Abstract
The past decade has seen substantial advances in cardiovascular pharmacogenomics. Genetic determinants of response to clopidogrel and warfarin have been defined, resulting in changes to the product labels for these drugs that suggest the use of genetic information as a guide for therapy. Genetic tests are available, as are guidelines for incorporation of genetic information into patient-care decisions. These guidelines and the literature supporting them are reviewed herein. Significant advances have also been made in the pharmacogenomics of statin-induced myopathy and the response to β-blockers in heart failure, although the clinical applications of these findings are less clear. Other areas hold promise, including the pharmacogenomics of antihypertensive drugs, aspirin, and drug-induced long-QT syndrome (diLQTS). The potential value of pharmacogenomics in the discovery and development of new drugs is also described. In summary, pharmacogenomics has current applications in the management of cardiovascular disease, with clinically relevant data continuing to mount.
Uncovering the causes of interpatient variability in drug response, and then using that information for the benefit of patients, is at the heart of clinical pharmacology. Although the term “pharmacogenetics” was coined in the 1950s, only in the past decade has an explosion occurred in research focused on discovering the genetic basis for variations in drug efficacy, toxicity, and dose requirements. Pharmacogenomic research on cardiovascular drugs has been among the more active areas of investigation within this field. The past decade has seen substantial advances in our understanding of the genetic determinants of response to two commonly used cardiovascular drugs—clopidogrel and warfarin—such that the data on these drugs can now be used in the clinical setting. We highlight the data surrounding these examples along with other areas of active research in cardiovascular pharmacogenomics that—although they have not yet reached the stage of translation into practice—hold promise. The data arising from research on cardiovascular pharmacogenomics have not only led to potential clinical applications but have advanced our understanding of the metabolism and/or pharmacological mechanisms of a number of drugs. We also highlight the potential for research on pharmacogenomics to influence discovery and development of new drugs.
CARDIOVASCULAR PHARMACOGENOMICS AND LABELING
Despite the growing appreciation of pharmacogenomic markers influencing the response to cardiovascular drugs, only limited examples have labeling approved by the U.S. Food and Drug Administration (FDA). According to the FDA’s cataloging of labels, pharmacogenomic biomarkers are included in eight cardiovascular drug or drug-combination labels (atorvastatin, carvedilol, clopidogrel, isosorbide/hydralazine, metoprolol, propafenone, propranolol, and warfarin (Table 1). The types of information included in labels are variable and are of potential value in informing clinical decisions. For example, the information ranges from the effect of genetically influenced metabolism on drug exposure (e.g., the labels for hydralazine, carvedilol, and metoprolol) to information on disease (but not drug) genetics (e.g., atorvastatin) to more clinically practical dosing information (e.g., warfarin and clopidogrel). Notably, the relative importance of the information seems to be reflected in the location on the label. For example, the potential for therapeutic failure of clopidogrel in cytochrome P450 (CYP)-2C19 (CYP2C19) poor metabolizers is shown in a boxed warning (among other locations), whereas for other drugs the information is in the clinical pharmacology section.
Table 1.
Cardiovascular drugs with pharmacogenomic labeling (as of may 2011)
| Drug | Gene/biomarker | Label sections |
|---|---|---|
| Atorvastatin | LDLR | Warnings and precautions; clinical pharmacology; clinical studies |
| Carvedilol | CYP2D6 | Drug interactions; clinical pharmacology |
| Clopidogrel | CYP2C19 | Boxed warning; dosage and administration; warnings and precautions; drug interactions; clinical pharmacology |
| Isosorbide dinitrate/hydralazine | NAT1; NAT2 | Clinical pharmacology |
| Metoprolol | CYP2D6 | Precautions; clinical pharmacology |
| Propafenone | CYP2D6 | Clinical pharmacology |
| Propranolol | CYP2D6 | Precautions; drug interactions; clinical pharmacology |
| Warfarin | CYP2C9; VKORC1 | Dosage and administration; precautions; clinical pharmacology |
Among the cardiovascular drugs with pharmacogenomic data included in product labeling, warfarin and clopidogrel contain the strongest labeling. The warfarin label was updated twice (in 2007 and 2010) to reflect the growing body of knowledge regarding the influence of variations in the CYP2C9 and vitamin-K-epoxide reductase complex-1 (VKORC1) genes on dose requirements. The first update did not contain actionable information, probably a function of the limited data on the clinical utility tests. The second update provided a dosing table, with expected dose requirements broken down by CYP2C9 and VKORC1 genotypes. The clopidogrel label has been updated three times since 2009 to reflect knowledge gained regarding the influence of CYP2C19 genotype on treatment outcomes. The most recent clopidogrel label update, in March 2010, includes a boxed warning specifically advising the avoidance of clopidogrel in patients with known genetic polymorphisms of CYP2C19 and states that physicians should “consider alternative treatment or treatment strategies in patients identified as CYP2C19 poor metabolizers.” The label goes on to state that tests are available to determine a patient’s CYP2C19 genotype, which can be used to aid in treatment decisions. Commercially available genetic tests are available for these two examples, in addition to guidelines from the Clinical Pharmacogenetics Implementation Consortium (CPIC);1,2 there is general agreement that they are the most actionable among the cardiovascular examples.
CLINICAL APPLICATION OF PHARMACOGENOMIC KNOWLEDGE: CLOPIDOGREL AND WARFARIN
Clopidogrel pharmacogenomics
Clopidogrel inhibits platelet function, thereby preventing recurrent cardiovascular events in patients with acute coronary syndromes (ACSs) or patients undergoing percutaneous coronary intervention. Enzymatic modification of clopidogrel, a thienopyridine prodrug, is required to produce its bioactive thiol metabolite (SR 26334), which irreversibly binds to the platelet P2Y12 receptor, resulting in inhibition of adenosine diphosphate–stimulated platelet aggregation for the duration of the platelet’s life span (~10 days). Wide interindividual variation of the clopidogrel response is well recognized, and recent investigations have shown that this response is highly heritable.3 Single-nucleotide polymorphisms (SNPs) in several genes critical for clopidogrel metabolism, transport, and signaling affect its pharmacokinetics, and its pharmacodynamic actions have been investigated in the context of the genes for many enzymes, including those coding for several of the CYP enzymes (e.g., CYP1A2, CYP2C19, CYP3A4, and CYP3A5), P-glycoprotein (ABCB1), paraoxonase 1 (PON1), and P2Y12 (Table 2). Although inconsistent findings have been reported regarding the effects of polymorphisms in some of these genes on clopidogrel response, there is compelling evidence that variants of CYP2C19 significantly affect clopidogrel efficacy and recurrence rates of cardiovascular events.4,5
Table 2.
Level of evidence linking candidate-gene genotype to clopidogrel response
| Gene | Chromosome | Genetic variant | Effect on protein structure | Effect on protein function or expression | Level of evidencea |
|---|---|---|---|---|---|
| CYP2C19 | 10 | rs4244285 (*2) | Splicing defect | Decreased | High |
| CYP2C19 | 10 | rs4986893 (*3) | W212X | Decreased | Moderate |
| CYP2C19 | 10 | rs12248560 (*17) | None | Increased | Moderate |
| CYP1A2 | 15 | rs762551 (*1F) | None | Increased | Weak |
| CYP2C9 | 10 | rs1057910 (*3) | I359L | Decreased | Weak |
| CYP3A4 | 7 | rs2242480 (*1G) | None | Increased | Weak |
| CYP3A5 | 7 | rs776746 (*3) | Splicing defect | Decreased | Weak |
| ABCB1 | 7 | rs1045642 (C3435T) | I1145I | Decreased | Moderate |
| P2Y12 | 3 | H2 haplotype | None | Gain of function | Weak |
| PON1 | 7 | rs662 | Q192R | Increased | Weak |
Additional polymorphisms exist in each candidate gene. Selected genes were investigated in at least three independent publications.
Level of evidence is based on previously published criteria. Definitions: High, consistent evidence from several well-designed, well-conducted studies; Moderate, evidence is sufficient to determine effects, but the strength of evidence is limited by the number, quality, or consistency of the individual studies, generalizability, or indirect nature of the evidence; Weak, evidence is inconsistent and/or insufficient to assess the effects on health outcomes because of limited number or power of studies, important flaws in their design or conduct, gaps in the chain of evidence, or lack of information.
From ref. 1.
CYP2C19 encodes the protein CYP2C19, the hepatic enzyme responsible for the metabolism of many drugs, including clopidogrel, benzodiazepines, and some proton-pump inhibitors. Multiple polymorphisms in CYP2C19 have been identified that result in both decreased and increased functions.6 The most common of these variants include the loss-of-function CYP2C19*2 variant (c.681G>A; rs4244285) and the gain-of-function CYP2C19*17 variant (c.−806C>T; rs12248560). In populations of European, African, and East Asian ancestry, the frequency of the loss-of-function CYP2C19*2 allele is relatively high (15%, 15%, and 29%, respectively), as is the frequency of the gain-of-function CYP2C19*17 allele (21%, 16%, and 3%, respectively). In addition, the frequency of the loss-of-function CYP2C19*3 variant (c.636G>A; rs4986893) is considerable in Asian populations, ranging from 2 to 9%. Other functional variants in CYP2C19 are rare, generally having frequencies less than 1%.
Several recent studies have shown that polymorphisms in CYP2C19 alter the concentration of the active metabolite of clopidogrel, residual platelet reactivity, and the rate of cardiovascular events in patients with ACSs or patients undergoing percutaneous coronary intervention. For example, in a genome-wide association study (GWAS) in healthy subjects, CYP2C19*2 accounted for ~12% of the total variation in residual adenosine diphosphate–stimulated platelet aggregation after administration of standard-dose clopidogrel for 1 week.3 Studies with candidate genes have shown that CYP2C19*2 carriers have lower levels of the active metabolite of clopidogrel as compared with *1/*1 homozygotes.7 Similarly, a growing number of studies have shown that clopidogrel-treated ACS patients carrying the CYP2C19*2 allele have an increased risk of experiencing adverse recurrent cardiovascular outcomes.4,5 Mega and colleagues evaluated data from 9,685 patients from nine independent studies and reported that patients carrying one or two copies of the CYP2C19*2 allele had a significantly increased risk of experiencing a composite end point consisting of cardiovascular death, myocardial infarction, or stroke (hazard ratio (HR) = 1.55, 95% confidence interval (CI): 1.11–2.17, P = 0.01; and HR = 1.76, 95% CI: 1.24–2.50, P = 0.002, for CYP2C19*2 heterozygotes and homozygotes, respectively).4 Furthermore, in 5,894 patients evaluated for stent thrombosis, carriers of the CYP2C19*2 reduced-function allele had a significantly increased risk of stent thrombosis (HR = 2.67, 95% CI: 1.69–4.22, P < 0.0001; and HR = 3.97, 95% CI: 1.75–9.02, P = 0.001, for CYP2C19*2 heterozygotes and homozygotes, respectively). Overall, these data convincingly show that CYP2C19*2 affects the concentration of the active metabolite of clopidogrel, platelet reactivity, and risk of cardiovascular events in a gene- and dose-dependent manner in ACS and percutaneous coronary intervention patients treated with clopidogrel. Although fewer studies have investigated the less common loss-of-function CYP2C19*3 variant, it appears that this variant confers an increased risk, similar to CYP2C19*2.8
Despite the overall consistency of the association between CYP2C19*2 genotype and decreased clopidogrel response, some large-scale well-conducted studies failed to show an association. These studies included coronary artery disease patients with lower risk for cardiovascular events or other indications for clopidogrel, e.g., stroke, atrial fibrillation, or peripheral vascular disease.9,10 Therefore, the clinical utility of CYP2C19 genotyping may be limited to patients with coronary artery disease who are at high risk for recurrent events.11
Similar to the investigations of CYP2C19*2, several studies have evaluated the effect of the gain-of-function CYP2C19*17 variant on adenosine diphosphate–stimulated platelet inhibition and cardiovascular outcomes in response to clopidogrel therapy, albeit with less consistent results. Although some reports indicate that clopidogrel-treated carriers of CYP2C19*17 have less residual platelet aggregation as compared to noncarriers—i.e., a greater response12,13—others have shown no such effect.14 Similarly, inconsistent results have been reported regarding the impact of CYP2C19*17 on recurrent cardiovascular events. Some studies have shown no association between CYP2C19*17 and stent thrombosis15 or composite cardiovascular end points,3 although the former study did observe that CYP2C19*17 significantly increased bleeding risk in a gene- and dose-dependent manner (odds ratio (OR) = 1.80, 95% CI: 1.03–3.14, for CYP2C19*17 heterozygotes vs. noncarriers; and OR = 3.27, 95% CI: 1.33–8.10, for CYP2C19*17 homozygotes vs. noncarriers). Other studies have found a significant impact of CYP2C19*17 genotype on the rate of cardiovascular events. For example, in 928 high-risk patients with acute myocardial infarction, CYP2C19*17 carriers had a 37% reduction in clinically driven target-lesion revascularization and a 22% reduction in major adverse cardiovascular events as compared to noncarriers.16 More recently, in a large-scale study of patients with ACS, Paré and colleagues showed that CYP2C19*17 carriers have a significantly lower risk of experiencing a recurrent cardiovascular event as compared to noncarriers (7.7% vs. 10%, respectively).10 Some of the inconsistencies in the reported results regarding CYP2C19*17 may be due to the fact that the *17 and the *2 variants are in partial linkage disequilibrium such that individuals carrying the *17 variant are less likely to carry the *2 variant.17
As a result of these studies and several others, in 2009 the FDA decided that the available data provide compelling evidence that genetic variation of CYP2C19 is a significant predictor of pharmacokinetics, pharmacodynamics, and clinical response. On 12 March 2010, a boxed warning was issued stating that health professionals should be aware that some patients may be poor metabolizers of clopidogrel, that genetic tests are available to determine CYP2C19 status, and that alternative therapy should be considered in these individuals. Despite this warning, the FDA has not mandated genetic testing for CYP2C19 status before initiation of clopidogrel therapy, which has led to confusion among physicians regarding how to clinically implement this information and treat their patients most effectively. Furthermore, recent recommendations by the American College of Cardiology Foundation/American Heart Association state that, in the absence of prospective randomized trials of clinical outcomes, “the evidence base is insufficient to recommend either routine genetic or platelet function testing at the present time.”18
With the recent FDA approval of prasugrel and with ticagrelor available in Europe, neither of which require CYP2C19 for activation, it may be argued that all patients should be given these agents rather than clopidogrel, irrespective of genotype. However, this approach is not cost-effective because clopidogrel is due to come off patent in the near future; moreover, this approach is not desirable given the increased risk of fatal and nonfatal bleeding in patients taking prasugrel as compared to those on clopidogrel.
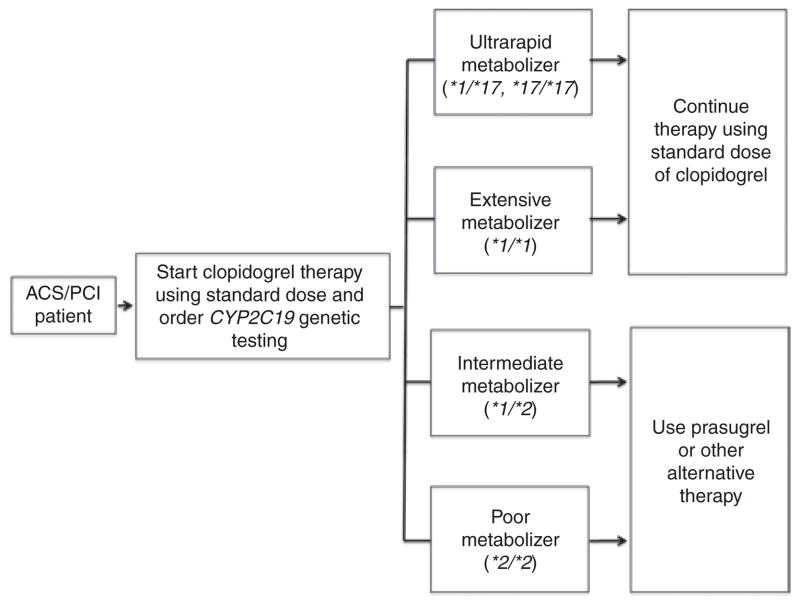
Although prospective randomized trials of genotype-directed antiplatelet therapy are currently being conducted, it will be years before the data become available. In the interim, the CPIC of the Pharmacogenomics Research Network has published guidelines for both the testing for CYP2C19 genotype and the ensuing clopidogrel therapy (Figure 1).1 This group is supported by the National Institutes of Health and is not affiliated with any commercial entity. The guidelines provide recommendations for the use of genetic information to guide clopidogrel therapy as well as a comprehensive review of the literature.1 The CPIC authors note that the most compelling evidence for a relation between CYP2C19 genotype and clopidogrel response exists in ACS patients who have undergone percutaneous coronary intervention.
Figure 1.
Clinical Pharmacogenetics Implementation Consortium guidelines for initiating antiplatelet therapy in patients with coronary disease based on cytochrome P450 2C19 genotype. ACS, acute coronary syndrome; CYP2C19, cytochrome P450 2C19; PCI, percutaneous coronary intervention. Adapted from ref. 1.
These high-risk patients can be genotyped and categorized as extensive (EM), intermediate (IM), or poor (PM) clopidogrel metabolizers based on *1/*1, *1/*2, and *2/*2 genotypes, respectively; patients who carry at least one copy of the CYP2C19*17 allele can be categorized as ultrarapid metabolizers (UMs).
The guidelines suggest that patients who are EMs (*1/*1) or UMs (*1/*17; *17/*17) receive the standard dose of clopidogrel and that patients who are IMs (*1/*2) or PMs (*2/*2) be considered for alternative antiplatelet therapy (e.g., prasugrel or ticagrelor) when not contraindicated (Figure 1). In addition to CYP2C19 metabolizer status, physicians should consider other factors, including age, body mass index, diabetes status, and use of proton-pump inhibitors (most notably omeprazole), which are associated with a high level of on-treatment platelet aggregation.19 At the time of this writing, there are not sufficient data to recommend higher-dose clopidogrel for IMs or PMs because some small-scale studies show benefits but others do not.
Although the guidelines proposed by the CPIC regarding genetic variations in CYP2C19 and antiplatelet therapy cannot possibly encompass the myriad different clinical situations presented to physicians, they do provide a framework within which to incorporate important and reproducible genetic data into effective individualized antiplatelet therapies. At the time of this writing, prospective randomized clinical trials, comparative effectiveness trials, and studies of pharmacoeconomics regarding genotype-directed antiplatelet therapies are under way and will probably result in future revisions to the guidelines. In the meantime, however, it seems both prudent and logical to take advantage of all sources of information to most effectively treat patients at high risk for recurrent cardiovascular events.
Pharmacogenomics of warfarin
Although in use for nearly 60 years, warfarin remains a difficult drug to manage because of its narrow therapeutic index and the wide interpatient variability in its dose requirements. Warfarin consistently ranks among the leading causes of serious drug-related adverse events, prompting a boxed warning in its labeling regarding bleeding risk. It interferes with the activation of vitamin K–dependent clotting factors (II, VII, IX, and X) by inhibiting VKORC1. Warfarin is usually initiated at a similar dose for all patients, typically 5 mg/day, with dose adjustment according to the international normalized ratio (INR). The problem with this trial-and-error dosing approach is that it often leads to over- or under-anticoagulation during the initial months of therapy, when the risk of bleeding is the greatest.20 Warfarin pharmacogenomics aims to enhance our understanding of patient-specific determinants of warfarin response in order to improve dosing accuracy and reduce the risk for adverse sequelae with warfarin therapy.
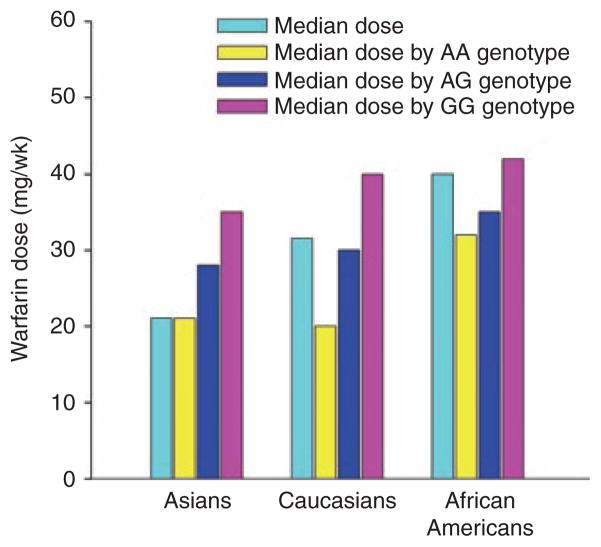
Table 3 highlights the genes/SNPs that have been most strongly associated with warfarin-dose variability. There are substantial and convincing data from numerous candidate-gene studies and GWASs showing that CYP2C9 and VKORC1 genotypes affect the dose requirements for warfarin.21–24 Specifically, the CYP2C9*2 (R144C; rs1799853) and *3 (I359L; rs1057910) alleles lead to 40–70% reductions in S-warfarin clearance and ~20–40% lower warfarin dose requirements, respectively. These alleles are also associated with increased bleeding risk.25,26 The VKORC1 rs9923231 −1639G>A (or rs9934438 1173C>T) SNP increases sensitivity to warfarin at its target site, further reducing dose requirements. The −1639G>A variant is in near-complete linkage disequilibrium with the 1173C>T variant in all major continental populations27 and therefore may be used to predict dose requirements for warfarin. In association with clinical factors (e.g., age, body size, and use of amiodarone), the CYP2C9*2 and *3 alleles and VKORC1 −1639G>A genotype explain 50–60% of the variability in maintenance dose of warfarin among Caucasians22,28 but only ~25% of the variability among African Americans.22,29,30 Prediction of decreased doses in African Americans is secondary to lower frequencies of the CYP2C9*2, CYP2C9*3, and VKORC1 −1639A alleles in this population.27 However, as shown in Figure 2, the racial/ethnic differences in warfarin dose requirements are almost completely explained by the varying frequencies of the VKORC1 allele between the major continental populations, such that within a VKORC1 genotype, doses are similar across population groups.
Table 3.
| Allele | Caucasians | Asians | African Americans |
|---|---|---|---|
| CYP2C9*2 rs1799853 | 0.12 | 0 | 0.02 |
| CYP2C9*3 rs1057910 | 0.06 | 0.03–0.04 | 0.01 |
| CYP2C9*5 rs28371686 | 0 | 0 | 0.01 |
| CYP2C9*6 rs9332131 | 0 | 0 | 0.01 |
| CYP2C9*8 rs7900194 | 0 | 0 | 0.06 |
| CYP2C9*11 rs28371685 | 0 | 0.01 | 0.02 |
| CYP2C9 rs7089580 | ND | ND | 0.23 |
| VKORC1 rs9923231 (−1639G>A) | 0.40 | 0.90–0.94 | 0.10 |
| VKORC1 rs61162043 | ND | ND | 0.47 |
| CYP4F2 rs2108622 (V433M) | 0.23 | 0.24–0.25 | 0.09 |
| CALU rs339097 | 0 | 0.01–0.02 | 0.16 |
ND, not determined.
Figure 2.
Median warfarin dose requirements in Asians, Caucasians, and African Americans, overall and by VKORC1 genotype; based on data from the International Warfarin Pharmacogenetics Consortium.29 The cyan bars indicate the median warfarin dose in each racial group, overall. The yellow, blue, and pink bars show the dose within each racial group by the VKORC1 −1639 AA, AG, and GG genotypes, respectively. As shown, the overall warfarin dose requirements are lower in Asians and higher in African Americans, compared with Caucasians. This racial difference in dose is largely explained by a higher frequency of the VKORC1 −1639 AA (low-dose) genotype in Asians, AG (intermediate-dose) genotype in Caucasians, and GG (high-dose) genotype in African Americans, resulting in similar doses by race within genotype. wk, week. Prepared from data in ref. 27.
The CYP4F2 V433M (rs2108622) variant explains an additional 1–2% of the variability in warfarin dose.31 CYP4F2 metabolizes vitamin K to hydroxy–vitamin K, thus limiting the quantity of vitamin K available for carboxylation of the clotting factor.32 The V433M variant is common in Caucasians and Asians and is associated with lower CYP4F2 activity and higher warfarin dose requirements; this has been replicated in several independent studies.24,33–35
Two GWASs have shown that the VKORC1 −1639G>A variant is the major genetic determinant of warfarin maintenance dose in Caucasians; CYP2C9*2 and *3 contribute to a lesser extent.21,24 A third GWAS, in Japanese patients, also showed that VKORC1 genotype is the strongest predictor of warfarin dose.34 After controlling for VKORC1 and CYP2C9 variants, the CYP4F2 V433M genotype emerged as a further predictor of dose requirements in two of these GWASs.24,34 Whether the VKORC1 and CYP2C9 variants are the primary contributors to warfarin dose requirements in African American populations is unknown. However, GWASs are under way to help address this question.
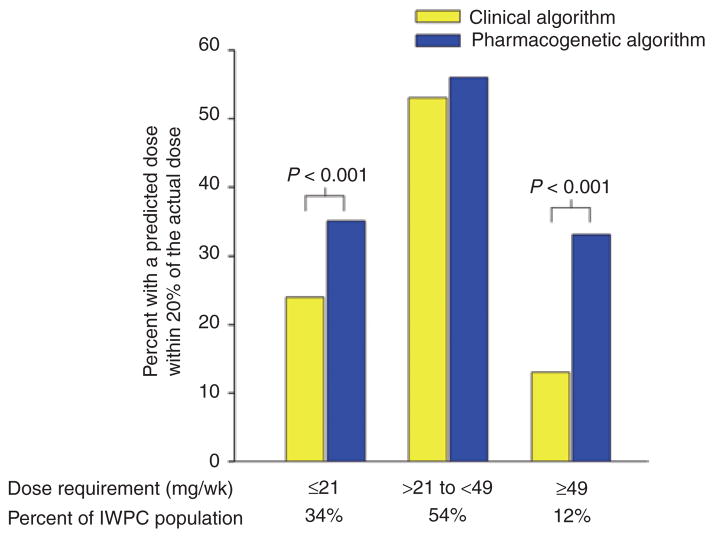
As noted above, the warfarin labeling was revised in August 2007 to include information on CYP2C9 and VKORC1 genotypes as predictors of dose response and to inform dose adjustment in patients with variant genotypes. In early 2010, the labeling was further revised to include specific dosing recommendations according to CYP2C9 and VKORC1 genotypes. Several validated dosing algorithms, including one from the International Warfarin Pharmacogenetics Consortium and one proposed by Gage and colleagues, are also available to assist clinicians with genotype-guided dosing.22,28 Both of these algorithms are available in a user-friendly tool at http://www.warfarindosing.org. In the Gage algorithm, dose prediction may be refined with input of previous INR and dose data.29 Figure 3 highlights the improvement in dose prediction achieved by using the pharmacogenetics algorithm in comparison to the clinical algorithm. The CPIC guidelines2 recommend the use of the Gage or International Warfarin Pharmacogenetics Consortium dosing algorithm as the preferred approach for incorporation of genetic information into warfarin-dose prediction, based on published data suggesting the superiority of this approach. When electronic access to these algorithms is not available, referring to the dosing table in the warfarin product label is the recommended alternative approach.
Figure 3.
Percentage of patients whose actual warfarin dose fell within 20% of the predicted dose according to either the clinical or the pharmacogenetic dosing algorithm derived by the International Warfarin Pharmacogenetics Consortium (IWPC).22 The pharmacogenetic algorithm was more predictive of the actual dose requirements for those requiring ≤21 mg/week or ≥49 mg/week of warfarin. For individuals requiring intermediate doses, the clinical and pharmacogenetic algorithms were similarly predictive of the dose requirements. The percentages of patients in each dosage group are shown at the bottom. wk, week. Adapted from ref. 22.
Despite the wealth of data supporting genetic determination of warfarin dose requirements, recent labeling changes, and the availability of both decision-support tools (dosing algorithms) and at least five FDA-cleared platforms for warfarin genotyping, genetic testing is not widely embraced in clinical practice. In fact, current consensus guidelines warn against the routine use of genetic data to guide dosing.20 This is a grade 2C recommendation, indicating that the evidence supporting the suggestion is limited and that patient values and preferences should be taken into account. Similarly, the American College of Medical Genetics does not endorse genetic testing except in cases of unusual warfarin response.36 As an additional impediment to clinical implementation of genetic testing for warfarin, the Centers for Medicare and Medicaid Services announced in 2009 that coverage for such testing would be denied unless testing is provided in the context of a controlled clinical study.
So why are clinicians and policy makers reluctant to embrace warfarin pharmacogenetics? Barriers to the clinical implementation of genetic testing generally include the need to establish the clinical validity, analytical validity, and clinical utility of testing. In the case of warfarin, the clinical validity is well established, as discussed above. The analytical validity is also well established. However, some variants that are not included on some genotyping platforms are worth mentioning given their implications for African Americans. The CYP2C9*5 (D360E), *6 (10601delA), *8 (R150H), and *11 (R335W) alleles occur almost exclusively in African Americans and are associated with reduced metabolism.37 The *8 allele is as common as other CYP2C9 alleles combined and is correlated with lower warfarin dose requirements in African Americans.38 Recently, the CYP2C9 rs7089580, VKORC1 rs61162043, and calumenin rs339097 gene variants were identified in African Americans; these are associated with higher maintenance doses in this population.30,39 GWASs in African Americans may show other variants with implications for warfarin dosing in this population.
Whether the clinical utility of genetic testing for warfarin has been established is a more debatable question. There is evidence for benefit with genotype-guided dosing from a small-scale clinical trial and a comparative effectiveness study.40,41 In the latter, patients were offered free genotyping for the CYP2C9*2, CYP2C9*3, and VKORC1 −1639G>A variants, with the results being provided to their physician. During the initial 6 months of therapy, those who underwent genetic testing had 30% fewer hospitalizations for any cause and for bleeding or thromboembolism as compared to historical controls.41 These data are tempered by findings from another clinical trial, in which patients were randomized to genotype-guided or clinical-based warfarin dosing. Because a large sample size would be required to demonstrate reductions in serious adverse events, investigators focused on INR values outside the therapeutic range as a marker of increased bleeding or thrombotic risk.42 The percentage of out-of-range INRs was similar between the two dose-strategy groups. An exploratory analysis showed a significant benefit with pharmacogenetic dosing for patients with either multiple variant alleles (who required 3–4 mg/day) or the wild-type genotype (who required 6–7 mg/day), whereas carriers of single variant alleles (who required about 5 mg/day) appeared to have no benefit from genotype-guided dosing. This is consistent with findings from the International Warfarin Pharmacogenetics Consortium, in which a pharmacogenetic algorithm more accurately predicted low (≤3 mg/week) and high (≥7 mg/week) warfarin doses than a clinical algorithm; however, the two algorithms were similarly predictive of intermediate doses (Figure 3).22 Based on these data, genotype-guided therapy may not be of benefit to carriers of a single variant allele (~40% of Caucasians), in whom a dose of 5 mg/day (the typical starting dose) would be predicted.
The National Heart, Lung, and Blood Institute–sponsored Clarification of Optimal Anticoagulation Through Genetics (COAG) trial is empowered to account for the potential lack of benefit with genotype-guided therapy in patients with a single variant. The COAG trial is a prospective, multicenter, randomized, double-blind trial that began in September 2009.43 It aims to determine whether the percentage of time spent within the therapeutic INR range (primary outcome) or the occurrence of an INR >4 or a serious event (secondary outcome) during the initial 4 weeks of therapy differs between the pharmacogenetic- and clinical-dosing strategies. The trial is expected to be completed in December 2012. Several other randomized prospective trials addressing the efficacy, safety, and economic implications of warfarin pharmacogenetics are under way in the United States and Europe (the Clinical and Economic Implications of Genetic Testing for Warfarin Management trial, the Genetics Informatics Trial of Warfarin to Prevent Deep Venous Thrombosis (GIFT), and the European Pharmacogenetics of Anticoagulant Therapy trial (EU-PACT) (ClinicalTrials.gov)).
The need for rapid genotyping is an often-cited barrier to the implementation of warfarin pharmacogenetics. Clinical laboratories often lack the personnel or equipment for rapid genotyping. If samples are sent to an outside facility, results may not be available for several days, at which time INR results are available to guide therapy. Nonetheless, there are data showing that even with a delay of 4–5 days, a pharmacogenetic algorithm incorporating previous INR results and warfarin doses provides a more accurate prediction of the warfarin maintenance dose than clinical factors alone.29 In addition, with continuing technological advances, time for obtaining genotyping results will continue to decrease, and it is anticipated that, eventually, the genetic information will be available in medical records.
In summary, the clinical and analytical validity of warfarin pharmacogenetics is well established, at least for Caucasians. Variants that best predict dose requirements in non-Caucasians are still being investigated. Clinical implementation of genotype-guided therapy is largely hindered by the lack of data from randomized clinical trials in a sufficient number of patients to prove its beneficial outcomes. However, the data to date provide clear evidence of the ability to better predict the stable warfarin dose requirement with the use of genetic information. Results from ongoing clinical trials will help to define the role of warfarin pharmacogenetics in clinical practice. In the meantime, the CPIC guidelines on warfarin provide guidance for incorporation of genetic information for the accurate prediction of warfarin dose when such information is available.
CLINICAL IMPLEMENTATION OF PHARMACOGENOMICS: PROGRAMS AND APPROACHES
One approach to the use of genotype data in clinical practice is to order the genetic test as other laboratory/diagnostic tests are ordered. However, as discussed above, this approach presents certain barriers, including the need for the clinician to remember to order the test, turnaround time, costs of the genetic test, and the clinician’s uncertainty about what to do with the genetic information. As genotyping and sequencing technologies advance, whole-genome sequencing is expected to eventually replace single genetic tests. The cost for whole-genome sequencing is expected to fall below the $1,000 mark in the next few years. Thus, the possibility of completion of whole-genome sequencing at a single point in a person’s life, with the data available for use thereafter, now appears feasible. An example of such an approach was recently published.44 Thus, an alternative vision is to embed genotypic information in an electronic medical record, to be accessed as needed, with point-of-care decision support provided when a prescription is written for a drug with responses known to be modulated by genetic variants. Clopidogrel and warfarin are examples of drugs for which, as a first step to enabling this long-term vision for genomic medicine, the PREDICT (Pharmacogenomic Resource for Enhanced Decisions in Care and Therapy) project of the Vanderbilt University Medical Center is focused on implementing preemptive genotyping for patients likely to receive drugs with responses known to be modulated by pharmacogenomic variants. The initial target population comprises patients scheduled for cardiac catheterization, 40% of whom ultimately receive clopidogrel. The program was launched in September 2010, and as of 28 April 2011, 1,769 patients had been genotyped; of these, 42 had the CYP2C19*2/*2 genotype and 342 possessed the CYP2C19*1/*2 genotype. Alerts are generated to the clinician ordering clopidogrel to provide guidance relative to the CYP2C19 genotype. Genotyping is conducted in an environment in accordance with the Clinical Laboratory Improvement Amendments—using the Illumina VeraCode platform (for 184 variants considered relevant to drug responses), allowing extension of the data to other drug–gene pairs, for use as the clinical need arises.
A similar implementation program is also under way as a network-wide project within the scope of the National Institutes of Health–funded Pharmacogenomics Research Network. The Translational Pharmacogenomics Project involves six institutions that are implementing programs similar to the PREDICT project at Vanderbilt; among other aims, the project will define the challenges and successes associated with a pharmacogenomics clinical implementation program. Large-scale implementation of pharmacogenomics will also require the engagement of regulatory agencies, policy makers, insurers, and other stakeholders
COMPELLING, BUT NOT CLINICALLY ACTIONABLE, EXAMPLES IN CARDIOVASCULAR PHARMACOGENOMICS
Heart-failure pharmacogenomics: opportunities in drug development?
There is little literature on the pharmacogenomics of the various treatment modalities for heart failure, with the exception of β-blockers, on which there are numerous studies.45 As noted in Table 1, the two most commonly used β-blockers in heart failure—metoprolol and carvedilol—have FDA labeling regarding polymorphisms in CYP2D6. The CYP2D6 enzyme catalyzes a major metabolic pathway for both of these drugs (more so with metoprolol than carvedilol). Data show differences in plasma drug concentration accounted for by CYP2D6 genotype; however, the data are limited, suggesting that different drug concentrations have clinical relevance. For example, a study of metoprolol succinate during the titration period in heart-failure patients showed the expected differences in plasma metoprolol concentration caused by CYP2D6 genotype. However, there were no differences in the number of patients with symptomatic worsening or decompensation during the titration phase among different CYP2D6 genotype gps.46 Therefore, although the evidence for a pharmacokinetic effect of the CYP2D6 genotype on these drugs is clear, this does not appear to translate into important differences in terms of responses or adverse effects.
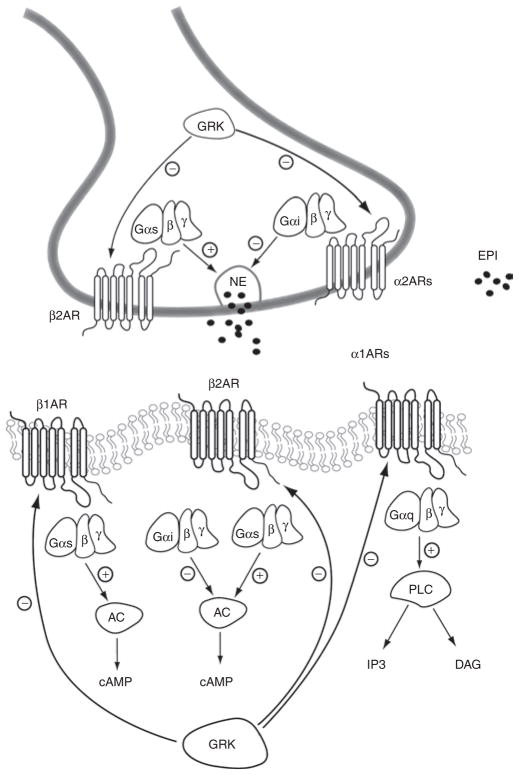
The most compelling data on β-blocker pharmacogenomics in heart failure fall neatly into the pathway–candidate gene paradigm, and the relevant pathway is highlighted in Figure 4. Significant genetic associations in this pathway arise from the β-1 and β-2-adrenergic receptor genes (ADRB1 and ADRB2), the α-2C-adrenergic receptor gene (ADRA2C), and the G-protein-coupled receptor kinase-5 gene (GRK5).
Figure 4.
Schematic representation of adrenergic receptor signaling in the heart. α1AR, α1-adrenergic receptors; α2AR, α2-adrenergic receptors; AC, adenylyl cyclase; β1AR, β1-adrenergic receptors; β2AR, β2-adrenergic receptors; cAMP, cyclic adenosyl monophosphate; DAG, diacylglycerol; EPI, epinephrine; Gαs, G-protein α subunit, stimulatory; Gαi, G-protein α subunit, inhibitory; GαqG-protein a subunit q; GRK, G-protein receptor kinase; IP3, inositol triphosphate; NE, norepinephrine; PLC, phospholipase C. From ref. 47.
The best studied among these is ADRB1, which has two common, nonsynonymous polymorphisms (Ser49Gly and Arg389Gly), which have been documented in numerous in vitro mutagenesis, ex vivo, and human studies as being functional.47 The strongest literature is related to Arg389Gly, where the Arg form of the receptor couples more efficiently to G-protein, leading to greater downstream signaling. Some, but not all, studies on heart failure suggest that Arg389Arg patients have greater improvement in left-ventricular ejection fraction with β-blocker treatment.45,47 Studies on the association of this polymorphism with clinical outcomes also suggest an association. The β-Blocker Evaluation of Survival Trial (BEST), which compared bucindolol to placebo, found that Arg389Arg patients had significant benefits from bucindolol (reduced mortality and hospitalizations), whereas in Gly389 carriers no significant benefit was observed in comparison with placebo.48 A population cohort study found significantly better outcomes in Arg389Arg patients treated with a high dose of β-blocker vs. those on low doses or no β-blocker. Other cohort studies, however, have not observed significant differences in outcomes by genotype.47 An important difference in the studies with positive associations vs. those showing absence of association is the approach to analysis. Those with no observed association included cohorts in which all, or nearly all, patients were treated with a β-blocker, and comparisons across ADRB1 genotypes showed no differences. However, studies comparing treated vs. untreated patients within a genotype have more consistently shown differences in outcomes.47 This suggests that the Arg389Arg genotype may have negatively influenced the outcomes, with the negative effect of this genotype offset by the β-blocker.
A four-amino-acid insertion–deletion polymorphism in ADRA2C, which influences norepinephrine release through negative-feedback mechanisms,47 has also been associated with β-blocker efficacy. In BEST, ADRA2C deletion carriers did not obtain the benefit from bucindolol that was observed in insertion homozygotes.49 It is postulated that the negative effect of the deletion allele on the efficacy of bucindolol is related to greater sympatholysis in deletion carriers; sympatholytic effects are unique to bucindolol among the β-blockers tested for heart failure.49 By contrast, a study of metoprolol, which is a β-1-selective blocker, found that patients who were ADRB1 Arg389Arg and ADRA2C Del carriers had the greatest improvement in ejection fraction, a surrogate for improved outcomes.50 These seemingly disparate findings in fact align well when one considers the functional mechanism of the polymorphism and the differences in pharmacological properties between metoprolol and bucindolol.
Finally, several studies have documented a pharmacogenomic effect of the Leu41Gln variant of GRK5. A series of elegant in vitro and animal studies suggest that the Leu41 allele blunts the effects of catecholamines; it has therefore been called an endogenous β-blocker.51 The first study of this SNP in humans showed that Leu41 carriers not treated with β-blockers had outcomes that were significantly better than those of Gln41Gln β-blocker-untreated patients and similar to Gln41Gln β-blocker-treated patients.51 Studies of two other populations suggested that the benefit of administration of β-blockers was confined to Gln41Gln individuals.51,52
Collectively, these studies indicate that genetic variability in the adrenergic signaling pathway has an important influence on the benefits of β-blockers in heart failure. They also suggest that certain genotype groups derive minimal benefit from β-blocker therapy. However, the consensus guideline–driven use of β-blockers in all patients with systolic heart failure makes clinical application of this information difficult because it would mean withholding β-blocker treatment, and, unlike with clopidogrel, there are no alternatives.
However, these data may prove beneficial in other ways in heart failure—specifically, in drug development. Several promising drug classes have failed to show documentable efficacy in heart failure in the past 10–15 years in late phase III clinical trials.45 This suggests the need to target drug development in heart failure to those most likely to benefit from a therapy that is an addition to the standard regimen consisting of an angiotensin-converting enzyme–inhibitor, a β-blocker, a diuretic, and digoxin. Pharmacogenomic data may provide one way to target therapy during the development of drugs for heart failure. One example might be a study that enrolls only patients who are ADRB1 Gly or GKR5 Leu41 carriers and then tests the novel therapy against placebo in the background of standard therapies. Because the data suggest that these genotype groups might derive minimal benefit from β-blockers, it should be easier to document the benefit from the novel therapy. By contrast, those with genotypes responsive to β-blockers might already be obtaining the maximal benefit possible from pharmacological therapies, and therefore their inclusion in trials impairs the ability to document drug efficacy. This approach is not one that has been employed to date; however, it has been discussed.
These pharmacogenomic data are, however, being used in another way in the development of bucindolol, which does not have FDA approval. Specifically, ARCA Biopharma (Broomfield, CO) made a new drug application to the FDA seeking approval for use of bucindolol in ADRB1 Arg389Arg patients. The FDA denied the initial new drug application and requested a randomized controlled trial. The company has plans to launch, in late 2011, a superiority trial in 3,200 ADRB1 Arg389Arg patients who will be randomized to metoprolol succinate or bucindolol.53 To our knowledge, this represents the first example of pharmacogenomically guided drug development for the treatment of cardiovascular disease.
SERIOUS ADVERSE DRUG EFFECTS AND PHARMACOGENOMICS
The idea that risk for an unusual, rare, and serious adverse drug effect could have a genetic component goes back to the identification of hemolytic anemia during treatment with antimalarials in World War II. In this case, the risk for this unexpected drug effect was much higher among African-American subjects and was associated with a deficiency of glucose-6-phosphate dehydrogenase. Other adverse effects are “expected” in that they represent sensitivity to a drug’s known pharmacologic effects, such as serious bleeding during anticoagulant therapy. Notably, even these adverse effects may have a genomic component. In recent years, there have been a number of examples of serious adverse drug events that had been described as idiosyncratic now known to have a clear genetic component. These include the hypersensitivity reaction to abacavir, severe skin reactions to carbamazepine, and drug-induced liver injury resulting from lumiracoxib administration—all of which are linked to markers in human leukocyte antigen.54
Serious adverse drug events associated with administration of cardiovascular drugs or those affecting the cardiovascular system (e.g., statin-induced myopathy and rhabdomyolysis; drug-induced long-QT syndrome and torsade de pointes), however, do not appear to be linked to human leukocyte antigen.
Muscle toxicity during treatment with inhibitors of 3-hydroxy-3-methyl-glutaryl-coenzyme-A reductase (statins)
Rhabdomyolysis is a very rare effect of statins, although varying degrees of muscle aches and creatine kinase elevations are more common. All of these symptoms are known to be more common with higher (vs. lower) statin doses and therefore presumably represent drug concentration–related adverse effects. Candidate-gene studies examined the role of both drug metabolism and transport pathways and suggested that variants in CYP3A555 and the uptake transporter organic anion–transporting polypeptide 1B1 (encoded by SLCO1B1)56 modulate the corresponding risk. The Study of the Effectiveness of Additional Reductions in Cholesterol and Homocysteine (SEARCH) consortium examined the creatine kinase values of more than 12,000 subjects administered low- or high-dose simvastatin (20 and 80 mg/day, respectively). There were 8 possible cases of muscle toxicity in the 6,033 patients treated with the low dose but 98 possible cases among the 6,031 assigned the high dose. A GWAS that compared 85 patients in the high-dose arm showing muscle toxicity with 90 controls in the high-dose arm not developing toxicity identified a single SNP (rs4363657) of genome-wide significance in SLCO1B1; this SNP tags a known nonsynonymous variant (Val174Ala).57 Among the patients homozygous for the CC risk allele (2.1% of the study population), 18.6% developed muscle toxicity over a period of 5 years, as compared to 0.63% in the low-risk (TT) group, which comprised 73% of the population; the OR was 16.9, and risk in heterozygotes was intermediate. This result was replicated in a separate cohort in the original publication and has been replicated in two other studies,58,59 including one that used simply staying on statins as an end point. In the latter study, Val174Ala also predicted the discontinuation of atorvastatin (but not of pravastatin). These findings emphasize how environmental factors (notably, drug dose and duration of therapy) and genetics interact to produce a drug-response phenotype.
This represents an example of a pharmacogenomic marker that apparently modulates risk; yet its predictive value, and therefore clinical utility, remains to be defined.
Drug-induced long-QT syndrome
Congenital long-QT syndrome (LQTS) is a rare genetic disease recognized first in the 1950s and ‘60s. Affected individuals displayed a marked prolongation of the QT interval, recurrent syncope, and risk for sudden death due to torsade de pointes (TdP), a morphologically distinctive polymorphic ventricular tachycardia. Exposure to certain drugs can also produce similar electrocardiographic findings, and this adverse drug effect has therefore been termed drug-induced LQTS (diLQTS). QT-prolonging antiarrhythmics are by far the most commonly implicated drugs, and 1–5% of patients exposed to sotalol, dofetilide, or quinidine develop diLQTS.60 This adverse effect is seen, much less frequently, with drugs used for noncardiovascular indications, including antihistamines, antibiotics, anti-psychotics, and methadone. A common feature of diLQTS is that risk is increased with higher doses or higher plasma concentrations of most culprit drugs. In some cases, genetically determined variable drug metabolism has been implicated; the risk may be higher among CYP2D6 poor metabolizers treated with thioridazine, a CYP2D6 substrate. The thioridazine product label describes this risk. Similarly, CYP-based drug interactions can increase risk; a very prominent example was diLQTS during treatment with the antihistamine terfenadine. Terfenadine prolongs the QT interval but ordinarily undergoes extensive presystemic CYP3A4-mediated clearance to its antihistaminic metabolite fexofenadine, which does not prolong QT. When CYP3A4 is inhibited (e.g., by ketoconazole or similar strong inhibitors) or in patients taking an overdose of terfenadine, the parent drug appears in plasma and prolongs QT, which can cause TdP. Because of this adverse effect, and after the development of fexofenadine as a non-QT-prolonging antihistamine, terfenadine was withdrawn from the US market in 1997. The condition of diLQTS can occur at low doses and low concentrations in susceptible individuals; in addition, diLQTS is more common at low, rather than high, doses of quinidine. The explanation is thought to be that at low doses the drug produces arrhythmogenic effects on cardiac repolarization, whereas at higher doses it produces electrophysiologic effects that inhibit repolarization-related arrhythmias.
Mutations in 13 different genes encoding ion channels or proteins modulating ion-channel function have been identified in families with congenital LQTS. Although initial studies of the congenital syndrome focused on patients with obvious QT prolongation and recurrent episodes of syncope, subsequent studies in susceptible families have identified the clinical phenotype of incomplete penetrance. Thus, one hypothesis being explored is that patients with diLQTS represent subclinical cases of the congenital syndrome in which drug exposure exposes the full-blown clinical phenotype.
A number of small-scale studies (involving up to ~100 patients) have examined the frequency of subclinical congenital LQTS gene mutations in patients with drug-induced TdP.61,62 Small-scale studies have identified mutations in the five major congenital LQTS-disease genes in 10–20% of subjects with diLQTS. More recently, one small-scale study in Japan suggested a higher incidence, ~40%,63 and a study using targeted next-generation sequencing to screen all 13 congenital LQTS-disease genes and other arrhythmia susceptibility genes in 31 patients identified rare variants predicted to be deleterious to protein function in 20 patients (64.5%).64 Therefore, there seems to be little doubt that congenital LQTS mutations contribute to the risk for diLQTS; however, the extent to which they explain the risk is uncertain. Arrhythmias in the congenital syndromes are often adrenergically triggered; nevertheless, polymorphisms in the β-1- and β-2-adrenergic receptor genes are not associated with diLQTS.64
Preliminary data using intensive candidate-gene and GWAS approaches for analysis of risk for drug-induced TdP have been reported. The cases were selected from multiple US and European sites using common definitions, and controls were either large-scale healthy populations or patients starting QT-prolonging antiarrhythmics for clinical indications and not developing LQT intervals. The candidate-gene study examined ~1,500 SNPs in 18 candidate genes and identified variants in the genes of the IKs channel as risk variants. The GWAS identified multiple associated genomic regions, none of which included obvious candidate genes.64
Thus, there have been substantial efforts to identify the genes that place patients at risk for diLQTS. There are few notable findings beyond the genes associated with congenital LQT, and at this point no genomic markers are sufficiently robust for predicting the risk of development of diLQTS.
PROMISING AREAS IN CARDIOVASCULAR PHARMACOGENOMIC RESEARCH
Pharmacogenomics of antihypertensive drugs
Several areas of research in cardiovascular pharmacogenomics hold promise, but they do not yet have the level of replicated data as cited in the aforementioned examples. The pharmacogenomics of antihypertensive drugs is one of these, and studies range from testing genetic associations with the lowering of blood pressure (BP), adverse metabolic effects, and long-term sequelae of hypertension such as death, stroke, and myocardial infarction.
Among the first-line antihypertensive drug classes, those with the greatest body of literature are the β-blockers and thiazide diuretics. Similar to the data on β-blockers in heart failure, there are interesting β-blocker pharmacogenomics data in hypertension, particularly for the ADRB1 gene. Consistent with the data in heart failure, the Arg389Gly and Ser49Gly polymorphisms have been associated with differential BP-lowering levels in a number of studies, with the Ar389Arg genotype or the Ser49/Arg389 haplotype associated with the best antihypertensive response.47,65 The Ser49/Arg389 haplotype has also been associated with improved outcomes (particularly lower death rate) in atenolol-treated patients with hypertension (as compared to those treated with verapamil).65 This finding is also consistent with the literature on heart failure, in which the genotype/haplotype appears to be associated with risk, and β-blockers offset this risk.
Interesting data are also available for thiazide diuretics. One of the polymorphisms most consistently associated with thiazide response is the ADD1 Gly460Trp polymorphism, a documented functional polymorphism.64,66,67 Although not all the studies have documented an ADD1 association with thiazide response, work in this area has led to the development of a novel antihypertensive drug class that targets α-adducin (the protein encoded by ADD1) and ouabain; moreover, the data obtained in a phase II study on the BP-lowering efficacy of the drug are quite impressive.68 This highlights not only the possibility of defining genetic determinants of drug response through pharmacogenomics research but also identification of novel drug targets.
Another candidate gene of interest in the context of thiazides is NEDD4L, which contains a documented functional SNP, plays a role in sodium reabsorption, and has been associated with BP response and clinical outcomes after thiazide treatment.66,69 Finally, the only published GWAS for antihypertensive response discovered a SNP on chromosome 12 associated with the thiazide response in African Americans,70 which was recently replicated in an independent cohort. There are no known genes in the region involved in the thiazide response. However, an interesting candidate is FRS2, whose encoded protein is involved in fibroblast growth factor signaling, which plays a role in vascular smooth muscle cell regulation. Because the vascular mechanisms for lowering BP with thiazides are not well defined, this highlights the potential for pharmacogenomics research to identify novel mechanisms of action. It may also represent a novel drug target.
There is a limited, but interesting, body of data on calcium-channel blockers. The majority of studies have focused on outcomes associated with calcium-channel blocker therapy relative to calcium-signaling genes, including the target protein’s gene, CACNA1C, along with CACNB2 and KCNMB1.71–73 These findings require replication; however, they involve strong biological candidates, apart from the fact that CACNB2 has been documented from GWASs to be a hypertension gene. Of interest, despite a relatively large number of studies with angiotensin-converting enzyme inhibitors (or angiotensin receptor blockers), there are few examples of convincing genetic associations with response to these drugs.
Aspirin pharmacogenomics
Dual antiplatelet therapy with aspirin and clopidogrel is the most commonly used antiplatelet regimen for patients with ACSs. Although considerable advances in clopidogrel pharmacogenomics have resulted in potential clinical applications (see above), the pharmacogenomics of aspirin response remains a promising, yet poorly understood, area of investigation. The variability in aspirin response is well documented, and heritability estimates suggest that genetic factors contribute moderately to residual platelet reactivity after aspirin treatment.74 Despite these data and a large number of candidate-gene studies, genetic variations in relatively few genes have been reproducibly associated with aspirin response. Some of the more intensively studied candidate genes include those for cyclooxygenase 1, the purinergic platelet receptors P2Y1 and P2Y12, and several platelet glycoproteins (e.g., GPIIb-IIIa, GPVI, GPIa, and GPIb). Variants in few, if any, of these genes have been reproducibly shown to be associated with aspirin response. A recent systematic review by Goodman and colleagues75 of 31 studies evaluating 11 genes showed that the GPIIIa PIA1/A2 polymorphism was associated with aspirin resistance in healthy subjects (P = 0.009; OR = 2.36, 95% CI: 1.24–4.48) but not in the combined group of healthy subjects and patients with cardiovascular disease (P = 0.40; OR = 1.14, 95% CI: 0.84–1.54). Polymorphisms in the genes encoding cyclooxygenase 1, P2Y1, P2Y12, and GP1a were not associated with aspirin resistance.75
Several factors contribute to inconsistencies among studies. Assessment of aspirin resistance has been difficult not only because of varying methodologies used in measuring platelet function but also because of differences in the definition of aspirin resistance itself. The problem of phenotype-assessment heterogeneity is compounded by small sample sizes and incomplete coverage of variations in the candidate genes studied. Future studies must focus on genome-wide approaches that are more comprehensive and that are conducted in large numbers of well-phenotyped individuals.
Statin pharmacogenomics
There has been extensive work on the genetic predictors of lowering of low-density lipoprotein (LDL) cholesterol, but few genes have emerged, and they generally explain relatively small percentages of the LDL response. The strongest data are available for genes encoding the protein target 3-hydroxy-3-methyl-glutaryl-coenzyme-A reductase (HMGCR) and the LDL receptor (LDLR), which are both very strong biological candidates.64 A nonsynonymous SNP in the gene for kinesin family member 6 (KIF6) has been suggested as a predictor of outcomes with statin therapy, and a commercially available test is available. However, unlike most of the examples discussed in this review, the potential role of KIF6 in coronary disease or statin response is unknown, and recent data have called this association into question. Overall, with the exception of the predictors of statin-induced muscle toxicity, it appears that identification of SNPs that are sufficiently predictive of statin response to have clinical utility will remain a challenge.
CONCLUSION
Research into the pharmacogenomics of cardiovascular drugs has led to the elucidation of examples with clinically actionable findings (warfarin and clopidogrel), which not only have the potential to improve management of patients prescribed these drugs but have also advanced our understanding of their pharmacokinetics and pharmacodynamics. Guidelines for the use of genetic information to guide warfarin and clopidogrel therapy have been published, and in the future, genetic information may be available within the medical record. This will obviate the need to order specific genetic tests and possibly enhance the pace of translation into practice in cardiovascular pharmacogenomics. A drug-transporter polymorphism has been strongly implicated in statin-induced myopathy, and, although this may not be predictive enough to be used clinically, it has enhanced our understanding of this potentially serious adverse drug event. Research on β-blocker pharmacogenomics in heart failure has clearly implicated various components of the adrenergic signaling pathway, and these data have the potential to influence the future development of drugs to treat heart failure. Areas of active investigation that hold promise include the pharmacogenomics of antihypertensives and aspirin. The clinical utilization of pharmacogenomics in cardiovascular disease holds promise, and clinical implementation has begun in certain centers. The potential approaches for implementation are described here, and certain challenges are discussed. The findings from the vanguard centers that are leading clinical implementation will provide important insight into the challenges and future directions for pharmacogenomics.
Acknowledgments
This work was supported in part by grants from the National Institutes of Health Pharmacogenomics Research Network: U01 GM074492 (J.A.J.), U19 HL065962 (D.M.R.), U01 HL105198 (A.R.S.), K23 HL091120 (A.L.B.), and AHA 10GRNT3750024 (L.H.C.).
Footnotes
CONFLICT OF INTEREST
J.A.J. is on an advisory board for Medco. A.R.S. is a consultant for Bristol–Myers Squibb/Sanofi–Aventis Pharmaceuticals Partnership. D.M.R. is a consultant for Merck, Novartis, Dai-Ichi, and Astellas. D.M.R. also receives royalties for a patent on a “Method of Screening for Susceptibility to Drug-Induced Cardiac Arrhythmia.” The other authors declared no conflict of interest.
References
- 1.Scott SA, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for Cytochrome P450-2C19 (CYP2C19) Genotype and Clopidogrel Therapy. Clin Pharmacol Ther. 2011;90:328–332. doi: 10.1038/clpt.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson JA, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing. Clin Pharmacol Ther. doi: 10.1038/clpt.2011.185. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shuldiner AR, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA. 2009;302:849–857. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mega JL, et al. Reduced-function CYP2C19 genotype and risk of adverse clinical outcomes among patients treated with clopidogrel predominantly for PCI: a meta-analysis. JAMA. 2010;304:1821–1830. doi: 10.1001/jama.2010.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sofi F, Giusti B, Marcucci R, Gori AM, Abbate R, Gensini GF. Cytochrome P450 2C19(*)2 polymorphism and cardiovascular recurrences in patients taking clopidogrel: a meta-analysis. Pharmacogenomics J. 2011;11:199–206. doi: 10.1038/tpj.2010.21. [DOI] [PubMed] [Google Scholar]
- 6.Xie HG, Zou JJ, Hu ZY, Zhang JJ, Ye F, Chen SL. Individual variability in the disposition of and response to clopidogrel: pharmacogenomics and beyond. Pharmacol Ther. 2011;129:267–289. doi: 10.1016/j.pharmthera.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 7.Brandt JT, et al. Common polymorphisms of CYP2C19 and CYP2C9 affect the pharmacokinetic and pharmacodynamic response to clopidogrel but not prasugrel. J Thromb Haemost. 2007;5:2429–2436. doi: 10.1111/j.1538-7836.2007.02775.x. [DOI] [PubMed] [Google Scholar]
- 8.Hwang SJ, et al. The cytochrome 2C19*2 and *3 alleles attenuate response to clopidogrel similarly in East Asian patients undergoing elective percutaneous coronary intervention. Thromb Res. 2011;127:23–28. doi: 10.1016/j.thromres.2010.10.021. [DOI] [PubMed] [Google Scholar]
- 9.Wallentin L, et al. Effect of CYP2C19 and ABCB1 single nucleotide polymorphisms on outcomes of treatment with ticagrelor versus clopidogrel for acute coronary syndromes: a genetic substudy of the PLATO trial. Lancet. 2010;376:1320–1328. doi: 10.1016/S0140-6736(10)61274-3. [DOI] [PubMed] [Google Scholar]
- 10.Paré G, et al. Effects of CYP2C19 genotype on outcomes of clopidogrel treatment. N Engl J Med. 2010;363:1704–1714. doi: 10.1056/NEJMoa1008410. [DOI] [PubMed] [Google Scholar]
- 11.Fuster V, Sweeny JM. Clopidogrel and the reduced-function CYP2C19 genetic variant: a limited piece of the overall therapeutic puzzle. JAMA. 2010;304:1839–1840. doi: 10.1001/jama.2010.1566. [DOI] [PubMed] [Google Scholar]
- 12.Frére C, Cuisset T, Gaborit B, Alessi MC, Hulot JS. The CYP2C19*17 allele is associated with better platelet response to clopidogrel in patients admitted for non-ST acute coronary syndrome. J Thromb Haemost. 2009;7:1409–1411. doi: 10.1111/j.1538-7836.2009.03500.x. [DOI] [PubMed] [Google Scholar]
- 13.Sibbing D, et al. Isolated and interactive impact of common CYP2C19 genetic variants on the antiplatelet effect of chronic clopidogrel therapy. J Thromb Haemost. 2010;8:1685–1693. doi: 10.1111/j.1538-7836.2010.03921.x. [DOI] [PubMed] [Google Scholar]
- 14.Simon T, et al. Genetic determinants of response to clopidogrel and cardiovascular events. N Engl J Med. 2009;360:363–375. doi: 10.1056/NEJMoa0808227. [DOI] [PubMed] [Google Scholar]
- 15.Sibbing D, et al. Cytochrome 2C19*17 allelic variant, platelet aggregation, bleeding events, and stent thrombosis in clopidogrel-treated patients with coronary stent placement. Circulation. 2010;121:512–518. doi: 10.1161/CIRCULATIONAHA.109.885194. [DOI] [PubMed] [Google Scholar]
- 16.Tiroch KA, et al. Protective effect of the CYP2C19 *17 polymorphism with increased activation of clopidogrel on cardiovascular events. Am Heart J. 2010;160:506–512. doi: 10.1016/j.ahj.2010.06.039. [DOI] [PubMed] [Google Scholar]
- 17.Gurbel PA, Tantry US, Shuldiner AR. Letter by Gurbel et al regarding article, “Cytochrome 2C19*17 allelic variant, platelet aggregation, bleeding events, and stent thrombosis in clopidogrel-treated patients with coronary stent placement”. Circulation. 2010;122:e478. doi: 10.1161/CIRCULATIONAHA.110.943548. author reply e479. [DOI] [PubMed] [Google Scholar]
- 18.Holmes DR, Jr, Dehmer GJ, Kaul S, Leifer D, O’Gara PT, Stein CM. ACCF/AHA clopidogrel clinical alert: approaches to the FDA “boxed warning”: a report of the American College of Cardiology Foundation Task Force on clinical expert consensus documents and the American Heart Association endorsed by the Society for Cardiovascular Angiography and Interventions and the Society of Thoracic Surgeons. J Am Coll Cardiol. 2010;56:321–341. doi: 10.1016/j.jacc.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 19.Gilard M, et al. Influence of omeprazole on the antiplatelet action of clopidogrel associated with aspirin: the randomized, double-blind OCLA (Omeprazole CLopidogrel Aspirin) study. J Am Coll Cardiol. 2008;51:256–260. doi: 10.1016/j.jacc.2007.06.064. [DOI] [PubMed] [Google Scholar]
- 20.Ansell J, Hirsh J, Hylek E, Jacobson A, Crowther M, Palareti G. Pharmacology and management of the vitamin K antagonists: American College of Chest Physicians Evidence-Based Clinical Practice Guidelines (8th Edition) Chest. 2008;133:160S–198S. doi: 10.1378/chest.08-0670. [DOI] [PubMed] [Google Scholar]
- 21.Cooper GM, et al. A genome-wide scan for common genetic variants with a large influence on warfarin maintenance dose. Blood. 2008;112:1022–1027. doi: 10.1182/blood-2008-01-134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein TE, et al. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360:753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rieder MJ, et al. Effect of VKORC1 haplotypes on transcriptional regulation and warfarin dose. N Engl J Med. 2005;352:2285–2293. doi: 10.1056/NEJMoa044503. [DOI] [PubMed] [Google Scholar]
- 24.Takeuchi F, et al. A genome-wide association study confirms VKORC1, CYP2C9, and CYP4F2 as principal genetic determinants of warfarin dose. PLoS Genet. 2009;5:e1000433. doi: 10.1371/journal.pgen.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanderson S, Emery J, Higgins J. CYP2C9 gene variants, drug dose, and bleeding risk in warfarin-treated patients: a HuGEnet systematic review and meta-analysis. Genet Med. 2005;7:97–104. doi: 10.1097/01.gim.0000153664.65759.cf. [DOI] [PubMed] [Google Scholar]
- 26.Limdi NA, et al. Influence of CYP2C9 and VKORC1 1173C/T genotype on the risk of hemorrhagic complications in African-American and European-American patients on warfarin. Clin Pharmacol Ther. 2008;83:312–321. doi: 10.1038/sj.clpt.6100290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Limdi NA, et al. Warfarin pharmacogenetics: a single VKORC1 polymorphism is predictive of dose across 3 racial groups. Blood. 2010;115:3827–3834. doi: 10.1182/blood-2009-12-255992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gage BF, et al. Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther. 2008;84:326–331. doi: 10.1038/clpt.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lenzini P, et al. Integration of genetic, clinical, and INR data to refine warfarin dosing. Clin Pharmacol Ther. 2010;87:572–578. doi: 10.1038/clpt.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perera MA, et al. The missing association: sequencing-based discovery of novel SNPs in VKORC1 and CYP2C9 that affect warfarin dose in African Americans. Clin Pharmacol Ther. 2011;89:408–415. doi: 10.1038/clpt.2010.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Caldwell MD, et al. CYP4F2 genetic variant alters required warfarin dose. Blood. 2008;111:4106–4112. doi: 10.1182/blood-2007-11-122010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McDonald MG, Rieder MJ, Nakano M, Hsia CK, Rettie AE. CYP4F2 is a vitamin K1 oxidase: an explanation for altered warfarin dose in carriers of the V433M variant. Mol Pharmacol. 2009;75:1337–1346. doi: 10.1124/mol.109.054833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Borgiani P, et al. CYP4F2 genetic variant (rs2108622) significantly contributes to warfarin dosing variability in the Italian population. Pharmacogenomics. 2009;10:261–266. doi: 10.2217/14622416.10.2.261. [DOI] [PubMed] [Google Scholar]
- 34.Cha PC, et al. Genome-wide association study identifies genetic determinants of warfarin responsiveness for Japanese. Hum Mol Genet. 2010;19:4735–4744. doi: 10.1093/hmg/ddq389. [DOI] [PubMed] [Google Scholar]
- 35.Pautas E, et al. Genetic factors (VKORC1, CYP2C9, EPHX1, and CYP4F2) are predictor variables for warfarin response in very elderly, frail inpatients. Clin Pharmacol Ther. 2010;87:57–64. doi: 10.1038/clpt.2009.178. [DOI] [PubMed] [Google Scholar]
- 36.Flockhart DA, et al. Pharmacogenetic testing of CYP2C9 and VKORC1 alleles for warfarin. Genet Med. 2008;10:139–150. doi: 10.1097/GIM.0b013e318163c35f. [DOI] [PubMed] [Google Scholar]
- 37.Allabi AC, Gala JL, Horsmans Y. CYP2C9, CYP2C19, ABCB1 (MDR1) genetic polymorphisms and phenytoin metabolism in a black Beninese population. Pharmacogenet Genomics. 2005;15:779–786. doi: 10.1097/01.fpc.0000174787.92861.91. [DOI] [PubMed] [Google Scholar]
- 38.Cavallari LH, et al. Genetic and clinical predictors of warfarin dose requirements in African Americans. Clin Pharmacol Ther. 2010;87:459–464. doi: 10.1038/clpt.2009.223. [DOI] [PubMed] [Google Scholar]
- 39.Voora D, et al. A polymorphism in the VKORC1 regulator calumenin predicts higher warfarin dose requirements in African Americans. Clin Pharmacol Ther. 2010;87:445–451. doi: 10.1038/clpt.2009.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Caraco Y, Blotnick S, Muszkat M. CYP2C9 genotype-guided warfarin prescribing enhances the efficacy and safety of anticoagulation: a prospective randomized controlled study. Clin Pharmacol Ther. 2008;83:460–470. doi: 10.1038/sj.clpt.6100316. [DOI] [PubMed] [Google Scholar]
- 41.Epstein RS, et al. Warfarin genotyping reduces hospitalization rates results from the MM-WES (Medco-Mayo Warfarin Effectiveness study) J Am Coll Cardiol. 2010;55:2804–2812. doi: 10.1016/j.jacc.2010.03.009. [DOI] [PubMed] [Google Scholar]
- 42.Anderson JL, et al. Randomized trial of genotype-guided versus standard warfarin dosing in patients initiating oral anticoagulation. Circulation. 2007;116:2563–2570. doi: 10.1161/CIRCULATIONAHA.107.737312. [DOI] [PubMed] [Google Scholar]
- 43.French B, et al. Statistical design of personalized medicine interventions: the Clarification of Optimal Anticoagulation through Genetics (COAG) trial. Trials. 2010;11:108. doi: 10.1186/1745-6215-11-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ashley EA, et al. Clinical assessment incorporating a personal genome. Lancet. 2010;375:1525–1535. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Davis HM, Johnson JA. Heart failure pharmacogenetics: past, present, and future. Curr Cardiol Rep. 2011;13:175–184. doi: 10.1007/s11886-011-0181-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Terra SG, et al. beta-Adrenergic receptor polymorphisms and responses during titration of metoprolol controlled release/extended release in heart failure. Clin Pharmacol Ther. 2005;77:127–137. doi: 10.1016/j.clpt.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 47.Johnson JA, Liggett SB. Cardiovascular pharmacogenomics of adrenergic receptor signaling: clinical implications and future directions. Clin Pharmacol Ther. 2011;89:366–378. doi: 10.1038/clpt.2010.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liggett SB, et al. A polymorphism within a conserved beta(1)-adrenergic receptor motif alters cardiac function and beta-blocker response in human heart failure. Proc Natl Acad Sci USA. 2006;103:11288–11293. doi: 10.1073/pnas.0509937103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bristow MR, et al. An alpha2C-adrenergic receptor polymorphism alters the norepinephrine-lowering effects and therapeutic response of the beta-blocker bucindolol in chronic heart failure. Circ Heart Fail. 2010;3:21–28. doi: 10.1161/CIRCHEARTFAILURE.109.885962. [DOI] [PubMed] [Google Scholar]
- 50.Lobmeyer MT, et al. Synergistic polymorphisms of beta1 and alpha2C-adrenergic receptors and the influence on left ventricular ejection fraction response to beta-blocker therapy in heart failure. Pharmacogenet Genomics. 2007;17:277–282. doi: 10.1097/FPC.0b013e3280105245. [DOI] [PubMed] [Google Scholar]
- 51.Liggett SB, et al. A GRK5 polymorphism that inhibits beta-adrenergic receptor signaling is protective in heart failure. Nat Med. 2008;14:510–517. doi: 10.1038/nm1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cresci S, et al. Clinical and genetic modifiers of long-term survival in heart failure. J Am Coll Cardiol. 2009;54:432–444. doi: 10.1016/j.jacc.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.ARCA Biopharma. ARCA announces special protocol assessment agreement with FDA for bucindolol development in genotype-defined heart failure population. http://phx.corporate-ir.net/phoenix.zhtml?c=109749&p=irol-newsArticle&ID=1427666&highlight=
- 54.Pirmohamed M. Pharmacogenetics of idiosyncratic adverse drug reactions. In: Vetrecht J, editor. Adverse Drug Reactions. Vol. 196. Springer; Berlin, Germany: 2010. pp. 477–491. [DOI] [PubMed] [Google Scholar]
- 55.Wilke RA, Moore JH, Burmester JK. Relative impact of CYP3A genotype and concomitant medication on the severity of atorvastatin-induced muscle damage. Pharmacogenet Genomics. 2005;15:415–421. doi: 10.1097/01213011-200506000-00007. [DOI] [PubMed] [Google Scholar]
- 56.Pasanen MK, Neuvonen M, Neuvonen PJ, Niemi M. SLCO1B1 polymorphism markedly affects the pharmacokinetics of simvastatin acid. Pharmacogenet Genomics. 2006;16:873–879. doi: 10.1097/01.fpc.0000230416.82349.90. [DOI] [PubMed] [Google Scholar]
- 57.Link E, et al. SLCO1B1 variants and statin-induced myopathy–a genomewide study. N Engl J Med. 2008;359:789–799. doi: 10.1056/NEJMoa0801936. [DOI] [PubMed] [Google Scholar]
- 58.Donnelly LA, et al. Common nonsynonymous substitutions in SLCO1B1 predispose to statin intolerance in routinely treated individuals with type 2 diabetes: a go-DARTS study. Clin Pharmacol Ther. 2011;89:210–216. doi: 10.1038/clpt.2010.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Voora D, et al. The SLCO1B1*5 genetic variant is associated with statin-induced side effects. J Am Coll Cardiol. 2009;54:1609–1616. doi: 10.1016/j.jacc.2009.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Roden DM. Long QT syndrome: reduced repolarization reserve and the genetic link. J Intern Med. 2006;259:59–69. doi: 10.1111/j.1365-2796.2005.01589.x. [DOI] [PubMed] [Google Scholar]
- 61.Paulussen AD, et al. Genetic variations of KCNQ1, KCNH2, SCN5A, KCNE1, and KCNE2 in drug-induced long QT syndrome patients. J Mol Med. 2004;82:182–188. doi: 10.1007/s00109-003-0522-z. [DOI] [PubMed] [Google Scholar]
- 62.Yang P, et al. Allelic variants in long-QT disease genes in patients with drug-associated torsades de pointes. Circulation. 2002;105:1943–1948. doi: 10.1161/01.cir.0000014448.19052.4c. [DOI] [PubMed] [Google Scholar]
- 63.Itoh H, et al. Latent genetic backgrounds and molecular pathogenesis in drug-induced long QT syndrome. Circ Arrhythm Electrophysiol. 2009;2:511–523. doi: 10.1161/CIRCEP.109.862649. [DOI] [PubMed] [Google Scholar]
- 64.Roden DM, et al. Cardiovascular pharmacogenomics. Circ Res. doi: 10.1161/CIRCRESAHA.110.230995. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pacanowski MA, et al. Beta-adrenergic receptor gene polymorphisms and beta-blocker treatment outcomes in hypertension. Clin Pharmacol Ther. 2008;84:715–721. doi: 10.1038/clpt.2008.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Manunta P, et al. Physiological interaction between alpha-adducin and WNK1-NEDD4L pathways on sodium-related blood pressure regulation. Hypertension. 2008;52:366–372. doi: 10.1161/HYPERTENSIONAHA.108.113977. [DOI] [PubMed] [Google Scholar]
- 67.Sciarrone MT, et al. ACE and alpha-adducin polymorphism as markers of individual response to diuretic therapy. Hypertension. 2003;41:398–403. doi: 10.1161/01.HYP.0000057010.27011.2C. [DOI] [PubMed] [Google Scholar]
- 68.Lanzani C, et al. Adducin- and ouabain-related gene variants predict the antihypertensive activity of rostafuroxin, part 2: clinical studies. Sci Transl Med. 2010;2:59, ra87. doi: 10.1126/scitranslmed.3001814. [DOI] [PubMed] [Google Scholar]
- 69.Svensson-Färbom P, et al. A functional variant of the NEDD4L gene is associated with beneficial treatment response with β-blockers and diuretics in hypertensive patients. J Hypertens. 2011;29:388–395. doi: 10.1097/HJH.0b013e3283410390. [DOI] [PubMed] [Google Scholar]
- 70.Turner ST, et al. Genomic association analysis suggests chromosome 12 locus influencing antihypertensive response to thiazide diuretic. Hypertension. 2008;52:359–365. doi: 10.1161/HYPERTENSIONAHA.107.104273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Beitelshees AL, et al. KCNMB1 genotype influences response to verapamil SR and adverse outcomes in the International Verapamil SR/Trandolapril Study (INVEST) Pharmacogenet Genomics. 2007;17:719–729. doi: 10.1097/FPC.0b013e32810f2e3c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Beitelshees AL, et al. CACNA1C gene polymorphisms, cardiovascular disease outcomes, and treatment response. Circ Cardiovasc Genet. 2009;2:362–370. doi: 10.1161/CIRCGENETICS.109.857839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Niu Y, et al. Genetic variation in the beta2 subunit of the voltage-gated calcium channel and pharmacogenetic association with adverse cardiovascular outcomes in the INternational VErapamil SR-Trandolapril STudy GENEtic Substudy (INVEST-GENES) Circ Cardiovasc Genet. 2010;3:548–555. doi: 10.1161/CIRCGENETICS.110.957654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Faraday N, et al. Heritability of platelet responsiveness to aspirin in activation pathways directly and indirectly related to cyclooxygenase-1. Circulation. 2007;115:2490–2496. doi: 10.1161/CIRCULATIONAHA.106.667584. [DOI] [PubMed] [Google Scholar]
- 75.Goodman T, Ferro A, Sharma P. Pharmacogenetics of aspirin resistance: a comprehensive systematic review. Br J Clin Pharmacol. 2008;66:222–232. doi: 10.1111/j.1365-2125.2008.03183.x. [DOI] [PMC free article] [PubMed] [Google Scholar]