811- | D815 | +0.3 | +0.0 |
| H | E334 | E786Q | D811- | D815 | −0.2 | +1.0 |
The “-” symbol denotes the acidic residue that are deprotonated. The ion selectivity in the binding sites ΔΔGna,k is defined as , where the first term is the free energy difference in the binding sites and the second term is the free energy difference in bulk water evaluated by FEP/MD (by definition, ΔΔGNa,K is positive if the site is selective for K+ over Na+, and negative otherwise). Unless specified otherwise, all calculations are based on the 2ZXE crystal structure; results based on the 3B8E are given in parentheses for comparison.
The pKa calculations based on the two crystal structures indicate that Asp811 should be deprotonated and Glu334 and Glu786 should be protonated under physiological conditions. The results are more ambiguous in the case of Asp815, with different predictions based on the 3B8E and 2ZXE X-ray structures. The discrepancy is most likely due to the uncertainties with the empirical pKa calculations based on MCCE and PROPKA (see METHODS). To obtain one additional estimate of the pKa of Asp815, all-atom FEP/MD simulations with explicit solvent molecules were carried out. To sample more effectively the configuration of the polar and charged residues in the binding sites, the pKa shift of Asp815 was computed using alchemical FEP/MD simulations with applied restraining potentials together with umbrella sampling potential of mean force (PMF) calculations (see Supplementary Methods, Explicit solvent pKa calculations). The combined FEP/MD-PMF calculation yields an estimated pKa of about 9.4, indicating that Asp815 should be protonated under physiological conditions (see Supplementary Figure 1). The calculated protonation states for all the key acidic residues in the cation binding sites I and II of the Na/K pump are in broad agreement with previous results obtained for the homologous residues of the sarco(endo)plasmic reticulum Ca2+ pump (SERCA) in the corresponding functional states9–11. The pH-dependence of the C=O infrared bands of Ca2+ free phosphoenzyme E2·Pi state of SERCA probed by Fourier transform infrared (FTIR) difference spectroscopy has provided direct evidence for the protonation of carboxyl groups upon Ca2+ release12,13. Although SERCA transports Ca2+ and H+ while the Na/K pump transports Na+ and K+, the binding sites have high homology5–7 and comparisons can be helpful.
The calculated relative free energies for K+ to Na+, ΔΔGNa,K, reveal that ion selectivity is extremely sensitive to the protonation states of those four acidic side-chains. When all four acidic residues are deprotonated, the binding of Na+ over K+ to sites I and II is considerably more favorable (Table 2, simulation A). In contrast, the computations with protonated Glu334, Glu786 and Asp815 yield binding sites that are very selective for K+ over Na+, with calculated relative free energies that compare quantitatively well with experimental estimates (Table 2, simulation B)14. In interpreting these results, it is important to emphasize that alchemical FEP/MD calculations do not provide information about the individual absolute ion binding affinities, but only about the relative binding free energy of Na+ and K+.
Structural stability of the binding sites
The trends from the FEP/MD simulations are also reflected in the structural stability of the K+ binding sites observed in the MD simulations (see Supplementary Methods, Stability of the binding sites and ion coordination). As shown in Figure 1, when Glu334, Glu786, and Asp815 are protonated, the average root-mean-square deviation (RMSD) of the non-hydrogen atoms is only about 0.5–1.0 Å with respect to the X-ray structure. However, large distortions from the X-ray structure occur when the binding site residues are deprotonated (Supplementary Figures 2–3). This result reinforces the conclusions from FEP/MD that those acidic side-chains must be protonated to yield structurally stable K+-selective binding sites. The K+ ion bound at site I is well coordinated by three carbonyl oxygens, three hydroxyl oxygens and one carboxylic group (two oxygens), while the K+ ion bound at site II is coordinated by six carbonyl oxygens and one carboxylic group (two oxygens). The average K+ coordination numbers for sites I and II are 6.3 and 7.4, respectively, with 1.4 water molecules within the first coordination shell of the K+ ion bound in site I. Consistent with the high resolution X-ray structure 2ZXE, no water molecule is coordinating the ion at site II. In contrast, both sites become more extensively hydrated when all four acidic residues are deprotonated, with 3.4 and 2.4 water molecules coordinating the two K+ ions. Under those conditions, the average coordination number of K+ at sites I and II are 7.6 and 7.0, respectively (Supplementary Figures 4–6). This analysis shows that the protonation states of Glu334, Glu786, and Asp815 play an integral role in ensuring the structural stability of the cation binding sites in E2·Pi.
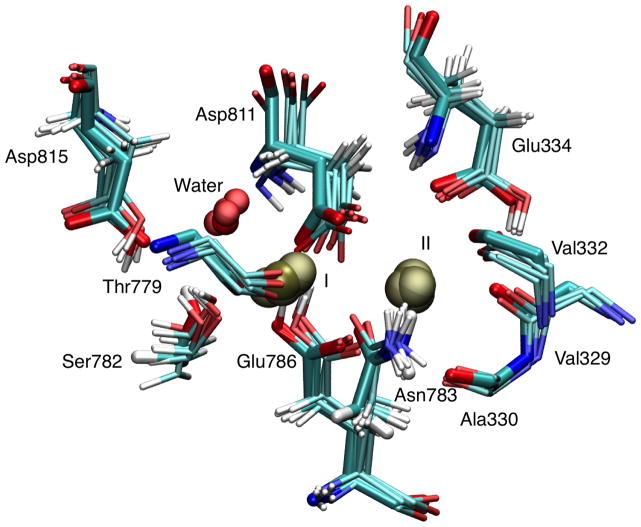
Figure 1.
Superposition of instantaneous configurations from MD simulations of the Na/K pump taken at 5ns, 8ns, 11ns, 14ns, 17ns, and 20ns from Simulation B in Table 1 with the protonation states of the binding site residues assigned according to the theoretical prediction (thin lines) superimposed with the 2ZXE crystal structure (thick lines). The average heavy-atom RMSD are 0.5 Å for Glu334, 0.4 Å for Glu786, 0.8 Å for Asp811, and 0.8 Å for Asp815.
FEP calculations based on simplified reduced models
Ion selectivity can be exquisitely sensitive to multiple microscopic factors, and for this reason, the results from computations must be considered with caution. For example, even slight re-arrangements in side-chain orientations might be sufficient for modulating the selectivity of the different states of the Na/K pump15. This raises the question of whether meaningful conclusions can possibly be drawn about K+ selectivity from computations in view of the limited resolution of currently available X-ray structures and the various approximations involved in MD simulations. Nevertheless, further analysis indicates that the marked effects of protonation noted here are strongly predetermined by the character of the cation binding sites. In particular, similar results are obtained from FEP/MD for both X-ray structures (2ZXE and 3B8E), despite the structural differences (Table 2, simulations A and B). In addition, an analysis of the factors governing ion selectivity in the K+ binding sites based on simplified reduced models16 confirms the robustness of the present results (see Supplementary Methods, Reduced binding site models). If the acidic residues are deprotonated, then both cation binding sites become strongly Na+ selective over K+ (see Supplementary Figure 7). The assertion seems incontrovertible: protonation of the some of the “high-field” carboxylate side chains is absolutely required for K+ selectivity16,17. In the case of site I, the incorrect selectivity from deprotonated side chains cannot be reversed, even it is assumed that the X-ray structure is extremely stiff and virtually rigid. In the case of site II, the incorrect selectivity can be reversed, but only if the structural stiffness of the sites is increased beyond what can be realistically achieved for a protein. This study of reduced models shows that the trends observed in the FEP/MD simulations are not overly sensitive to slight structural changes or the approximations involved in MD simulations because they are inherently governed by the number and chemical nature of the coordinating ligands rather than by the precise molecular geometry of the binding sites. This view is consistent with the recent finding that the organic cation acetamidinium can substitute for K+ and be imported by the pump, suggesting that the shared sites must be able to achieve their high specificity via a surrounding molecular architecture that is able to remain fairly flexible18, and is in broad accord with previous studies of ion selectivity for other membrane proteins19–21.
To summarize the main results from the computations, four lines of evidence indicate that Glu334, Glu786, and Asp815 are likely to be protonated at the E2·Pi state of the Na/K pump. First, this is a direct outcome of pKa calculations (Table 1). Second, MD simulations exhibit small deviations relative to the X-ray structure only when these side-chains are protonated (Figure 1). Third, it is possible for the two cation binding sites to become selective for K+ over Na+ only when these side-chains are protonated (Table 2). Fourth, such a trend is very robust and cannot be reversed by small structural details according to an analysis of reduced models (Supplementary Figure 7). On the basis of these results, one can conclude with confidence that the protonation of acidic residues at the binding sites I and II is an important determinant of the K+ selectivity of the E2·Pi state.
Impact of external pH on K+ selectivity
A prediction from the present computational analyses is that the ion selectivity at the sites I and II will be affected by extracellular pH. Raising the extracellular pH should undermine the ability of the shared sites to properly select K+ over Na+ (n.b., the high pH is not meant to reflect physiological function but is designed to increase the probability of the deprotonated state of the acidic residues while the pump is in an outwardly open state). To experimentally test this prediction, we studied electrophysiologically the function of the Na/K pump expressed in Xenopus oocytes. Physiologically, the outward-facing state that selects between Na+ and K+ is actually E2P, i.e., the state that precedes E2·Pi in the forward pump cycle. The calculations are based on the E2·Pi state captured by the X-ray structure, though both states must select K+ over Na+ with a mechanism that is expected to be similar. In order to enhance the population of the E2P state, the intracellular Na+ concentration was increased as described in METHODS. To evaluate the impact of external pH on selectivity, the apparent affinity for external K+ (K0.5) to activate Na/K pump currents was measured at different external pH in the presence and absence of external Na+.
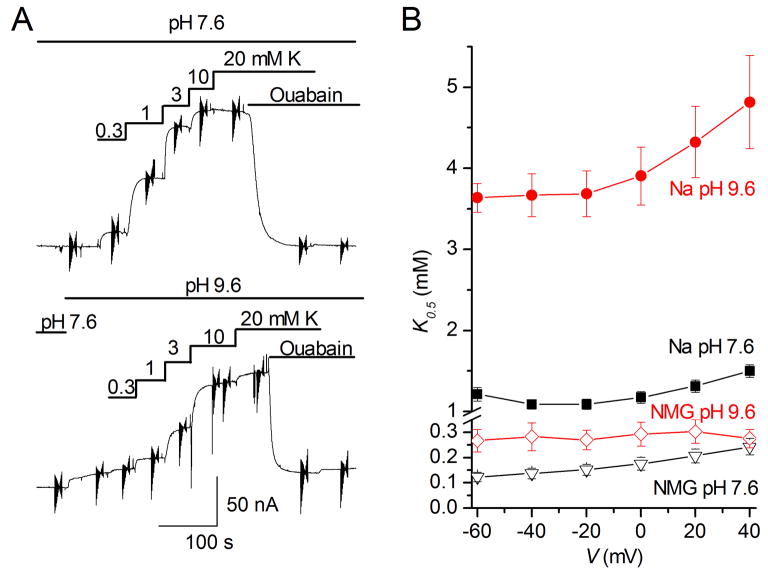
Figure 2a illustrates the titration experiment performed in an oocyte bathed by 125 mM external Na+ at two different external pH. The continuous recordings at a holding potential (Vh)= −50 mV clearly show that a lower external K+ concentration is needed to activate the currents at pH 7.6 (top trace) than at pH 9.6 (bottom). Vertical deflections in the current trace correspond to application of brief, 50-ms square V pulses to measure the current induced by external K+ at different voltages (V). From these measurements, the Hill equation was fitted as a function of K+ concentration for each voltage to extract the apparent affinity K0.5. The results plotted in Figure 2b show that there is a 3–4 fold reduction in apparent affinity for external K+ at positive voltages when the external pH is increased from 7.6 to 9.6 with external Na+ (filled symbols). This corresponds to an effective change in relative K+/Na+ free energy of ~0.8 kcal·mol−1. While the net impact on K0.5 is small, this is expected because the observed change in relative free energy reflects a weighted average over the protonated and deprotonated states (see Supplementary Methods, Apparent selectivity and pH). Accordingly, the loss of selectivity for K+ over Na+ as the pH is increased is expected to be less than the change in ΔΔGNa,K with protonated and unprotonated Asp815 (see Supplementary Figure 8).
Figure 2.
a. K+-induced mediated outward pump currents from a single Na+-loaded oocyte (see METHODS), held at -50 mV in the presence of 125 mM Na+, at two different external pH (7.6, top trace and 9.6, bottom trace). Vertical deflection of the current trace represents 50-ms long voltage pulses (in a compressed time scale) used to obtain the half maximally activating [K+] (from Hill fits to the [K+]-dependence of outward current). Application of 10 mM ouabain inhibits the K+-induced outward Na/K pump current. b. Voltage dependence of the K0.5 for K+ activation of outward pump currents at two different pH, in the presence and absence of Na+ in the external solution. The measurements were performed at pH 7.6 (circles) and pH 9.6 (triangles) in the presence of 125 mM external Na+ (red) or in the presence of 125 mM NMG (black). The voltage range chosen avoids influence of the non-competitive voltage dependent binding of external Na+ to the Na-exclusive site-III, on the apparent affinity for external K+. Data points are mean standard deviations from 5 oocytes where titration at both pH was evaluated.
One possible interpretation of the experimental results shown in Figure 2a and b is that the probability of the protonated state of the carboxylate side chains decreases slightly when the binding sites are exposed to the high extracellular pH in the outwardly open state, giving rise to a small shift in the apparent selectivity. However, the change in apparent K0.5 as a function of external pH could be due to an increased affinity of the sites for K+, or a reduced affinity for Na+, or both. To address this issue, we evaluated the change in apparent affinity K0.5 induced by external pH in the absence of competition from external Na+ (the latter is replaced by NMG in the experiments). Here, the K0.5 becomes smaller (due to the lack of competition with Na+), and is considerably less sensitive to changes in the external pH (Figure 2b, open symbols). These results demonstrate that external K+ competes less well with external Na+ for the sites I and II when the external pH is increased, as predicted by the computations. The experimental observations can be correlated with the pKa calculations (Table 1). The latter indicate that Glu334 and Glu786 are unlikely to become deprotonated, while Asp811 remains deprotonated under most experimentally accessible conditions. This leaves Asp815, which appears to be the side-chain most susceptible to be affected by the extracellular pH. Interestingly, FEP/MD simulations based on a model in which Glu334 and Glu786 are protonated and Asp811 and Asp815 are deprotonated indicate that site II remains selective for K+ and that only site I becomes selective for Na+ (Table 2, simulation C). Thus, only a partial loss of K+ selectivity is predicted to occur at high pH, in accord with experimental observations (Figure 2). Analysis of the ion coordination indicates that the binding site I becomes more water accessible when Asp815 is deprotonated (see Supplementary Figures 5–6). The coordination number increased from 6.3 to 7.0, and the number of water molecules within the first coordination shell goes from 1.4 to 2.6. Additional FEP/MD simulations show that the selectivity of the two sites for K+ over Na+ is lost with deprotonated Glu334 (Table 2, simulation D) as well as with deprotonated Glu786 (Table 2, simulation E).
Comparison with previous site-directed mutational studies
The present results are consistent with previous observations describing the impact of pH on the Na/K pump activity22–26. Specifically, Skou and Esmann23 noted that “protons increase the apparent affinity for K+” and “decrease in protons increases the apparent affinity for Na+”, consistent with the present results. However, the observed sensitivity to pH has traditionally been interpreted by assuming that protons affect the relative stability of the E1/E2 conformational states or the concentration of ions at the access channel entrance26. According to this view, the apparent ion selectivity of the pump is indirectly affected by pH: increasing the proton concentration favors the conformation that binds K+, and lowering the proton concentration favors the conformation that binds Na+. Although such an explanation cannot be ruled out on the sole basis of currently available experimental observations, the critical importance of the ionization state of the acidic side-chain directly involved in ion coordination revealed by the FEP/MD simulations sheds a new light on these previous observations. Furthermore, additional pKa calculations indicate that the pH sensitivity is unlikely to arise from some arbitrary extracellular-facing titratable residue (see Supplementary Table 3).
The conclusion that in the E2·Pi state, Glu334, Glu786 and Asp815 are protonated and neutral while Asp811 is deprotonated and negatively charged suggests that the pump should display normal K+/Na+ selectivity with the charge-conserving mutants E334Q, E786Q and D815N, but not with D811N. Previous mutational studies are broadly consistent with these observations, although reported experiments were not always optimized to address the question of ion selectivity in energetic terms. The mutant E334Q is able to transport K+ (ref. 27). Similarly, the mutant E786Q behaves like the wild-type pump28. These results are consistent with the computational modeling predicting that these two residues should be protonated and neutral. Similarly, based on the I–V curves reported28, the mutant E789Q appears to behave like the wild-type pump in terms of the external K+/Na+ selectivity of the shared sites. In sharpt contrast, electrogenic cation transport for the D811N mutant is severely impaired and produces no pump currents (c.f. Fig. 3 in ref 28). This position appears to be particularly sensitive because even the charge-conserving mutant D811E yields a decreased Na+ affinity at the extracellular binding sites28,29. This is qualitatively consistent with the computational analysis predicting that this residue must be deprotonated and negatively charged. The behavior of the D815N mutant is more complicated. It shows a decreased apparent K+ affinity, changes from 0.79 mM to 2.68 mM28. This may appear to be inconsistent with the computational modeling predicting that this residue should be neutral and protonated. However, mutations at this position display complex functional effects. For example, even the conservative mutant D815E displays a decreased apparent K+ affinity from 0.79 mM to 3.20 mM, larger than for D815N. Our own attempts to measure the pH dependence of K0.5 in oocytes for the D815N mutant were unsuccessful, consistent with the reported reduced K+-induced current in this mutant28.
Additional FEP/MD simulations were carried out to estimate the changes in ΔΔGNa,K caused by mutations (see Supplementary Figure 9). Although the calculations with mutated residues are more uncertain than those based on the X-ray structure of the wild-type protein, they indicate that the mutation D815N (Table 2, simulation F) maintains K+ selectivity, and that the mutations E334Q (Table 2, simulation G) and E786Q (Table 2, simulation H) affect K+ selectivity, but to a much smaller extent than deprotonation of the side-chains. While the results appear to be in qualitative accord with experiments, they suggest that even seemingly simple charge-conserving side-chain substitutions can have complex effects and must be interpreted with caution (see Supplementary Methods, Simulations of mutants). In particular, the experimentally observed function of site-directed mutants might be altered by a host of factors, including the absolute ion binding affinities or the relative stability of the E1 and E2 state, which are not probed by the FEP/MD calculations.
DISCUSSION
The combined results from computations and experiments show that the protonation states of key acidic residues impact both structurally and energetically on the function of the Na/K pump. According to our computations, protonation of Glu334, Glu786, and Asp815 is absolutely required to both ensure the structural integrity of the binding sites I and II of the Na/K pump in the K+-loaded E2·Pi state, and to robustly establish the selectivity of those sites for K+ over Na+. Although the present study was exclusively concerned with the K+ selectivity of the E2·Pi and E2P states, these findings beg the question of whether the same factors play a corresponding role in the Na+-selectivity of the E1 state. In this regards, it is worth noting some interesting analogies between the transport cycle of SERCA and of the Na/K pump concerning the protonation states of the acidic residues forming the binding sites. For instance, in the case of the E2·Pi state of SERCA, which is functionally equivalent to the K+-loaded E2·Pi state of the Na/K pump12,13,9,30, the corresponding acidic residues of the binding sites also become protonated. On the other hand, in the case of the Ca2+-loaded E1 state of SERCA, which is functionally equivalent to the Na+-loaded E1 state of the Na/K pump, the ion-coordinating side-chains are thought to be deprotonated31. Consistent with the suggestion that a deprotonated Glu786 might be required to support Na+ selectivity in the E1 state of the Na/K pump, it is observed that the E786Q mutant displays an apparent affinity for K+ that is similar to wild-type, but a 2.6-fold decrease in affinity for Na+ (ref. 32).
Of particular interest, a study by Poulsen et al33 recently concluded that two cytoplasmic protons enter the vacant site III to protonate Asp933 and Glu961 while the Na/K pump is in the E2 state, and return to the cytoplasm via a transient aqueous pathway when the K+ ions are released internally in the state E1 (PROPKA predicts that the pKa of Asp933 and Glu961 in the K+-loaded E2·Pi state are 8.9 and 10.3, respectively). According to the present results, three additional acidic residues, Glu334, Glu786, and Asp815, ought also to be protonated in order to support the K+ selectivity of the shared sites I and II in the E2P state. However, the observation that only Na+ and K+ ions are actively transported during the normal cycle of the pump imposes strict constraints on the sidedness of the source and sink for the protons implicated in any proposed mechanism. One possibility is that all these protons come from and return to the cytoplasmic side, in close correspondence with the mechanism described by Poulsen et al33. The deprotonated (high-field) carboxylates coordinating the cations in the binding sites I, II, and III would then guarantee a robust Na+ selectivity16,17. But alternative mechanistic scenarios are conceivable. For example, some of the acidic side chains coordinating the ions might remain protonated in all states if only a subset of protons undergo such transient movements in and out of the cytoplasm during the pump cycle. The presence of a few protonated carboxylates would not necessarily undermine the ability to establish Na+ selectivity of the binding sites of the state E1, as indicated by free energy calculations based on model systems17. These considerations suggest the hypothesis that the Na/K pump might control the chemistry of ion coordination by modulating the protonation states of key residues forming the cation binding sites to achieve the K+ and Na+ selectivities required at various stages of the functional cycle. However, without high resolution X-ray structures of the Na+-loaded E1 state, it is difficult to definitively elucidate the microscopic basis of selectivity of those ion binding sites at this time. Further work will be required to resolve these issues.
METHODS
Computations
All the simulations were carried out using the program CHARMM34 with the PARAM27 force field35 (see Supplementary Table 1). The functional state considered in all the computations, E2·Pi, implies a bound but not covalently attached phosphate to the pump. For the FEP/MD computations of ion selectivity and of the pKa of Asp815, the General Solvent Boundary Potential (GSBP)36 setup was adopted to simulate a spherical region of 18 Å radius centered on the cation binding sites of the pump with explicit water molecules. By definition, the K+/Na+ selectivity was calculated as from the FEP/MD simulations; i.e., a positive implies that the site is selective for K+ over Na+. The GSBP spherical inner region of 18 Å was centered at the mid-point of the position of the two K+ in the crystal structures. There is a total of about 60 water molecules in the inner region after equilibration. The long-range interactions were taken into account in the form of the solvent-shielded static field and the solvent-induced reaction field via 400 harmonic functions in the multipole expansion. The solvent-shielded static field and the reaction field matrix were calculated once with finite-difference Poisson-Boltzmann (PB), assuming dielectric constants of 1.0 inside the protein, immersed in a solvent with dielectric 78.5. The atomic Born radii for protein atoms used to setup the dielectric boundaries in the PB calculations were determined by FEP/MD simulations with explicit solvent39. The water molecules within the inner region are confined by a non-polar cavity potential to prevent entry into the surrounding dielectric continuum. Coordination numbers were evaluated on the basis of a proximity criterion, with an ion-ligand distance of 3.5 Å. All computational details and parameters are given in Supplementary Methods, Atomic systems and simulations. The pKa values of the ion binding site residues were calculated with Multi-Conformation Continuum Electrostatics (MCCE)37 and PROPKA (version 2.0 and 3.0)38 based on the two crystal structures6,7. The K+ ions were present in all the pKa computations. MCCE samples residue protonation and side-chain conformation as a function of pH and conformational flexibility is modeled by multiple side-chain conformations, which are constructed systematically by rotating rotatable side-chain bonds37. Conformational and protonation states of the relevant residues are sampled in a Monte Carlo sampling, assuming a Boltzmann distribution of states that yields the occupancy of each residue state as a function of pH. To investigate the effect of the protein dielectric constant, two different (4.0 and 8.0) were used. In contrast, PROPKA adopts a very fast empirical energy function which takes into account the desolvation effects and intra-protein interactions38. It has been shown that PROPKA and MCCE are more accurate for surface and buried residues, respectively40. The pKa shift of Asp815 was also calculated with explicit solvent using a combined FEP/MD umbrella sampling potential of mean force (PMF) method. For additional theoretical development and technical details, see Explicit solvent pKa calculations in Supplementary Methods. While the effects of solvation were empirically calibrated against experimental data for soluble proteins in PROPKA and MCCE, computations based on FEP/MD simulations including explicit water molecules can account more readily for the inherent flexibility of complex cation binding sites.
Electrophysiology experiments
Electrophysiological experiments to examine the extracellular pH effects on the function of the Na/K pump expressed in Xenopus oocytes were carried out according to standard protocols established previously18. Oocytes were enzymatically isolated as described41, injected with an equimolar mixture of the ouabain resistant Xenopus (Q120RN131D mutant, 25 ng) and Xenopus β3 cRNAs, and kept at 16ºC in SOS solution containing (in mM): 100 NaCl, 2 KCl, 1.8 CaCl2, 1 MgCl2 and 5 HEPES, supplemented with horse serum (SIGMA), and mixture of antimicotic and antibiotic (anti-anti, GIBCO) for 2–4 days until use. The double-mutant α1-Q120R-N131D was used due to its lower ouabain sensitivity (IC50<100μM) that allows reversible inhibition of exogenous pumps during the experiment and distinction from ouabain-sensitive endogenous pumps which remained continuously inhibited after preincubation with ouabain. Oocytes were Na-loaded by a 1 hr incubation in a solution containing (in mM): 100 HEPES, 90 NaOH 20 TEAOH 0.2 EGTA. (pH 7.6) and then transferred to a K+-free OR2 solution containing 10–20 mM ouabain and (in mM) 82.5 NaCl, 2 KCl, 1.8 CaCl2, 1 MgCl2 and 5 HEPES and, until recording. External solutions used during recording were composed of (in mM) 125 NaOH (or N-methyl D-glucamine, NMG), 10 HEPES, 10 MES, 10 TAPS, 10 CAPS, 5 Ba(OH)2, 1 Mg(OH)2, 0.5 Ca(OH)2 (pH 7.6 with methanesulfonic acid, MS). To measure K+-activated currents, K+ (up to 20 mM) was added to the external solutions from a 3 M K+-MS stock. Two-electrode voltage-clamp (TEVC) was made using an OC-725C amplifier (Warner Instruments, Hamden, CT), a Digidata 1440 A/D board, a Minidigi 1A and pClamp 10 software (Molecular Devices, Sunnyvale, CA). Signals were filtered at 2 kHz and digitized at 10 kHz. Resistance of current and voltage microelectrodes were 0.5–1 MΩ and 1–2 MΩ, respectively (filled with 3M KCl).
Supplementary Material
Acknowledgments
This work was supported by the NIH grant GM062342 (H.Y. and B.R.) and by the AHA grant BGIA2140172 (P.A.).
Footnotes
Competing Interests
The authors declare that they have no competing financial interests.
Author contributions
H.Y. and B.R. designed the computations and H.Y. carried out the computations. P.A. and B.R. set the overall aim of the experiments. The electrophysiological experiments were designed by P.A. and carried out by I.R. and P.A.. H.Y., P.A. and B.R. wrote the manuscript.
References
- 1.Jorgensen PL, Hakansson K+O, Karlish SJD. Structure and mechanism of Na+,K+-ATPase: Functional sites and their interactions. Annu Rev Physiol. 2003;65:817–849. doi: 10.1146/annurev.physiol.65.092101.142558. [DOI] [PubMed] [Google Scholar]
- 2.Post RL, Sen AK, Rosenthal AS. A phosphorylated intermediate in adenosine triphosphate-dependent sodium and potassium transport across K+idney membranes. J Biol Chem. 1965;240:1437–1445. [PubMed] [Google Scholar]
- 3.Jardetzky O. Simple allosteric model for membrane pumps. Nature. 1966;211:969–970. doi: 10.1038/211969a0. [DOI] [PubMed] [Google Scholar]
- 4.Albers RW. Biochemical aspects of active transport. Annu Rev Biochem. 1967;36:727–756. doi: 10.1146/annurev.bi.36.070167.003455. [DOI] [PubMed] [Google Scholar]
- 5.Ogawa H, Toyoshima C. Homology modeling of the cation binding sites of Na+/K+-ATPase. Proc Natl Acad Sci U S A. 2002;99:15977–15982. doi: 10.1073/pnas.202622299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morth JP, et al. Crystal structure of the sodium-potassium pump. Nature. 2007;450:1043–U6. doi: 10.1038/nature06419. [DOI] [PubMed] [Google Scholar]
- 7.Shinoda T, Ogawa H, Cornelius F, Toyoshima C. Crystal structure of the sodium-potassium pump at 2.4 Å resolution. Nature. 2009;459:446–U167. doi: 10.1038/nature07939. [DOI] [PubMed] [Google Scholar]
- 8.Perrin CL, Nielson JB. “Strong” hydrogen bonds in chemistry and biology. Annu Rev Phys Chem. 1997;48:511–544. doi: 10.1146/annurev.physchem.48.1.511. [DOI] [PubMed] [Google Scholar]
- 9.Obara K, et al. Structural role of countertransport revealed in Ca pump crystal structure in the absence of Ca. Proc Natl Acad Sci U S A. 2005;102:14489–14496. doi: 10.1073/pnas.0506222102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sugita Y, Miyashita N, Ikeguchi M, Kidera A, Toyoshima C. Protonation of the acidic residues in the transmembrane cation-binding sites of the Ca pump. J Am Chem Soc. 2005;127:6150–6151. doi: 10.1021/ja0427505. [DOI] [PubMed] [Google Scholar]
- 11.Hauser K, Barth A. Side-chain protonation and mobility in the sarcoplasmic reticulum Ca-ATPase: Implications for proton countertransport and Ca release. Biophys J. 2007;93:3259–3270. doi: 10.1529/biophysj.107.109363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andersson J, Hauser K, Karjalainen EL, Barth A. Protonation and hydrogen bonding of Ca site residues in the E2P phosphoenzyme intermediate of sarcoplasmic reticulum Ca-ATPase studied by a combination of infrared spectroscopy and electrostatic calculations. Biophys J. 2008;94:600–611. doi: 10.1529/biophysj.107.114033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weidemuller C, Hauser K. Ion transport and energy transduction of P-type ATPases: Implications from electrostatic calculations. Biochim Biophys Acta-Bioenerg. 2009;1787:721–729. doi: 10.1016/j.bbabio.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 14.Lauger P. Electrogenic ion pumps. 1 Sinauer Associates; Sunderland, MA: 1991. [Google Scholar]
- 15.Bublitz M, Poulsen H, Morth J, Nissen P. In and out of the cation pumps: P-Type ATPase structure revisited. Curr Opin Struct Biol. 2010;20:431–439. doi: 10.1016/j.sbi.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 16.Yu H, Noskov SY, Roux B. Two mechanisms of ion selectivity in protein binding sites. Proc Natl Acad Sci USA. 2010;107:20329–20334. doi: 10.1073/pnas.1007150107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roux B. Exploring the ion selectivity properties of a large number of simplified binding site models. Biophys J. 2010;98:2877–2885. doi: 10.1016/j.bpj.2010.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ratheal IM, et al. Selectivity of externally facing ion-binding sites in the Na+/K+ pump to alkali metals and organic cations. Proc Natl Acad Sci USA. 2010;107:18718–18723. doi: 10.1073/pnas.1004214107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noskov SY, Bernèche S, Roux B. Control of ion selectivity in potassium channels by electrostatic and dynamic properties of carbonyl ligands. Nature. 2004;431:830–834. doi: 10.1038/nature02943. [DOI] [PubMed] [Google Scholar]
- 20.Noskov SY, Roux B. Importance of hydration and dynamics on the selectivity of the K+csA and NaK channels. J Gen Physiol. 2007;129:135–143. doi: 10.1085/jgp.200609633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noskov SY, Roux B. Control of ion selectivity in LeuT: Two Na+ binding sites with two different mechanisms. J Mol Biol. 2008;377:804–818. doi: 10.1016/j.jmb.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Skou JC. Effects of ATP on the intermediary steps of the reaction of the Na+/K+-ATPase. IV Effect of ATP on K0.5 for Na+ and on hydrolysis at different pH and temperature. Biochim Biophys Acta. 1979;567:421–435. doi: 10.1016/0005-2744(79)90128-1. [DOI] [PubMed] [Google Scholar]
- 23.Skou JC, Esmann M. Effects of ATP and protons on the Na:K+ selectivity of the Na+/K+-ATPase studied by ligand effects on intrinsic and extrinsic fluorescence. Biochim Biophys Acta. 1980;601:386–402. doi: 10.1016/0005-2736(80)90543-x. [DOI] [PubMed] [Google Scholar]
- 24.Breitwieser GE, Altamirano AA, Russell JM. Effects of pH changes on sodium pump fluxes in squid giant axon. Am J Physiol. 1987;253:C547–C554. doi: 10.1152/ajpcell.1987.253.4.C547. [DOI] [PubMed] [Google Scholar]
- 25.Salonikidis PS, Kirichenko SN, Tatjanenko LV, Schwarz W, Vasilets LA. Extracellular pH modulates K+inetics of the Na+/K+-ATPase. Biochim Biophys Acta. 2000;1509:496–504. doi: 10.1016/s0005-2736(00)00356-4. [DOI] [PubMed] [Google Scholar]
- 26.Milanick MA, Arnett KL. Extracellular protons regulate the extracellular cation selectivity of the sodium pump. J Gen Physiol. 2002;120:497–508. doi: 10.1085/jgp.20028573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuntzweiler TA, Wallick ET, Johnson CL, Lingrel JB. Glutamic acid 327 in the sheep alpha 1 isoform of Na(+),K(+)-ATPase stabilizes a K+(+)-induced conformational change. J Biol Chem. 1995;270:2993–3000. doi: 10.1074/jbc.270.7.2993. [DOI] [PubMed] [Google Scholar]
- 28.Koenderink JB, et al. Electrophysiological analysis of the mutated Na+,K+-ATPase cation binding pocket. J Biol Chem. 2003;278:51213–51222. doi: 10.1074/jbc.M306384200. [DOI] [PubMed] [Google Scholar]
- 29.Koenderink JB, Swarts HG, Hermsen HP, Willems PH, De Pont JJ. Mutation of aspartate 804 of Na(+),K(+)-ATPase modifies the cation binding pocket and thereby generates a high Na(+)-ATPase activity. Biochemistry. 2000;39:9959–9966. doi: 10.1021/bi0001168. [DOI] [PubMed] [Google Scholar]
- 30.Toyoshima C, Nomura H, Tsuda T. Lumenal gating mechanism revealed in calcium pump crystal structures with phosphate analogues. Nature. 2004;432:361–368. doi: 10.1038/nature02981. [DOI] [PubMed] [Google Scholar]
- 31.Toyoshima C. Structural aspects of ion pumping by Ca2+-ATPase of sarcoplasmic reticulum. Arch Biochem Biophys. 2008;476:3–11. doi: 10.1016/j.abb.2008.04.017. [DOI] [PubMed] [Google Scholar]
- 32.Feng J, Lingrel JB. Functional consequences of substitutions of the carboxyl residue glutamate 779 of the Na+,K+-ATPase. Cell Mol Biol Res. 1995;41:29–37. [PubMed] [Google Scholar]
- 33.Poulsen H, et al. Neurological disease mutations compromise a C-terminal ion pathway in the Na+/K+-ATPase. Nature. 2010;467:99–102. doi: 10.1038/nature09309. [DOI] [PubMed] [Google Scholar]
- 34.Brooks BR, et al. CHARMM: The biomolecular simulation program. J Comp Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.MacKerell AD, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 36.Im W, Berneche S, Roux B. Generalized solvent boundary potential for computer simulations. J Chem Phys. 2001;114:2924–2937. [Google Scholar]
- 37.Song YF, Mao JJ, Gunner MR. MCCE2: Improving protein pK(a) calculations with extensive side chain rotamer sampling. J Comput Chem. 2009;30:2231–2247. doi: 10.1002/jcc.21222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Robertson AD, Jensen JH. Very fast empirical prediction and rationalization of protein pKa values. Proteins. 2005;61:704–721. doi: 10.1002/prot.20660. [DOI] [PubMed] [Google Scholar]
- 39.Nina M, Beglov D, Roux B. Atomic radii for continuum electrostatics calculations based on molecular dynamics free energy simulations. J Phys Chem B. 1997;101:5239–5248. [Google Scholar]
- 40.Davies MN, Toseland CP, Moss DS, Flower DR. Benchmarking pKa prediction. BMC Biochemistry. 2006;7:18. doi: 10.1186/1471-2091-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yaragatupalli S, Olivera JF, Gatto C, Artigas P. Altered Na+ transport after an intracellular alpha-subunit deletion reveals strict external sequential release of Na+ from the Na+/K+ pump. Proc Natl Acad Sci USA. 2009;106:15507–15512. doi: 10.1073/pnas.0903752106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.