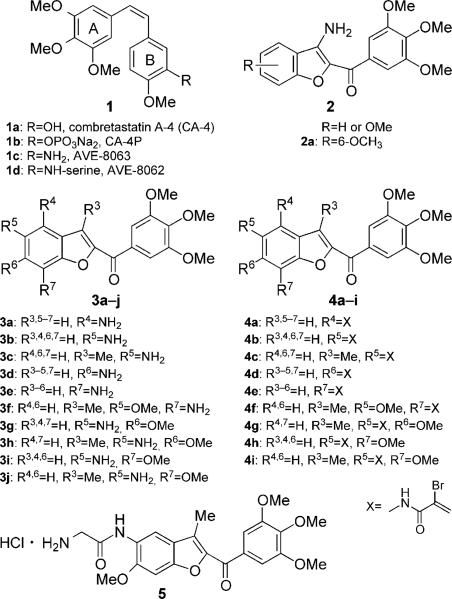
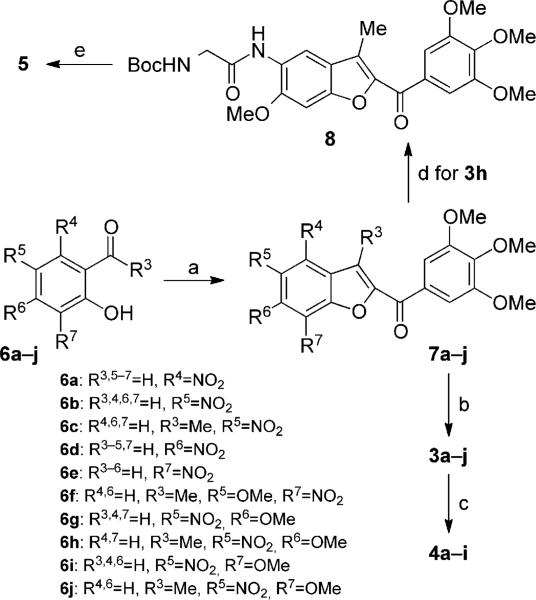
Abstract
Induction of apoptosis is a promising strategy that could lead to the discovery of new molecules active in cancer chemotherapy. This property is generally observed when cells are treated with agents that target microtubules, dynamic structures that play a crucial role in cell division. Small molecules such as benzo[b]furans are attractive as inhibitors of tubulin polymerization. A new class of inhibitors of tubulin polymerization based on the 2-(3′,4′,5′-trimethoxybenzoyl)benzo[b]furan molecular skeleton, with the amino group placed at different positions on the benzene ring, were synthesized and evaluated for antiproliferative activity, inhibition of tubulin polymerization, and cell-cycle effects. The methoxy substitution pattern on the benzene portion of the benzo[b]furan moiety played an important role in affecting antiproliferative activity. In the series of 5-amino derivatives, the greatest inhibition of cell growth oc curred if the methoxy substituent is placed at the C6 position, whereas C7 substitution decreases potency. The most promising compound in this series is 2-(3′,4′,5′-trimethoxybenzoyl)-3-methyl-5-amino-6-methoxybenzo[b]furan (3h), which inhibits cancer cell growth at nanomolar concentrations (IC50=16–24 nm), and interacts strongly with tubulin by binding to the colchicine site. Sub-G1 apoptotic cells in cultures of HL-60 and U937 cells were observed by flow cytometric analysis after treatment with 3h in a concentration-dependent manner. We also show that compound 3h induces apoptosis by activation of caspase-3, -8, and -9, and this is associated with cytochrome c release from mitochondria. The introduction of an α-bromoacryloyl group increased antiproliferative activity with respect to the parent amino derivatives.
Keywords: antiproliferative agents, apoptosis, caspase activation, colchicine binding site, tubulin
Introduction
Apoptosis, programmed cell death, plays a critical role in normal cell development and is a well-regulated mechanism to eliminate damaged and dysfunctional cells.[1] This process was recently recognized as an important phenomenon for successful cancer chemotherapy[2] and identification of compounds that activate apoptosis is recognized as a promising strategy for the discovery of potential anticancer agents.[3]
Antimitotic agents are a major class of cytotoxic drugs for cancer treatment, and tubulin is the target for numerous small natural and synthetic molecules that inhibit the formation of the mitotic spindle.[4] The microtubule system of eukaryotic cells is a critical element in a variety of essential cellular processes in addition to mitotic spindle assembly, including determination and maintenance of cell shape, regulation of motility, cell signaling, secretion, and intracellular transport.[5]
Among the natural microtubule depolymerizing agents, combretastatin A-4 (CA-4, 1a, Figure 1) is one of the more studied compounds. CA-4, isolated from the bark of the South African tree Combretum caffrum,[6] strongly inhibits the polymerization of tubulin by binding to the colchicine site.[7] CA-4 inhibits cell growth at low- to mid-nanomolar concentrations. The water-soluble sodium phosphate prodrug of CA-4 (CA-4P, fosbretabulin, or Zybrestat, 1b) has yielded promising results in human clinical trials.[8] Because of its structural simplicity, along with its ability to selectivity damage tumor neovasculature, numerous CA-4 analogues have been developed and evaluated in structure–activity relationship (SAR) studies.[9]
Figure 1.
Lead structures of tubulin polymerization inhibitors.
The 3′-hydroxy group of the CA-4 B ring can be replaced by an amine to furnish the 3′-amino derivative 1c, named AVE-8063 or AC-7739, with no substantial loss of activity. This compound is a potent inhibitor of tubulin assembly and also showed significant cytotoxicity against human cancer cell lines.[10] The corresponding amido serine prodrugs (1d, AVE-8062 or Ambrabulin) and CA-4P are potent vascular disrupting agents (VDA) and have been demonstrated to cause vascular shutdown in established tumors in vivo.[11] AVE-6062 was found to have more potent activity compared with CA-4P and is currently undergoing phase III clinical trials for the treatment of solid tumors.[12]
As a part of our continuing search for synthetic small molecule tubulin inhibitors, we previously reported a series of methoxy-substituted 2-(3′,4′,5′-trimethoxybenzoyl)-3-aminobenzo[b]furan derivatives, with general structure 2, as a new class of antimitotic agents.[9a] In this series of compounds, potent activity was highly dependent on the presence and position of four methoxy substituents.[13] A fairly dramatic difference was observed between C4/5 and C6/7-methoxy substituted compounds. The greatest activity occurred when the methoxy group was located at the 6- or 7-position, whereas 4- and 5-methoxy derivatives were inactive. Compound 2a, with a methoxy group at the 6-position, displayed potent growth inhibition of different cancer cell lines. This agent inhibited tubulin polymerization and caused G2/M phase arrest of the cell cycle. These results prompted us to study this class of compounds in further detail.
Herein we describe the synthesis and SAR of new 2-(3′,4′,5′-trimethoxybenzoyl)benzo[b]furan derivatives, some of which are potent antitubulin agents. In this series, the 3′,4′,5′-trimethoxyphenyl of the 2-benzoyl moiety was kept unchanged because it is the characteristic structural requirement for activity in numerous inhibitors of tubulin polymerization, such as colchicine, CA-4, and podophyllotoxin.[14]
In the first series of new derivatives described herein, the amino group of the compound with general structure 2 was moved from the 3-position to each of the four possible positions of the benzene portion of the benzo[b]furan skeleton, to obtain compounds 3a–e, with the concomitant presence of an additional methoxy group at the 5- (3 f), 6- (3g and 3h), or 7-(3i and 3j) positions.
With the aim of optimizing the potency of this first class of compounds, we also synthesized a second series of molecules (4a–i), characterized by the presence of a Michael acceptor, corresponding to an α-bromoacryloyl alkylating moiety of low chemical reactivity, an unusual feature for cytotoxic compounds.[15] The reactivity of the α-bromoacryoyl moiety has been hypothesized to be based on a first-step Michael-type nucleophilic attack, followed by a further reaction of the former vinylic bromo substituent alpha to the carbonyl, leading successively either to a second nucleophilic substitution or to beta elimination.[16]
An approach to increase water solubility is the preparation of amino acid prodrugs, that release the parent drug following enzymatic cleavage.[17] In the series of compounds described herein, the presence of an amino moiety provides a site for preparation of amino acid prodrugs, similar to what was successfully done in the case of AVE-8062. Thus, we synthesized the glycine amino acid prodrug of compound 3h, compound 5.
Results and Discussion
Chemistry
The nitro 2-(3′,4′,5′-trimethoxybenzoyl)benzo[b]furan derivatives 7a–j were synthesized in good yield by a “one-step” cyclization reaction of the corresponding, variously substituted nitrosalicylaldehydes 6a, b, d, e, g, and i or 2-hydroxyacetophenones 6c, f, h, and j with 2-bromo-1-(3′,4′,5′-trimethoxyphenyl)ethanone and anhydrous potassium carbonate in acetone at reflux (Scheme 1).[18] Subsequent reduction of the aromatic nitro group with iron in a mixture of 37% HCl in water and ethanol at reflux afforded the corresponding amino derivatives 3a–j. The hybrid compounds 4a–i were prepared in acceptable yields by the condensation of α-bromoacrylic acid with amino benzo[b]furan derivatives 4a–f and 4h, i, and j using an excess (2 equiv) of both EDCI and 1-HOBt in dry DMF as solvent at room temperature and with identical reaction times (18 h). The glycine prodrug 5 was synthesized by coupling the amino derivative 3h with N-Bocglycine using EDCI and HOBt as coupling agents, followed by cleavage of the N-Boc protecting group with 3m HCl in EtOAc.
Scheme 1.
Synthesis of compounds 3a–j, 4a–i, and 5. Reagents and conditions: a) 1-(3,4,5-trimethoxyphenyl)-2-bromoethanone, K2CO3, (CH3)2CO, reflux, 18 h; b) Fe, HCl (37% in H2O), EtOH, reflux, 3 h; c) α-bromoacrylic acid, EDCI, HOBt, DMF, RT, 18 h; d) N-Boc-glycine, EDCI, HOBt, DMF, RT, 12 h; e) 3m HCl in EtOAc, RT, 3 h.
Biology
Table 1 summarizes the growth inhibitory effects of 2-(3′,4′,5′-trimethoxy)benzo[b]furan derivatives 3a–j, 4a–i, and 5 against murine leukemia (L1210), murine mammary carcinoma (FM3A), human T-lymphoblastoid (Molt/4 and CEM), and human cervix carcinoma (HeLa) cells, with CA-4 (1) and 2a employed as reference compounds. The 3-methyl-5-amino-6-methoxy derivative 3h was the most active compound identified in this study, inhibiting the growth of L1210, FM3A, Molt/4, CEM, and HeLa cells with IC50 values of 19, 24, 22, 22, and 16 nm, respectively. Compound 3h was twofold more potent than CA-4 against FM3A cells. The two compounds were equipotent against Molt/4 cells, but 3h was much less active than CA-4 against L1210, CEM, and HeLa cells. Moving the amino group from the 3- to the 5-position (compound 2a versus 3g) led to increased activity against L1210, FM3A, and Molt/4 cells, whereas these derivatives had similar activity against CEM cells. These effects were more evident in the 3-methyl counterpart 3h, which was 4–20-fold more active than 2a. Thus, the 3-amino position cannot be considered essential for activity. SAR information derived from comparing the 3-unsubstituted derivatives 3b, g, and i with their 3-methyl congeners 3c, h, and j showed that introduction of the methyl almost always increased antiproliferative activity against these five cell lines.
Table 1.
In vitro inhibitory effects of compounds 2a, 3a–j, 4a–i, and CA-4 against the proliferation of murine leukemia (L1210), murine mammary carcinoma (FM3A), human T-lymphocyte (Molt/4 and CEM), and human cervix carcinoma (HeLa) cells.
| Compd | L1210 | FM3A/0 | IC50 [nm][a] Molt4/C8 | CEM/0 | HeLa |
|---|---|---|---|---|---|
| 3a | 6700 ± 600 | 9200 ± 800 | ND | 5400 ± 1200 | 2200 ± 600 |
| 3b | > 10 000 | > 10 000 | > 10 000 | > 10 000 | > 10 000 |
| 3c | 2500 ± 100 | 3600 ± 150 | ND | 2400 ± 400 | 5500 ± 530 |
| 3d | > 10 000 | > 10 000 | > 10 000 | > 10000 | 7500±200 |
| 3e | > 10 000 | > 10 000 | > 10 000 | > 10 000 | > 10 000 |
| 3f | > 10 000 | > 10 000 | > 10 000 | > 10 000 | > 10 000 |
| 3g | 73 ± 24 | 73 ± 2 | 66 ± 6.7 | 59 ± 4.1 | 42 ± 3.8 |
| 3h | 19 ± 2 | 24 ± 6 | 22 ± 4 | 22 ± 5 | 16 ± 1 |
| 3i | 600 ± 90 | 560 ± 90 | 870 ± 20 | 450 ± 10 | 370 ± 30 |
| 3j | 67 ± 4 | 140 ± 13 | 360 ± 20 | 120 ± 10 | 490 ± 54 |
| 4a | 78 ± 2.8 | 200 ± 90 | ND | 110 ± 20 | 1000 ± 80 |
| 4b | 150 ± 8 | 320 ± 30 | 230 ± 1 | 390 ± 30 | 1100 ± 90 |
| 4c | 320 ± 17 | 520 ± 29 | ND | 310 ± 80 | 430 ± 90 |
| 4d | 420 ± 90 | 1200 ± 90 | ND | 600 ± 37 | 1200 ± 80 |
| 4e | 800 ± 51 | 780 ± 62 | 1100 ± 90 | 970 ± 55 | 1000 ± 100 |
| 4f | 1200 ± 40 | 5900 ± 380 | ND | 3200 ± 240 | 5900 ± 900 |
| 4g | 430 ± 23 | 960 ± 78 | ND | 340 ± 20 | 1000 ± 100 |
| 4h | 120 ± 14 | 250 ± 15 | 300 ± 40 | 240 ± 38 | 780 ± 54 |
| 4i | 390 ± 20 | 1100 ± 100 | 920 ± 91 | 770 ± 15 | 1400 ± 120 |
| 5 | 14 ± 1 | 10 ± 8 | 18 ± 1 | 20 ± 2 | 14 ± 1 |
| 2a | 430 ± 40 | 280 ± 16 | 140 ± 20 | 87 ± 22 | ND |
| CA-4 | 2.8 ± 1.1 | 42 ± 6.0 | 16 ± 1.4 | 1.9 ± 1.6 | 1.9 ± 1.6 |
Compound concentration required to inhibit tumor cell proliferation by 50%; data are expressed as the mean ± SE from the dose–response curves of at least three independent experiments; ND: not determined.
In the series of amino monosubstituted derivatives 3a, 3b, 3d, and 3e, only the 4-amino isomer 3a exhibited antiproliferative activity below 10 μm (IC50=2.2–9.2 μm). Starting from the 4-amino derivative 3b, addition of a C3 methyl group to furnish 3c, caused an increase in activity. The activity of these compounds was further influenced by the presence and position of a methoxy group in the benzene portion of the benzo[b]furan skeleton. In comparing the C6 and C7 methoxy derivatives, the greatest activity occurred with the methoxy group located at C6 (compare the activities of C6 methoxy derivatives 3g and h with those of C7 methoxy congeners 3i and j). As predicted from the results described above, the 3-methyl analogues 3h and j were more potent than their un-substituted counterparts 3g and i. In keeping with the findings described above, derivative 3h, with a C6 methoxy group, was two- to fourfold more potent than the 3-unsubstituted analogue 3g and 3–30-fold more active than 3j, with a C7 methoxy group. Starting from 3j, switching the positions of the amino and methoxy groups, to yield 3 f, resulted in a dramatic loss of activity. Comparing compounds 3g and 3i further confirmed the superior antiproliferative activity of having a C6 methoxy group, as in 3g, as opposed to a C7 methoxy group, as in 3i.
Further SAR information that can be derived from 3g and 3h is that the ortho relationship between the amino and the methoxy groups plays an essential role in maximal activity, perhaps mimicking the B ring of aminocombretastatin 1c. In conclusion, the introduction of an amino group at C5 of 2-(3′,4′,5′-trimethoxybenzoyl)benzo[b]furan and a methoxy group at C6 are important for maximal cytotoxicity.
Starting with compounds 3a–f, linking the α-bromoacryloyl moiety to the amino group furnished derivatives 4a–f and led to a substantial increase in antiproliferative activity in all cases. However, starting with the methoxy substituted analogues 3h, i, and j, the introduction of the α-bromoacryloyl moiety to produce 4g, h, and i, the effects were less dramatic. In fact, 4g and 4i had less activity than their congeners 3h and 3j, whereas the conversion of 3i to 4h led to only small increases in activity in four cell lines. For the unsubstituted α-bromoacryloylamido derivatives 4a, b, d, and e, the greatest activity occurred when the α-bromoacryloylamido moiety was located at C4 or C5, the least at C6 or C7 except that the four compounds had similar activity against HeLa cells. Finally, there are no differences in antiproliferative activities in all cell lines between 3h and its glycine hydrochloride prodrug 5.
To investigate whether the antiproliferative activities of these compounds were related to an interaction with the microtubule system, the most active compounds, 3g, 3h, 3i, 3j, and 4a, were evaluated for their in vitro inhibition of tubulin polymerization and for their inhibitory effects on the binding of [3H]colchicine to tubulin (in the former assay, tubulin was at 10 μm; in the latter assay, tubulin was at 1 μm, whereas the compounds and colchicine were at 5 μm).[19] CA-4 was examined in simultaneous experiments as a reference compound (Table 2). The derivative 3h, with an IC50 value of 0.56 μm, exhibited antitubulin activity approximately twofold greater than that of CA-4 (1.0 μm), whereas 3g and 3j, with IC50 values of 1.4 and-1.6 μm, respectively, were slightly less potent than CA-4. Compound 4a did not greatly alter tubulin assembly at a concentration as high as 40 μm. Compound 3i was half as active as 3j. Thus, it is probable that the mechanism of action of the most potent α-bromoacryloylamido derivative 4a did not involve tubulin.
Table 2.
Inhibition of tubulin polymerization and colchicine binding by compounds 3g, 3h, 3i, 3j, 4a, and CA4.
| Compd | IC50 ± SD [μm][a] | Colch. bind. ± SD [%][b] |
|---|---|---|
| 3g | 1.4 ± 0.01 | 72 ± 0.5 |
| 3h | 0.56 ± 0.08 | 96 ± 0.09 |
| 3i | 3.1 ± 0.2 | 56 ± 6 |
| 3j | 1.6 ± 0.1 | 49 ± 0.5 |
| 4a | > 40 | ND |
| CA-4 (1a) | 1.0 ± 0.1 | 99 ± 3 |
Inhibition of tubulin polymerization; the concentration of tubulin was 10 μm.
Inhibition of [3H]colchicine binding; tubulin, colchicine, and test compound were at 1, 5, and 5 μm, respectively
ND: not determined.
The order of inhibitory effects on tubulin assembly was 3h > CA-4 > 3g > 3j > 3i ≫ 4a, which was consistent with the results of the antiproliferative assays, except that 4a was more cytostatic than 3i. The most potent compound in this series was compound 3h, with a tubulin polymerization IC50 value of 0.56 μm. This is in agreement with 3h being the compound with the greatest antiproliferative activity.
In the colchicine binding studies, compound 3h strongly inhibited the binding of [3H]colchicine to tubulin as 96% inhibition occurred when both this agent and colchicine were at a concentration of 5 μm. This derivative was as active as CA-4, which in these experiments inhibited colchicine binding by 99%. These data indicate that 3h strongly binds to the colchicine site on tubulin. These data are consistent with the conclusion that tubulin is the intracellular target of compounds 3g–j.
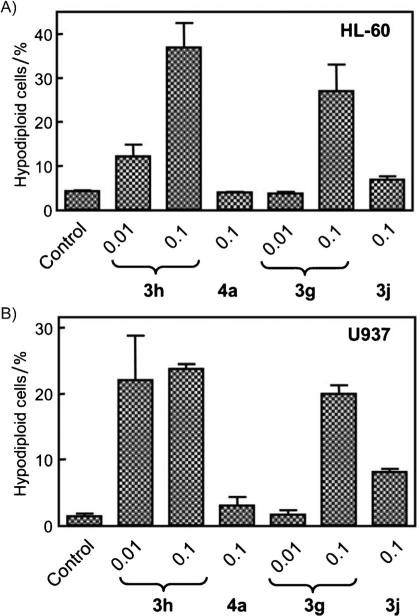
Once the antiproliferative and antitubulin activities were determined, compounds 3g, 3h, 3j, and 4a were selected for further evaluation on their effects on human myeloid leukemia HL-60 and U937 cell-cycle distribution by flow cytometric analysis. Table 3 shows, for each compound, the percentage of cells in each phase of the cell cycle, whereas Figure 2 summarizes the fractions of cells in the sub-G1 peak, which represents apoptotic cells. Cells were cultured for 24 h with each compound at 100 nm, and the two most active agents (3g and 3h) were also examined at 10 nm. These data show that 100 nm 4a had no effect on cell-cycle distribution, consistent with the conclusion above that the compound did not target tubulin. With 100 nm 3j, only a modest increase in apoptotic cells was observed. Compounds 3g and 3h had different cell-cycle effects in the two cell lines.
Table 3.
Effect of compounds 3g, 3h, 3j, and 4a on cell-cycle phase distribution of human myeloid leukemia HL-60 and U937 cells.
| Compd | Conc. [μm] | Cell-cycle phase [%] | |||
|---|---|---|---|---|---|
| Sub-G1 | G1 | S | G2/M | ||
| HL-60 [a] | |||||
| Control | – | 4.2 ± 0.2 | 45.8 ± 0.2 | 12.7 ± 0.8 | 33.3 ± 3.1 |
| 3g | 0.01 | 3.7 ± 0.3 | 45.5 ± 0.1 | 12.3 ± 2.0 | 34.8 ± 3.0 |
| 3g | 0.1 | 27.0 ± 6.0 | 25.9 ± 9.7 | 9.8 ± 0.7 | 20.8 ± 3.2 |
| 3h | 0.01 | 12.1 ± 2.7 | 35.0 ± 0.8 | 11.9 ± 0.3 | 32.3 ± 7.6 |
| 3h | 0.1 | 36.9 ± 5.6 | 30.0 ± 6.0 | 10.0 ± 4.0 | 24.0 ± 5.0 |
| 3j | 0.1 | 6.8 ± 0.8 | 41.9 ± 3.2 | 13.1 ±1.3 | 33.1 ± 7.0 |
| 4a | 0.1 | 3.9 ± 0.1 | 43.0 ± 1.0 | 12.5 ± 1.3 | 36.8 ± 1.7 |
| U937 [a] | |||||
| Control | – | 1.4 ± 0.4 | 47.2 ± 2.5 | 24.7 ± 0.6 | 26.8 ± 4.1 |
| 3g | 0.01 | 1.6 ± 0.7 | 42.9 ± 2.4 | 24.5 ± 2.3 | 32.3 ± 1.1 |
| 3g | 0.1 | 20.0 ± 1.3 | 13.8 ± 0.3 | 10.5 ± 0.6 | 49.6 ± 0.9 |
| 3h | 0.01 | 22.1 ± 6.8 | 26.2 ± 11.8 | 17.2 ± 1.6 | 32.3 ± 5.4 |
| 3h | 0.1 | 23.8 ± 0.7 | 17.2 ± 0.7 | 14.5 ± 1.8 | 38.3 ± 2.0 |
| 3j | 0.1 | 8.1 ± 0.4 | 44.0 ± 1.4 | 19.7 ± 0.2 | 28.6 ± 1.5 |
| 4a | 0.1 | 3.0 ± 1.3 | 49.9 ± 3.8 | 23.5 ± 1.5 | 24.5 ± 1.5 |
Cells were cultured with the indicated concentrations of test compounds for 24 h, and the cell-cycle phase distribution was determined by flow cytometry; values represent the mean ± SE of two independent experiments with three determinations in each.
Figure 2.
A) HL-60 and B) U937 cells were treated with the indicated concentrations (in μm) of compounds 3g, 3h, 3j, and 4a for 24 h, and apoptosis was evaluated by flow cytometry. Values represent the mean ±SE of three independent experiments, each performed in triplicate.
In HL-60 cells, 3g caused an increase in apoptotic cells only at 100 nm, whereas 3h was effective at both concentrations. Neither compound, however, caused the increase in G2/M cells frequently observed with antitubulin agents. In fact, the increase in apoptotic cells seemed to come partially at the expense of cells in the G2/M phase, as well as cells in the G1 phase of the cell cycle.
In the U937 cells, there were concentration-dependent increases in both the apoptotic and G2/M fractions with both 3g and 3h. These increases were accompanied by decreases in both the G1 and S fractions. In this cell line, the changes were those expected for treatment with antitubulin agents.
The detection of a significant increase in sub-G1 peak upon incubation with compounds 3g and 3h in a concentration-dependent manner in both cell lines, suggested that they exert their growth inhibiting effect by induction of apoptosis. We then examined further the apoptotic effects of compounds 3g, 3h, 3j, and 4a in the two cell lines.
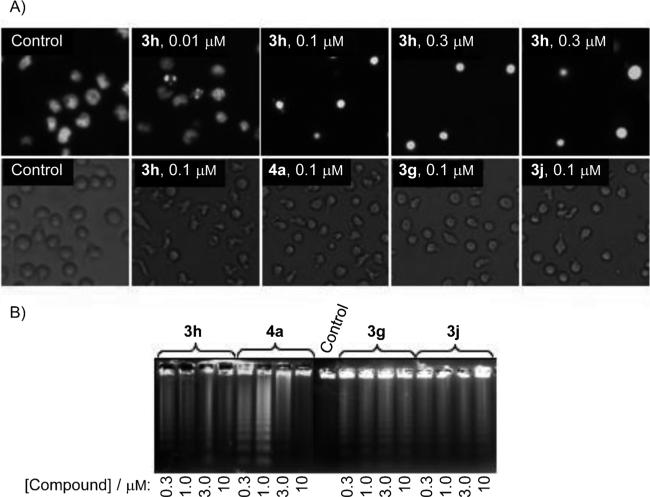
As shown in Figure 3A (upper row), using fluorescence microscopy to analyze the morphological changes of U937 cells, compound 3h induced the appearance of condensed and fragmented chromatin characteristic of apoptotic cell death at various concentrations.[20] Control U937 cells showed normal features, with the nuclei round and homogeneous. Also compounds 3g, 3j, and 4a induced important morphological changes characteristic of apoptotic cells, as visualized by phase-contrast microscopy (lower row).
Figure 3.
Morphological changes and apoptosis induction by compounds 3g, 3h, 3j, and 4a on human leukemia cells. A) Upper row: photomicrographs of representative fields of U937 cells stained with Hoechst 33258 to evaluate nuclear chromatin condensation (apoptosis) after treatment with the indicated concentrations of compound 3h for 6 h. Lower row: U937 cells were incubated with medium alone (control) or the indicated compounds; images of cells in culture were obtained with an inverted phase-contrast microscope. B) HL-60 cells were incubated with the indicated compounds at the indicated concentrations for 6 h, and genomic DNA was extracted, separated on an agarose gel, and visualized under UV light by ethidium bromide staining.
Degradation of DNA into a specific fragmentation pattern is a characteristic feature of apoptosis.[21] We also examined whether these compounds induced chromosomal DNA fragmentation, which is considered the end point of the apoptotic pathway resulting from activation of caspase-activated endonuclease. As we expected, when human HL-60 cells were incubated at the indicated concentrations of compounds 3g, 3h, 3j, and 4a, the DNA showed the typical fragmentation patterns formed by internucleosomal hydrolysis of chromatin, which then led to the appearance of DNA ladders when separated by agarose gel electrophoresis (Figure 3B).
Apoptosis is mediated by the activation of a family of cysteine aspartyl-specific proteases known as caspases, expressed as inactive pro-enzymes in living cells that are sequentially activated by specific proteolytic cleavage.[22] Caspase-8 is the main executer of the extrinsic pathway initiated at the plasma membrane by activation of cell surface death receptors, whereas caspase-9 mediates the mitochondrial pathway. Both caspases, called apical caspases, are usually the first to be activated in the apoptotic process, and they subsequently activate downstream caspase-3, which finally causes apoptosis.[23]
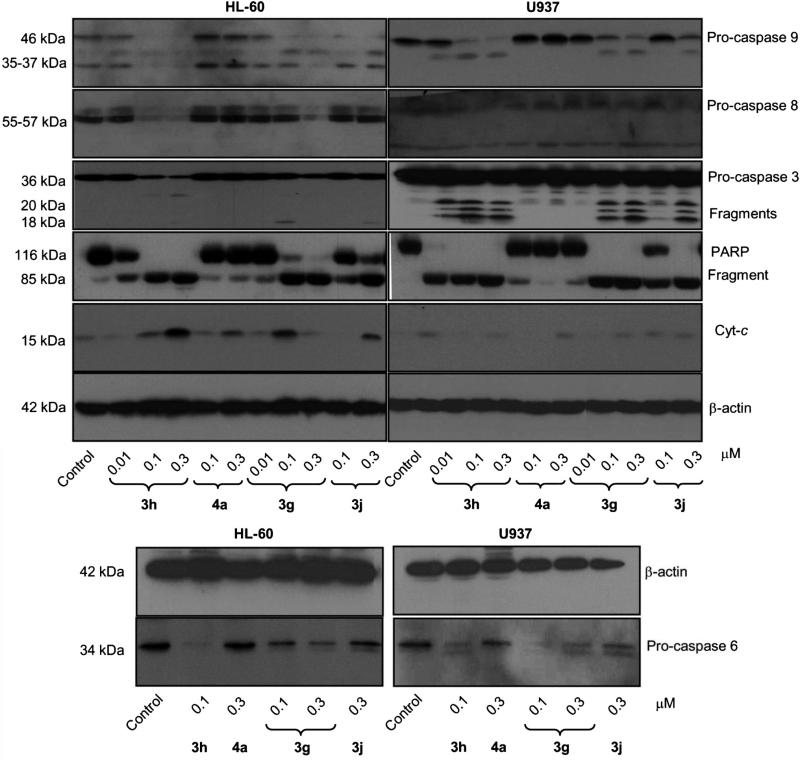
As the proteolytic processing of caspases is an important event in caspase-dependent apoptotic cell death, we evaluated the effect of 3g, 3h, 3j, and 4a on caspases. To this end, HL-60 and U937 cells were treated with the compounds, and the initiator (caspase-8 and -9) and executioner (caspase-3 and -6) caspases were determined by Western blot analysis (Figure 4). We found that 3g, 3h, 3j, and 4a stimulated the cleavage of inactive procaspase-9 to the active 37 kDa fragment, and also, that concentrations of compounds 3g and 3h as low as 0.3 μm significantly promoted procaspase-8 hydrolysis. As expected, compounds 3g and 3h significantly promoted the cleavage of inactive pro-caspases-3 and -6 in both cell lines (Figure 4). Among the different caspases, caspase-3 is considered to be the key mediator of apoptosis induced by cytotoxic agents. The executioner caspase-3 is responsible for the physiological (for example, cleavage of the DNA repair enzyme poly(ADP-ribose) polymerase-1, nuclear lamins, and cytoskeleton proteins) and morphological changes (DNA-strand breaks, nuclear membrane damage, and membrane blebbing) that occur in apoptosis.[23a]
Figure 4.
Involvement of caspases in the induction of apoptosis in human leukemia cells. Cells were incubated in the presence of the indicated concentrations of compounds 3g, 3h, 3j, or 4a, and cell lysates were assayed by immunoblotting for the cleavage of pro-caspases-9, -8, -6, and -3, poly(ADP-ribose) polymerase (PARP), and cytochrome c release. β-Actin was used as a loading control.
To determine whether the procaspase-3 processing was associated with an increase in enzymatic activity, we also examined the induction of cleavage of poly(ADP-ribose) polymerase (PARP), a known nuclear enzyme substrate of caspase-3 that plays an important role in DNA repair.[24] Hydrolysis of PARP guarantees cellular disassembly and supports a role for this protein as a key regulator of apoptosis in cells treated with tested compounds.[25] As shown in Figure 4, a concentration-dependent hydrolysis of the 116 kDa PARP protein to the 85 kDa fragment was detected after treatment with compounds 3g and 3h. Compounds 3j and 4a were less potent than compounds 3g and 3h in inducing PARP cleavage. As PARP is a substrate of caspase-3, the appearance of the 85 kDa fragment coincided with the activation of caspase-3, as shown by the decreased level of the 36 kDa pro-enzyme and the increased levels of the 20 and 18 kDa cleaved procaspase-3.
Mitochondria play a key role in cell death when their membranes become permeabilized. Outer mitochondrial membrane permeabilization is regulated by different members of the Bcl-2 family, which constitutes a critical cellular checkpoint in the intrinsic apoptotic pathway.[26] Release of cytochrome c from the mitochondria to the cytosol is a central event in apoptotic signaling.[27] To evaluate whether apoptosis induced by compounds 3g, 3h, 3j, and 4a on HL-60 and U937 cells involves cytochrome c release from mitochondria to cytosol, dose-response experiments were performed, and cytosolic preparations were analyzed by immunoblotting. Membranes were stripped and re-probed with β-actin antibody as a loading control. We found a significant increase in the amount of cyto-chrome c in the cytosol in both cell lines (15 kDa band) (Figure 4).
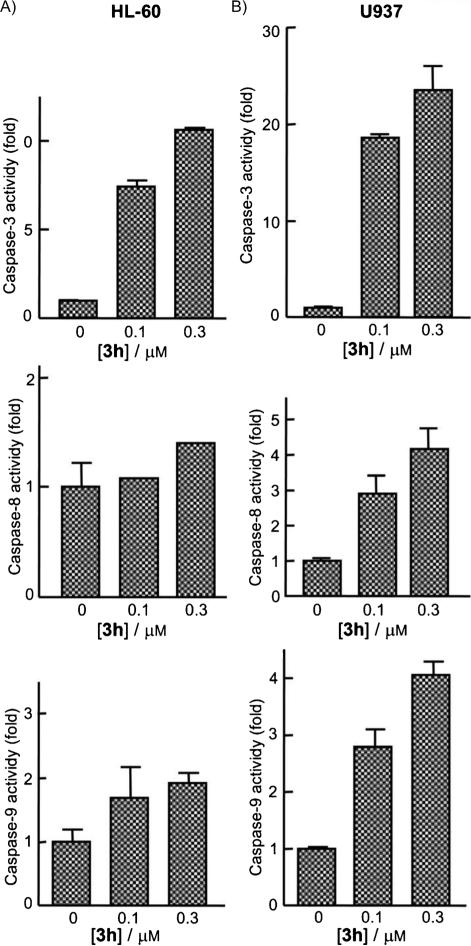
As caspase processing does not always correlate with activity, enzymatic activities of caspase-3-like proteases (caspase-3/7), and of caspase-8 and -9 were also investigated in extracts of control or compound 3h-treated HL-60 and U937 cells. Treatment was with 0.1 or 0.3 μm 3h for 24 h. Cell lysates were assayed for cleavage of the colorimetric tetrapeptide substrates DEVD-pNA, IETD-pNA, and LEHD-pNA as specific substrates for caspase-3/7, caspase-8, and caspase-9, respectively. As shown in Figure 5, increases in caspase-3, -8, and -9 activities occurred after 24 h of treatment and with a concentration as low as 0.1 μm. The enzymatic activities of caspase-3, -8, and -9 increased in U937 cells in a concentration-dependent manner, although this effect was less pronounced in HL-60 cells. In summary, compound 3h induced apoptosis in U937 and HL-60 cells through both the mitochondrial and death-receptor pathways.
Figure 5.
Kinetics of caspase-3/7, -8, and -9 activation in response to compound 3h. A) HL-60 and B) U937 cells were treated with the indicated concentrations of compound 3h and harvested at 24 h. Cell lysates were assayed for caspase-3/7, -8, and -9 activities using the DEVD-pNA, IETD-pNA, and LEHD-pNA colorimetric substrates, respectively. Results are expressed as fold increase in caspase activity relative to control. Values represent the mean ±S.E.; this histogram is representative of two independent experiments, each performed in triplicate.
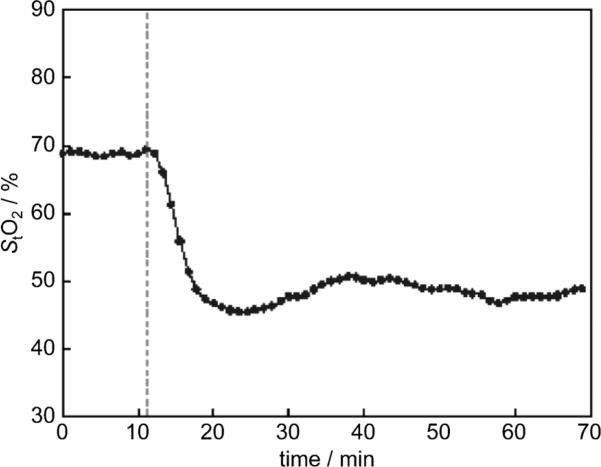
CA-4 and its analogues in clinical development have been shown to quickly and selectively shut down the blood flow of tumors.[28] These compounds are therefore called antivascular or vascular disrupting agents (VDAs). We tested the glycine prodrug of compound 3h, corresponding to derivative 5, for its ability to act as a VDA in an in vivo model. As an alternative method to using ultrasound and magnetic resonance imaging (MRI) to monitor blood flow in tumors, we used a new near-infrared optical technique called spatial frequency domain imaging (SFDI) to measure tissue oxygen saturation changes resulting from vascular disruption after administration of 5.[29] Previously, this technique had been applied to monitoring tumor responses to CA-4, which alters the hemodynamic parameters, such as blood volume, blood oxygenation, and blood flow, of tumors but not the surrounding normal tissues.[30] A rapid decrease in oxygen saturation is a result of vascular disruption and stoppage of blood flow in tumors. In this study, we measured the changes of oxygenated hemoglobin ([OHb]), deoxygenated hemoglobin ([RHb]), and total hemoglobin ([HbO2]) continuously by using an SFDI system in rat breast cancer tumors after administration of compound 5 via a bolus i.p. injection (30 mgkg-1). Within 10 min of administration, there was a rapid decrease in oxygen saturation in tumor tissues similar to what has been observed with CA-4 treatment (data not shown), confirming that derivative 5 causes vascular disruption in vivo (Figure 6).
Figure 6.
Compound 5 induced a rapid decrease in oxygen saturation in a rat breast cancer model. The spatial frequency domain imaging technique was used to obtain real-time images of tumors noninvasively. Compound 5 was given at the time point indicated by the dashed line, and monitoring was continued for 1 h. After data were collected and analyzed, the tumor tissue oxygen saturation (StO2) was plotted over time.
Molecular docking studies were also performed to investigate the possible binding mode of the most active compounds to tubulin. Results obtained for compound 3h (see figure S1 in Supporting Information) indicated that the amino group in C5 would establish hydrogen bonds with Thr179 and Ser178 and that the methoxy group at C6 would form a hydrogen bond with the side chain of Ser178. Compound 3j bound in a very similar orientation as 3h, with the trimethoxyphenyl moiety in contact with Cys241 and forming an extensive network of nonpolar interactions with the colchicine binding pocket. However, there was also a significant difference in the position of the benzo[b]furan ring, which shifted slightly to accommodate the C7 methoxy group. This movement did not allow the formation of all the hydrogen bonds observed for 3h, although the amino group was still within a distance compatible with a hydrogen bond with Thr179. The loss of the other hydrogen bonds could explain the observed loss in biological activity.
Conclusions
In the studies presented herein, we investigated molecules bearing an amino substituent at the 4-, 5-, 6-, and 7-positions of the benzene portion of the benzo[b]furan skeleton (compounds 3a–e), with the concomitant presence of a methoxy group at the 5- (3 f), 6- (3g and h), and 7- (3 and j) positions. Inhibition of cell growth was strongly dependent on the relative positions of the amino and methoxy moieties and on the presence of a methyl group at the 3-position of the benzo[b]-furan skeleton. Thus, for the 5-amino derivatives 3b and 3c, the introduction of a methoxy substituent at the C6 position resulted in the most active compounds (3g and 3h) of the series. The addition of a methoxy group at the C6 position resulted in a 100-fold increase in potency as compared with the parent compound 3c. For compound 3g, moving the methoxy group from the C6 to the C7 position, to furnish 3i, resulted in a tenfold decrease in activity, whereas the differences between C6 and C7 methoxy groups ranged from 3–30-fold for the 3-methyl derivative 3h. Comparing the 3-unsubstituted derivative 3g with the 3-methyl counterpart 3h, it was evident that introduction of a methyl at the C3 position led to a threefold increase in antiproliferative activity against all cell lines. For these latter compounds, the 5-amino-6-methoxy benzene portion of the benzo[b]furan skeleton mimicked the structure of the B ring of aminocombretastatin 1c. The high potency of compounds 3g and h allowed us to confirm that the ortho relationship between the 3-amino group and the carbonyl oxygen of the 2-(3′,4′,5′-trimethoxybenzoyl) moiety, as for 2a, was not required for activity.
All the unsubstituted α-bromoacryloylamido derivatives 4a, b, d, and e were 10–100-fold more active than their amino counterparts, demonstrating that the presence of an α-bromoacryloyl moiety significantly enhanced antiproliferative activity. With the exception of HeLa cells, shifting the α-bromoacryloylamido function from the C4/5 to the C6/7 position, led to a decrease in activity. Compounds 3h and 3g were able to induce apoptosis in HL-60 and U937 human leukemia cells, with 3h being the most effective derivative. Compound 3h at a concentration of 100 nm increased the percentage of apoptotic HL-60 and U937 cells by 9- and 17-fold, respectively. Morphological changes characteristic of apoptotic cells (fragmented and condensed chromatin) were demonstrated by fluorescence microscopy. Apoptosis induced by compound 3h was characterized by the activation of caspases-3, -8, and -9. Moreover, Western blot evaluation of extracts prepared from cells treated with these compounds demonstrated cleavage of 116 kDa PARP with the generation of the 85 kDa fragment usually observed in apoptotic cells. The potential glycine prodrug of compound 3h (compound 5) caused vascular disruption in vivo and thus represents a new vascular disrupting agent.
Experimental Section
Chemistry
General
1H and 13C NMR spectra were recorded on Bruker AC 200 and Varian 400 Mercury Plus spectrometers, respectively. Chemical shifts (δ) are given in ppm upfield from tetramethylsilane (TMS) as internal standard, and the spectra were recorded in appropriate deuterated solvents, as indicated. Melting points (mp) were determined on a Büchi–Tottoli apparatus and are uncorrected. All products reported showed 1H and 13C NMR spectra in agreement with the assigned structures. Elemental analyses were conducted by the Microanalytical Laboratory of the Chemistry Department of the University of Ferrara. Mass spectra were obtained by electrospray ionization (ESI) in positive mode using an ESI Micromass ZMD 2000 mass spectrometer. All reactions were carried out under an inert atmosphere of dry N2 gas, unless otherwise described. Standard syringe techniques were applied for transferring dry solvents. Reaction courses and product mixtures were routinely monitored by TLC on silica gel (pre-coated F254 Merck plates) and visualized with aqueous KMnO4. Flash chromatography was performed using 230–400 mesh silica gel and the indicated solvent system (petroleum ether (PE), bp: 40–60°C, was employed). Organic solutions were dried over anhydrous Na2SO4. CaCl2 was used in the distillation of DMF, and the distilled solvent was stored over molecular sieves (3 Å).
General procedure (A) for the synthesis of nitro-2-(3,4,5-trimethoxybenzoyl)benzo[b]furans 7a–j
2-Bromo-1-(3,4,5-trimethoxyphenyl)ethanone (289 mg, 1 mmol) and anhydrous K2CO3 (276 mg, 2 mmol) was added to a solution of the appropriate substituted nitrosalicylaldehyde or nitro-2-hydroxyacetophenone (1 mmol) in dry acetone (15 mL) while stirring, and the reaction mixture was held at reflux for 18 h. After cooling, the reaction mixture was evaporated and the residue was dissolved in a mixture of CH2Cl2 (15 mL) and H2O (5 mL). The organic layer was washed with brine, dried, and concentrated in vacuo to obtain a residue, which was purified by crystallization from PE.
General procedure (B) for the synthesis of amino 2-(3,4,5-trimethoxybenzoyl)benzo[b]furans 3a–j
A mixture of nitro derivative 7a–j (1 mmol) and iron powder (400 mg) was added to a mixture of EtOH (5 mL), acetic acid (3.5 mL), H2O (2 mL) and 35% HCl (2 drops). The suspension was held at reflux for 40 min, cooled, and filtered through Celite. The filtrate was diluted with H2O (25 mL) and extracted with CH2Cl2 (3×10 mL). The organic layers were combined, sequentially washed with 5% aqueous NaHCO3 (10 mL) and H2O (10 mL), dried over Na2SO4, and evaporated. The residue was purified by flash chromatography on silica gel.
(4-Aminobenzo[b]furan-2-yl)(3,4,5-trimethoxyphenyl)methanone (3a)
Following general procedure B, the residue was recrystallized with PE to give 3a as a yellow solid (235 mg, 72%): Rf=0.34 (PE/EtOAc, 1:1); mp: 67–69°C; 1H NMR (200 MHz, CDCl3): δ=3.94 (s, 3H), 3.96 (s, 6H), 4.10 (bs, 2H), 6.56 (d, J=8.4 Hz, 1H), 6.98 (d, J=8.4 Hz, 1H), 7.16 (s, 1H), 7.35 (s, 2H), 7.42 ppm (t, J=8.4 Hz, 1H); 13C NMR (100 MHz, CDCl3): δ=56.5, 56.7, 61.1, 102.4, 105.9, 107.2, 107.7, 113.6, 116.0, 129.7, 129.9, 132.5, 142.0, 142.4, 151.1, 153.1, 157.3, 182.9 ppm; MS (ESI): [M+1]+ =328.2; Anal. calcd for C18H17NO5: C 66.06, H 5.23, N 4.28, found: C 65.85, H 5.05, N 4.09.
(5-Aminobenzo[b]furan-2-yl)(3,4,5-trimethoxyphenyl)methanone (3b)
Following general procedure B, the residue was subjected to chromatography with EtOAc/PE (1:1) as eluent to give 3b as a yellow solid (255 mg, 78%): Rf=0.38 (PE/EtOAc, 1:1); mp: 108–110°C; 1H NMR (200 MHz, CDCl3): δ=3.66 (bs, 2H), 3.94 (s, 6H), 3.95 (s, 3H), 6.93 (d, J=8.6 Hz, 1H), 6.94 (s, 1H), 7.33 (s, 2H), 7.39 (s, 1H), 7.42 ppm (d, J=8.6 Hz, 1H); 13C NMR (100 MHz, CDCl3): δ=56.4, 56.7, 61.1, 102.5, 105.8, 107.3, 107.8, 113.4, 116.1, 129.6, 129.8, 132.4, 142.3, 142.6, 151.3, 153.2, 157.4, 182.6 ppm; MS (ESI): [M+1]+ =328.1; Anal. calcd for C18H17NO5: C 66.06, H 5.23, N 4.28, found: C 65.92, H 5.02, N 4.11.
(5-Amino-3-methylbenzo[b]furan-2-yl)(3,4,5-trimethoxyphenyl)-methanone (3c)
Following general procedure B, the residue was subjected to chromatography with EtOAc/PE (1:1) as eluent to give 3c as a yellow solid (188 mg, 55%): Rf=0.38 (PE/EtOAc, 1:1); mp: 162–164°C; 1H NMR (200 MHz, CDCl3): δ=2.56 (s, 3H), 3.64 (bs, 2H), 3.92 (s, 6H), 3.94 (s, 3H), 6.89 (d, J=9.0 Hz, 1H), 6.91 (s, 1H), 7.29 (d, J=9.0 Hz, 1H), 7.34 ppm (s, 2H); 13C NMR (100 MHz, CDCl3): δ=10.5, 56.4, 56.6, 61.2, 102.7, 105.4, 107.6, 107.9, 113.2, 116.6, 129.5, 129.7, 132.6, 142.2, 142.6, 151.5, 153.4, 157.6, 182.6 ppm; MS (ESI): [M+1]+ =343.6; Anal. calcd for C19H19NO5: C 66.85, H 5.61, N 4.10, found: C 66.72, H 5.38, N 4.01.
(6-Aminobenzo[b]furan-2-yl)(3,4,5-trimethoxyphenyl)methanone (3d)
Following general procedure B, the residue was subjected to chromatography with EtOAc/PE (3:7) as eluent to give 3d as a yellow solid (239 mg, 73%): Rf=0.34 (PE/EtOAc, 7:3); mp: 173–175°C; 1H NMR (200 MHz, CDCl3): δ=3.94 (s, 6H), 3.95 (s, 3H), 4.04 (bs, 2H), 6.88 (dd, J=8.4, 2.0 Hz, 1H), 6.85 (s, 1H), 7.27 (s, 2H), 7.42 (s, 1H), 7.47 ppm (dd, J=8.4, 2.0 Hz, 1H); 13C NMR (100 MHz, CDCl3): δ=56.1, 56.5, 61.4, 102.3, 105.4, 107.5, 107.9, 113.2, 116.2, 129.7, 130.0, 132.4, 142.2, 142.6, 151.3, 153.2, 157.5, 182.7 ppm; MS (ESI): [M+1]+ =328.1; Anal. calcd for C18H17NO5: C 66.06, H 5.23, N 4.28, found: C 65.91, H 5.00, N 4.11.
(7-Aminobenzo[b]furan-2-yl)(3,4,5-trimethoxyphenyl)methanone (3e)
Following general procedure B, the residue was subjected to chromatography with EtOAc/PE (3:7) as eluent to give 3e as a yellow solid (150 mg, 46%): Rf=0.34 (PE/EtOAc, 7:3); mp: 116–118°C; 1H NMR (200 MHz, CDCl3): δ=3.92 (s, 3H), 3.94 (s, 6H), 7.23 (m, 3H), 7.29 (s, 2H), 7.54 (s, 1H), 8.12 (d, J=8.4 Hz, 1H), 8.42 ppm (d, J=8.4 Hz, 1H); 13C NMR (100 MHz, CDCl3): δ=56.4, 56.8, 61.2, 102.6, 105.7, 107.4, 107.8, 113.4, 116.1, 129.6, 129.9, 132.3, 142.2, 142.5, 151.8, 153.3, 157.4, 182.6 ppm; MS (ESI): [M+1]+ =328.2; Anal. calcd for C18H17NO5: C 66.06, H 5.23, N 4.28, found: C 65.82, H 5.12, N 4.12.
(7-Amino-3-methyl-5-methoxybenzo[b]furan-2-yl)(3,4,5-trimethoxyphenyl)methanone (3 f)
Following general procedure B, the residue, purified by column chromatography with EtOAc/PE (4:6) as eluent, furnished 3 f as a yellow solid (241 mg, 65%): Rf=0.36 (PE/EtOAc, 1:1); mp: 193–194°C; 1H NMR (200 MHz, CDCl3): δ=2.57 (s, 3H), 3.84 (s, 6H), 3.92 (s, 3H), 3.95 (s, 3H), 3.98 (bs, 2H), 6.45 (s, 1H), 6.48 (s, 1H), 7.37 ppm (s, 2H); 13C NMR (100 MHz, CDCl3): δ=10.6, 55.9, 56.4, 56.6, 61.1, 91.7, 102.9, 107.4, 107.6, 127.7, 129.8, 132.8, 133.0, 139.5, 142.1, 148.6, 152.9, 153.2, 157.5, 184.5 ppm; MS (ESI): [M+1]+ =372.4; Anal. calcd for C20H21NO6: C 64.68, H 5.70, N 3.77, found: C 64.57, H 5.56, N 3.69.
(5-Amino-6-methoxybenzo[b]furan-2-yl)(3,4,5-trimethoxyphenyl)methanone (3g)
Following general procedure B, the residue was subjected to chromatography with EtOAc/PE (1:1) as eluent and furnished 3g as a yellow solid (221 mg, 62%): Rf=0.34 (PE/EtOAc, 7:3); mp: 138–139°C; 1H NMR (200 MHz, CDCl3): δ=3.94 (s, 6H), 3.95 (s, 3H), 3.96 (s, 3H), 4.04 (bs, 2H), 6.92 (s, 1H), 7.03 (s, 1H), 7.28 (s, 2H), 7.37 ppm (s, 1H); 13C NMR (100 MHz, CDCl3): δ=56.0, 56.2, 56.5, 61.1, 94.2, 105.1, 105.3, 107.0, 107.2, 116.6, 116.8, 119.9, 133.0, 133.2, 134.8, 150.6, 151.6, 153.1, 182.7 ppm; Anal. calcd for C19H19NO6: C 63.86, H 5.36, N 3.92, found: C 63.64, H 5.22, N 3.78.
(5-Amino-3-methyl-6-methoxybenzo[b]furan-2-yl)(3,4,5-trimethoxyphenyl)methanone (3h)
Following general procedure B, the residue was subjected to chromatography with EtOAc/PE (1:1) as eluent to give 3h as a yellow solid (293 mg, 79%): Rf=0.38 (PE/EtOAc, 7:3); mp: 135–136°C; 1H NMR (200 MHz, CDCl3): δ=1.88 (bs, 2H), 2.56 (s, 3H), 3.93 (s, 6H), 3.94 (s, 6H), 6.90 (s, 1H), 6.92 (s, 1H), 7.37 ppm (s, 2H); 13C NMR (100 MHz, CDCl3): δ=10.5, 56.1, 56.2, 56.4, 61.1, 94.0, 103.4, 103.7, 107.1, 107.4, 122.2, 127.6, 133.5, 134.2, 148.0, 149.5, 150.6, 152.8, 153.0, 183.9 ppm; MS (ESI): [M+1]+ =372.0; Anal. calcd for C20H21NO6: C 64.68, H 5.70, N 3.77, found: C 64.48, H 5.52, N 3.58.
(5-Amino-7-methoxybenzo[b]furan-2-yl)(3,4,5-trimethoxyphenyl)methanone (3i)
Following general procedure B, the residue was subjected to chromatography with EtOAc/PE (1:1) as eluent to give 3i as a yellow solid (200 mg, 56%): Rf=0.35 (PE/EtOAc, 7:3); mp: 140–142°C; 1H NMR (200 MHz, CDCl3): δ=3.78 (bs, 2H), 3.94 (s, 3H), 3.95 (s, 6H), 3.98 (s, 3H), 6.40 (d, J=2.2 Hz, 1H), 6.53 (d, J= 2.2 Hz, 1H), 7.40 (s, 1H), 7.42 ppm (s, 2H); 13C NMR (100 MHz, CDCl3): δ=56.1, 56.4, 56.6, 61.1, 97.9, 100.3, 100.8, 107.3, 107.4, 115.5, 115.9, 129.2, 132.2, 133.8, 144.0, 146.4, 151.1, 153.0, 182.8 ppm; MS (ESI): [M+1]+ =358.4; Anal. calcd for C19H19NO6: C 63.86, H 5.36, N 3.92, found: C 63.73, H 5.20, N 3.81.
(5-Amino-3-methyl-7-methoxybenzo[b]furan-2-yl)(3,4,5-trimethoxyphenyl)methanone (3j)
Following general procedure B, the residue, purified by column chromatography with EtOAc/PE (7:3) as eluent, furnished 3j as a yellow solid (156 mg, 42%): Rf=0.34 (PE/EtOAc, 7:3); mp: 179–180°C; 1H NMR (200 MHz, CDCl3): δ=2.57 (s, 3H), 3.76 (bs, 2H), 3.93 (s, 6H), 3.95 (s, 6H), 6.39 (d, J=1.0 Hz, 1H), 6.49 (d, J=1.0 Hz, 1H), 7.50 ppm (s, 2H); 13C NMR (100 MHz, CDCl3): δ=10.4, 56.2, 56.3, 56.5, 61.0, 96.5, 96.7, 100.8, 107.4, 107.6, 126.8, 131.4, 132.8, 138.8, 142.1, 143.6, 146.3, 148.9, 152.9, 187.6 ppm; [M+1]+ =372.4; Anal. calcd for C20H21NO6: C 64.68, H 5.70, N 3.77, found: C 64.51, H 5.53, N 3.61.
General Procedure (C) for the synthesis of compounds 4a–i
EDCI (383 mg, 2 mmol) and HOBt (270 mg, 2 mmol) were added to an ice-cooled solution of aminobenzo[b]furan (1 mmol) in dry DMF (5 mL), followed by α-bromoacrylic acid (2 mmol, 306 mg). The reaction was stirred at RT for 18 h and then concentrated in vacuo. The residue was dissolved with a mixture of CH2Cl2 (15 mL) and H2O (5 mL), and the organic phase was washed with brine (5 mL), dried over Na2SO4, and evaporated to dryness in vacuo. The resulting crude residue was purified by flash chromatography on silica gel.
2-Bromo-N-[2-(3,4,5-trimethoxybenzoyl)-1-benzo[b]furan-4-yl]acrylamide (4a)
Following general procedure C, the crude residue purified by flash chromatography using EtOAc/PE (3:7 v/v) as eluent furnished 4a as a white solid (202 mg, 44%): Rf=0.36 (PE/EtOAc, 7:3); mp: 152–154°C; 1H NMR (200 MHz, CDCl3): δ=3.95 (s, 6H), 3.97 (s, 3H), 6.21 (d, J=1.8 Hz, 1H), 7.18 (d, J=1.8 Hz, 1H), 7.39 (s, 2H), 7.43 (t, J=8.8 Hz, 1H), 7.49 (s, 1H), 7.60 (s, 1H), 7.72 (d, J=8.8 Hz, 1H), 8.74 ppm (bs, 1H); 13C NMR (100 MHz, CDCl3): δ=29.9, 56.2, 56.4, 61.1, 107.1, 107.3, 110.0, 110.2, 113.9, 115.9, 120.4, 122.1, 128.9, 129.6, 131.4, 131.9, 151.8, 153.2, 156.4, 158.8, 182.9 ppm; MS (ESI): [M]+ =460.4, [M+2]+ =462.6; Anal. calcd for C21H18BrNO6: C 54.80, H 3.94, N 3.04, found: C 54.62, H 3.68, N 2.88.
2-Bromo-N-[2-(3,4,5-trimethoxybenzoyl)-1-benzo[b]furan-5-yl]acrylamide (4b)
Following general procedure C, the crude residue purified by flash chromatography using EtOAc/PE (1:1 v/v) as eluent furnished 4b as a brown solid (235 mg, 51%): Rf=0.38 (PE/EtOAc, 1:1); mp: 165–166°C; 1H NMR (200 MHz, CDCl3): δ=3.95 (s, 6H), 3.97 (s, 3H), 6.18 (d, J=1.6 Hz, 1H), 7.17 (d, J=1.6 Hz, 1H), 7.33 (s, 2H), 7.58 (dd, J=8.8, 2.2 Hz, 1H), 7.61 (d, J=8.8 Hz, 1H), 8.22 (d, J=2.2 Hz, 1H), 8.26 (s, 1H), 8.52 ppm (bs, 1H); 13C NMR (100 MHz, CDCl3): δ=29.6, 56.4, 56.6, 61.1, 107.2, 107.4, 113.0, 113.2, 114.7, 116.1, 117.3, 121.9, 122.5, 127.5, 129.0, 132.1, 133.3, 153.2, 153.3, 159.0, 183.1 ppm; MS (ESI): [M]+ =460.4, [M+2]+ = 462.4; Anal. calcd for C21H18BrNO6: C 54.80, H 3.94, N 3.04, found: C 54.70, H 3.91, N 2.91.
2-Bromo-N-[3-methyl-2-(3,4,5-trimethoxybenzoyl)-1-benzo[b]furan-5-yl]acrylamide (4c)
Following general procedure C, the crude residue purified by flash chromatography using EtOAc/PE (4:6 v/v) as eluent furnished 4c as a white solid (313 mg, 66%): Rf=0.35 (PE/EtOAc, 1:1); mp: 99–100°C; 1H NMR (200 MHz, CDCl3): δ=2.62 (s, 3H), 3.93 (s, 6H), 3.96 (s, 3H), 6.18 (d, J=1.6 Hz, 1H), 7.17 (d, J=1.6 Hz, 1H), 7.40 (s, 2H), 7.52 (s, 2H), 8.14 (s, 1H), 8.51 ppm (bs, 1H); 13C NMR (100 MHz, CDCl3): δ=10.3, 29.8, 56.2, 56.4, 61.1, 107.2, 107.5, 112.6, 113.1, 121.8, 122.5, 126.9, 129.0, 129.8, 132.6, 132.9, 142.5, 149.3, 151.6, 153.0, 159.1, 184.5 ppm; MS (ESI): [M]+ = 474.5, [M+2]+ =476.5; Anal. calcd for C22H20BrNO6: C 55.71, H 4.25, N 2.95, found: C 55.58, H 4.11, N 2.77.
2-Bromo-N-[2-(3,4,5-trimethoxybenzoyl)-1-benzo[b]furan-6-yl]acrylamide (4d)
Following general procedure C, the crude residue purified by flash chromatography using EtOAc/PE (4:6 v/v) as eluent furnished 4d as a white solid (212 mg, 46%): Rf=0.38 (PE/EtOAc, 1:1); mp: 159–160°C; 1H NMR (200 MHz, CDCl3): δ=3.95 (s, 6H), 3.96 (s, 3H), 6.18 (d, J=1.6 Hz, 1H), 7.17 (d, J=1.6 Hz, 1H), 7.24 (s, 2H), 7.33 (d, J=8.8 Hz, 1H), 7.55 (s, 1H), 7.74 (d, J=8.8 Hz, 1H), 8.26 (s, 1H), 8.61 ppm (bs, 1H); 13C NMR (100 MHz, CDCl3): δ=29.4, 56.2, 56.5, 61.1, 107.2, 107.3, 115.9, 116.1, 117.2, 122.3, 123.6, 124.1, 129.3, 132.1, 137.4, 142.6, 153.1, 153.3, 156.4, 158.9, 182.7 ppm; MS (ESI): [M]+ =460.4, [M+2]+ =462.4; Anal. calcd for C21H18BrNO6: C 54.80, H 3.94, N 3.04, found: C 54.67, H 3.77, N 2.93.
2-Bromo-N-[2-(3,4,5-trimethoxybenzoyl)-1-benzo[b]furan-7-yl]acrylamide (4e)
Following general procedure C, the crude residue purified by flash chromatography using EtOAc/PE (3:7 v/v) as eluent furnished 4e as a brown solid (290 mg, 63%): Rf=0.34 (PE/EtOAc, 7:3); mp: 184–185°C; 1H NMR (200 MHz, CDCl3): δ=3.94 (s, 6H), 3.96 (s, 3H), 6.20 (d, J=1.8 Hz, 1H), 7.16 (d, J=1.8 Hz, 1H), 7.33 (s, 2H), 7.39 (t, J=8.0 Hz, 1H), 7.51 (d, J=8.0 Hz, 1H), 7.58 (s, 1H), 8.35 (d, J=8.0 Hz, 1H), 8.97 ppm (bs, 1H); 13C NMR (100 MHz, CDCl3): δ=29.8, 56.2, 56.6, 61.2, 107.1, 107.3, 116.1, 116.4, 118.9, 122.2, 123.2, 124.9, 127.4, 129.3, 132.0, 142.9, 146.5, 152.5, 153.2, 159.2, 182.8 ppm; MS (ESI): [M]+ =460.3, [M+2]+ =462.3; Anal. calcd for C21H18BrNO6: C 54.80, H 3.94, N 3.04, found: C 54.60, H 3.72, N 2.91.
2-Bromo-N-[3-methyl-5-methoxy-2-(3,4,5-trimethoxybenzoyl)-1-benzo[b]furan-7-yl]acrylamide (4 f)
Following general procedure C, the crude residue purified by flash chromatography using EtOAc/PE (4:6 v/v) as eluent furnished 4 f as a white solid (262 mg, 52%): Rf=0.38 (PE/EtOAc, 1:1); mp: 162–163°C; 1H NMR (200 MHz, CDCl3): δ=2.63 (s, 3H), 3.90 (s, 3H), 3.93 (s, 6H), 3.95 (s, 3H), 6.18 (d, J=1.8 Hz, 1H), 6.88 (d, J=1.8 Hz, 1H), 7.16 (d, J=2.2 Hz, 1H), 7.36 (s, 2H), 8.04 (d, J=2.2 Hz, 1H), 8.82 ppm (bs, 1H); 13C NMR (100 MHz, CDCl3): δ=10.3, 29.7, 56.2, 56.3, 56.6, 61.1, 99.2, 107.2, 107.5, 108.5, 121.9, 123.2, 127.6, 129.6, 129.7, 132.8, 140.0, 142.5, 148.8, 153.0, 156.9, 158.8, 183.2 ppm; MS (ESI): [M]+ =504.2, [M+2]+ =506.2; Anal. calcd for C23H22BrNO7: C 54.78, H 4.40, N 2.78, found: C 54.67, H 4.26, N 2.56.
2-Bromo-N-[3-methyl-6-methoxy-2-(3,4,5-trimethoxybenzoyl)-1-benzo[b]furan-5-yl]acrylamide (4g)
Following general procedure C, the crude residue purified by flash chromatography using EtOAc/PE (4:6 v/v) as eluent furnished 4g as a white solid (267 mg, 53%): Rf=0.38 (PE/EtOAc, 1:1); mp: 179–180°C; 1H NMR (200 MHz, CDCl3): δ=2.60 (s, 3H), 3.93 (s, 3H), 3.96 (s, 3H), 4.02 (s, 6H), 6.16 (d, J=1.6 Hz, 1H), 7.04 (s, 1H), 7.14 (d, J=1.6 Hz, 1H), 7.36 (s, 2H), 8.74 (s, 1H), 9.24 ppm (bs, 1H); 13C NMR (100 MHz, CDCl3): δ= 10.5, 29.8, 56.2, 56.4, 56.7, 61.1, 107.1, 107.4, 111.3, 122.2, 123.0, 124.7, 128.3, 128.5, 133.2, 140.0, 142.1, 148.5, 150.8, 151.7, 152.9, 158.7, 184.0 ppm; MS (ESI): [M]+ =504.5, [M+2]+ =506.5; Anal. calcd for C23H22BrNO7: C 54.78, H 4.40, N 2.78, found: C 54.62, H 4.21, N 2.58.
2-Bromo-N-[7-methoxy-2-(3,4,5-trimethoxybenzoyl)-1-benzo[b]-furan-5-yl]acrylamide (4h)
Following general procedure C, the crude residue purified by flash chromatography using EtOAc/PE (6:4 v/v) as eluent furnished 4h as a yellow solid (255 mg, 52%): Rf=0.33 (PE/EtOAc, 1:1); mp: 170–172°C; 1H NMR (200 MHz, CDCl3): δ=3.95 (s, 6H), 3.97 (s, 3H), 4.04 (s, 3H), 6.17 (d, J=1.8 Hz, 1H), 7.16 (d, J=1.8 Hz, 1H), 7.42 (s, 2H), 7.53 (s, 1H), 7.55 (s, 1H), 7.56 (s, 1H), 8.45 ppm (bs, 1H); 13C NMR (100 MHz, CDCl3): δ=29.8, 56.2, 56.4, 56.6, 61.1, 103.8, 106.0, 107.2, 107.4, 115.3, 115.8, 128.4, 129.0, 131.9, 134.0, 146.1, 149.2, 153.1, 153.6, 159.0, 161.2, 182.4 ppm; MS (ESI): [M]+ =490.2, [M+2]+ =492.2; Anal. calcd for C22H20BrNO7: C 53.89, H 4.11, N 2.86, found: C 53.69, H 3.88, N 2.65.
2-Bromo-N-[3-methyl-7-methoxy-2-(3,4,5-trimethoxybenzoyl)-1-benzo[b]furan-5-yl]acrylamide (4i)
Following general procedure C, the crude residue purified by flash chromatography using EtOAc/PE (4:6 v/v) as eluent furnished 4i as a white solid (262 mg, 52%): Rf=0.34 (PE/EtOAc, 4:6); mp: 162–163°C; 1H NMR (200 MHz, CDCl3): δ=2.62 (s, 3H), 3.94 (s, 6H), 3.96 (s, 3H), 4.02 (s, 3H), 6.18 (d, J=1.6 Hz, 1H), 7.18 (d, J=1.6 Hz, 1H), 7.49 (s, 1H), 7.51 (s, 2H), 7.56 (s, 1H), 8.51 ppm (bs, 1H); 13C NMR (100 MHz, CDCl3): δ= 10.4, 29.6, 56.1, 56.2, 56.6, 61.3, 107.1, 107.3, 111.2, 122.0, 123.2, 124.5, 128.1, 128.6, 133.3, 140.2, 142.3, 148.5, 150.6, 151.5, 152.7, 158.6, 182.4 ppm; MS (ESI): [M]+ =504.2, [M+2]+ =506.2; Anal. calcd for C23H22BrNO7: C 54.78, H 4.40, N 2.78, found: C 54.59, H 4.20, N 2.61.
tert-Butyl-(2-{[6-methoxy-3-methyl-2-(3,4,5-trimethoxybenzoyl)-1-benzo[b]furan-5-yl]amino}-2-oxoethyl)carbamate (8)
EDCI (192 mg, 1 mmol) and HOBt (135 mg, 1 mmol) were added to an ice-cooled solution of 3h (186 mg, 0.5 mmol) in dry DMF (5 mL), followed by the addition of N-(tert-butoxycarbonyl)glycine (0.6 mmol, 105 mg). The reaction was stirred at RT for 18 h and then concentrated in vacuo. The residue was dissolved with a mixture of CH2Cl2 (10 mL) and H2O (4 mL), and the organic phase was washed with brine (5 mL), dried over Na2SO4, and evaporated to dryness in vacuo. The resulting crude residue was purified by flash chromatography on silica gel with EtOAc/PE (6:4 v/v) as eluent to furnish 8 as a yellow powder (438 mg, 83%): Rf=0.36 (PE/EtOAc, 7:3); mp: 90–91°C; 1H NMR (200 MHz, CDCl3) δ=1.49 (s, 8H), 1.99 (m, 2H), 2.59 (s, 3H), 3.93 (s, 6H), 3.95 (s, 3H), 3.96 (s, 3H), 5.22 (bs, 1H), 7.00 (s, 1H), 7.34 (s, 2H), 8.51 (bs, 1H), 8.67 ppm (s, 1H); 13C NMR (100 MHz, CDCl3): δ=10.4, 28.4 (3C), 45.8, 56.0, 56.4, 56.6, 61.1, 83.4, 95.7, 107.1, 107.4, 108.5, 113.4, 113.6, 120.8, 124.3, 127.0, 132.7, 140.2, 143.9, 153.1, 154.1, 154.6, 168.2, 188.0 ppm; MS (ESI): [M+1]+=529.7.
N-[6-methoxy-3-methyl-2-(3,4,5-trimethoxybenzoyl)-1-benzo[b]-furan-5-yl]glycinamide hydrochloride (5)
The N-Boc-glycine derivative 8 (132 mg, 0.25 mmol) was added to a saturated solution of HCl in MeOH (10 mL). The mixture was stirred for 3 h at RT and evaporated at reduced pressure. The residue was suspended in Et2O (5 mL), and the mixture was stirred for 1 h and filtered to furnish the final compound as a grey solid (460 mg, >95%): Rf=0.28 (CH2Cl2/MeOH, 4:6); mp: 250–251°C; 1H NMR (200 MHz, [D6]DMSO) δ=2.58 (s, 3H), 3.74 (m, 1H), 3.78 (s, 6H), 3.87 (s, 3H), 3.96 (s, 3H), 7.32 (s, 2H), 7.53 (s, 1H), 8.19 (bs, 4H), 8.30 (s, 1H), 9.90 ppm (s, 1H); 13C NMR (100 MHz, CDCl3): δ=10.0, 41.0, 56.1, 56.2, 56.6, 60.2, 95.5, 107.0, 107.3, 113.4, 113.6, 120.8, 124.3, 127.0, 132.7, 141.4, 147.6, 151.7, 152.4, 152.6, 165.4, 183.0 ppm; MS (ESI): [M+1]+ = 428.8; Anal. calcd for C22H25ClN2O7: C 56.84, H 5.42, N 6.03, found: C 56.61, H 5.21, N 5.89.
Biology
Reagents
Stock solutions of the various compounds at 100 mm were made by dissolving them in dimethyl sulfoxide (DMSO), and aliquots were frozen at –20°C. Poly(vinylidene difluoride) (PVDF) membranes were purchased from Millipore (Billerica, MA, USA). Acrylamide, bisacrylamide, ammonium persulfate, and N,N,N′,N′-tetra-methylethylenediamine were from Bio-Rad (Hercules, CA, USA). Antibodies for poly(ADP-ribose) polymerase (PARP) and caspase-3 were purchased from Stressgen (Victoria, BC, Canada). An antibody against cytochrome c was purchased from BD PharMingen (San Diego, CA, USA). Anti-caspase-6 monoclonal antibody and antibodies against caspase-8 and caspase-9 were from Medical & Biological Laboratories (Nagoya, Japan). Secondary antibodies were from GE Healthcare (Little Chalfont, UK). All other chemicals were obtained from Sigma (St. Louis, MO, USA).
Cell growth inhibitory activity
Murine leukemia L1210, murine mammary carcinoma FM3A, human T-lymphocyte Molt/4 and CEM, and human cervix carcinoma (HeLa) cells were suspended at 300000–500000 cellsmL-1 of culture medium, and 100 μL cell suspension was added to 100 μL of an appropriate dilution of the test compounds in wells of 96-well microtiter plates. After incubation at 37°C for two days, cell number was determined using a Coulter counter. The IC50 value was defined as the compound concentration required to inhibit cell proliferation by 50%.
HL-60 and U937 cells were cultured in RPMI 1640 medium containing 10% (v/v) heat-inactivated fetal bovine serum, 100 unitsmL-1 penicillin and 100 mgmL-1 streptomycin at 37°C in a humidified atmosphere containing 5% CO2. Cell numbers was determined by counting in a hematocytometer, and cell viability was always >95% in all experiments as assayed by the trypan blue exclusion method. Further dilutions of stock solutions of compounds were made in culture media just before use. In all experiments, the final concentration of DMSO did not exceed 0.1% (v/v), a concentration which is nontoxic to the cells. The same concentration was present in control cultures.
Effects on tubulin polymerization and on colchicine binding to tubulin
Bovine brain tubulin was purified as described previously.[31] To evaluate the effect of the compounds on tubulin assembly in vitro, varying concentrations were pre-incubated with 10 μm tubulin in glutamate buffer at 30°C and then cooled to 0°C. After addition of GTP, the mixtures were transferred to 0°C cuvettes in a recording spectrophotometer and warmed to 30°C, and the assembly of tubulin was observed turbidimetrically. The IC50 value was defined as the compound concentration that inhibited the extent of assembly by 50% after a 20 min incubation. The ability of the test compounds to inhibit colchicine binding to tubulin was measured as described,[19b] except that the reaction mixtures contained 1 μm tubulin, 5 μm [3H]colchicine, and 5 μm test compound.
Evaluation of apoptosis
The rate of apoptotic cell death was analyzed by fluorescence microscopy and by flow cytometric analysis of propidium iodide (PI)-stained nuclei as described below.
Fluorescence microscopy analysis
Cells were harvested and fixed in 3% paraformaldehyde and incubated at RT for 10 min. The fixative was removed, and the cells were washed with phosphate-buffered saline (PBS), resuspended in 30–50 μL of PBS containing 20 μg mL-1 bis-benzimide trihydrochloride (Hoechst 33258), and incubated at RT for 15 min. Stained nuclei were visualized using a Zeiss fluorescence microscope.
Quantification of apoptosis by flow cytometry
To study changes in cell DNA content, histogram measurements of hypodiploid DNA formation was performed by flow cytometry using a Coulter EPICS™ cytometer (Beckman Coulter). Histograms were analyzed with the Expo 32 ADC Software™. Cells were collected and centrifuged at 500 g, washed with PBS, and resuspended in 50 μL of PBS. Following dropwise addition of 1 mL of ice-cold 75% EtOH, fixed cells were stored at –20°C for 1 h. Samples were then centrifuged at 500 g and washed with PBS before resuspension in 1 mL of PBS containing 50 μg mL-1 propidium iodide and 100 μg mL-1 RNase A. The cells were then incubated for 1 h at 37°C in the dark. The percentage of cells with decreased DNA staining, composed of apoptotic cells resulting from either fragmentation or decreased chromatin, was determined on a minimum of 10000 cells per experimental condition. Cell debris was excluded from analysis by selective gating based on anterior and right angle scattering.
Analysis of DNA fragmentation
A late biochemical hallmark of apoptosis is the fragmentation of the genomic DNA. It is an irreversible event and occurs before changes in plasma membrane permeability. DNA isolation and gel electrophoresis were performed as described previously.[32] Briefly, cells (1×105) were collected by centrifugation, washed with PBS, and incubated in 30 μL of lysis buffer [50 mm Tris-HCl (pH 8.0), 10 mm EDTA, 0.5% sodium dodecyl sulfate], containing 1 μg mL-1 RNase A at 37°C for 1 h. Then, 3 μL of proteinase K (10 μg mL-1) was added, and the mixture was incubated at 50°C for an additional 2 h. DNA was extracted with 100 μL of phenol-chloroform-isoamyl alcohol (24:24:1) and mixed with 5 μL of loading solution [10 mm EDTA, 1% (w/v) low melting-point agarose, 0.25% bromophenol blue, and 40% sucrose, pH 8.0]. Samples were separated by electrophoresis in 2% agarose gels in TAE buffer [40 mm Tris-acetate (pH 8.0), 1 mm EDTA], visualized by ultraviolet illumination after ethidium bromide (0.5 μg mL-1) staining, and the images were captured by a digital camera (Digi Doc system, Bio-Rad).
Western blot analysis
Cells (1×106 mL) were treated in the absence or presence of the indicated concentrations of compounds 3g, 3h, 3j, and 4a for 24 h and harvested by centrifugation at 500 g for 10 min. Cell pellets were resuspended in lysis buffer [20 mm Tris-HCl (pH 7.4), 2 mm EDTA, 137 mm NaCl, 10% glycerol, 1% Triton X-100, 2 mm tetrasodium pyrophosphate, 20 mm sodium β-glycero-phosphate, 10 mm sodium fluoride, 2 mm sodium orthovanadate], supplemented with protease inhibitors phenylmethylsulfonyl fluoride (PMSF, 1 mm), leupeptin, aprotinin, and pepstatin A (5 μg mL-1 each) for 15 min at 4°C. The cells were sonicated on ice (five times for 5 s at 40 W, with 20 s intervals between each sonication) with a Braun Labsonic 2000 microtip sonifier (Braun, Melsungen, Germany) and centrifuged at 11000 g for 10 min at 4°C. Protein concentration of supernatants was measured by the Bradford method,[33] and samples containing equal amounts of proteins were boiled in sodium dodecyl sulfate sample buffer for 5 min before loading on an sodium dodecyl sulfate-polyacrylamide gel (7.5% for PARP and 12.5% for caspases). Proteins were electrotransfered to PVDF membranes, blocked with 5% fat-free dry milk in Tris-buffered saline [50 mm Tris-HCl (pH 7.4), 150 mm NaCl] with 0.1% Tween 20, and then incubated with specific primary antibodies overnight at 4°C. After washing and incubation with an appropriate horseradish peroxidase-conjugated secondary antibody, the antigen-antibody complexes were visualized by enhanced chemiluminescence (Millipore) using the manufacturer's protocol.
Detection of cytochrome c
Release of cytochrome c from mitochondria was detected by Western blot analysis. After treatment, cells were washed twice with PBS and then resuspended in ice-cold buffer [20 mm HEPES (pH 7.5), 1.5 mm MgCl2, 10 mm KCl, 1 mm EDTA, 1 mm EGTA, 1 mm dithiothreitol, 0.1 mm phenylmethylsulfonyl fluoride and 5 μg mL-1 each of leupeptin, aprotinin, and pepstatin A] containing 250 mm sucrose. After a 15 min incubation on ice, cells were lysed by pushing them several times through a 22-gauge needle and the lysate spun down at 1000 g for 5 min at 4°C to eliminate nuclei and unbroken cells. The supernatant fraction was centrifuged at 15000 g for 20 min at 4°C, and the resulting pellet was designated as the mitochondrial fraction. The supernatant was further centrifuged at 105000 g for 45 min at 4°C and the resulting supernatant was designated as the cytosolic fraction. Cytosolic proteins (50 μg) were resolved on a 15% sodium dodecyl sulfate/polyacrylamide gel, and cytochrome c was detected by chemiluminescence as described above.
Assay of caspase activity
After treatments cells were harvested by centrifugation at 1000 g for 5 min at 4°C and washed with PBS, and the cell pellets were kept on ice. The cells were resuspended in cell lysis buffer (50 mm HEPES, pH 7.4, 1 mm dithiothreitol, 0.1 mm EDTA, 0.1% Chaps) and held on ice for 5 min. After centrifugation for 10 min at 17000 g at 4°C, the supernatants were analyzed for protein concentration by the Bradford assay and stored at –20°C until used to study caspase colorimetric enzymatic activity. Equal amounts of protein (≈20 μg) from different treatments were used, and the assays were%set up on ice. The net increase of absorbance at 405 nm after incubation at 37°C was indicative of enzyme activity. Specific labeled substrates for caspase-3, -8, and 9 activities were N-acetyl-Asp-Glu-Val-Asp-p-nitroaniline (DEVD-pNA), N-acetyl-Ile-Glu-Thr-Asp-p-nitroaniline (IETD-pNA), and N-acetyl-Leu-Glu-His-Asp-p-nitroaniline (LEHD-pNA) respectively.
Spatial frequency domain imaging to measure tissue oxygen saturation change as a result of vascular disruption
To monitor the dynamic physiological process associated with treatment in vivo, we used a fast, noncontact, near-infrared optical technique called spatial frequency domain imaging (SFDI). SFDI projects broadband light in sinusoidal patterns onto a tissue sample. The backscattered reflectance is then filtered by wavelength and used to analyze the optical content. A Monte Carlo forward model is then used to create a look up table for fitting these optical properties.[34] For this study, we projected the two spatial frequency patterns of NIR light with wavelengths from 700 to 850 nm at every 50 nm on the surface of breasts in a rat breast cancer model. After 10 min of baseline measurement, compound 5 was administered via a bolus i.p. injection (0.45 mL, 30 mg kg-1) and we acquired images for another 60 min to monitor the hemodynamic effects of compound 5 on tumor and normal breast tissue. Once the images at several wavelengths were obtained, quantitative fits for the concentration of oxy- ([OHb]), deoxy- ([RHb]), total ([THb]) hemoglobin, tissue oxygen saturation (% StO2=[OHb]/[THb]×100), and light scattering were carried out. The study was approved by the Institutional Animal Ethical Committee of the Department of Pathology, VA Medical Center, Long Beach, CA (USA).
Molecular modeling
All molecular modeling studies were performed on a MacPro dual 2.66 GHz Xeon running Ubuntu 10. The tubulin structure was downloaded from the Protein Data Bank (http://www.rcsb.org/; PDB ID: 1SA0).[35] Hydrogen atoms were added to the protein, using Molecular Operating Environment (MOE),[36] and minimized keeping all heavy atoms fixed until a RMSD gradient of 0.05 kcal mol-1 Å-1 was reached. Ligand structures were built with MOE and minimized using the MMFF94x force field until a RMSD gradient of 0.05 kcal mol-1 Å-1 was reached. The docking simulations were performed using PLANTS.[37]
Supplementary Material
Acknowledgements
This work was supported by a grant from the Ministry of Science and Innovation of Spain, from the European Regional Development Fund (SAF2010-21380 to F.E.), and the Instituto Canario de Investigaci n del C ncer (RED PRODNAT CANCER to J.B.). S.E. thanks the Ministry of Education of Spain for financial support. The authors thank Mar a Teresa Marrero, Dr. Alberto Casolari, Dr. Erika Marzola, and Dr. Elisa Durini for excellent technical assistance.
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/cmdc.201100279.
References
- 1.Thompson CB. Science. 1995;267:1456–1462. doi: 10.1126/science.7878464. [DOI] [PubMed] [Google Scholar]
- 2.Reed JC. Nat. Rev. Drug Discovery. 2002;1:111–121. doi: 10.1038/nrd726. [DOI] [PubMed] [Google Scholar]
- 3.Ghobrial IM, Witzig TE, Adjei AA. CA Cancer J. Clin. 2005;5:178–194. doi: 10.3322/canjclin.55.3.178. [DOI] [PubMed] [Google Scholar]
- 4.a Risinger AL, Giles FJ, Mooberry SL. Cancer Treat. Rev. 2009;35:255–261. doi: 10.1016/j.ctrv.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Carlson RO. Expert Opin. Invest. Drugs. 2008;17:707–722. doi: 10.1517/13543784.17.5.707. [DOI] [PubMed] [Google Scholar]
- 5.a Walczak CE. Curr. Opin. Cell Biol. 2000;12:52–56. doi: 10.1016/s0955-0674(99)00056-3. [DOI] [PubMed] [Google Scholar]; b Honore S, Pasquier E, Braguer D. Cell. Mol. Life Sci. 2005;62:3039–3056. doi: 10.1007/s00018-005-5330-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pettit GR, Singh SB, Hamel E, Lin CM, Alberts DS, Garcia-Kendall D. Experientia. 1989;45:209–211. doi: 10.1007/BF01954881. [DOI] [PubMed] [Google Scholar]
- 7.Lin CM, Ho HH, Pettit GR, Hamel E. Biochemistry. 1989;28:6984–6991. doi: 10.1021/bi00443a031. [DOI] [PubMed] [Google Scholar]
- 8.Patterson DM, Rustin GJS. Drugs Future. 2007;32:1025–1032. [Google Scholar]
- 9.a Tron GC, Pirali T, Sorba S, Pagliai F, Busacca S, Genazzani AA. J. Med. Chem. 2006;49:3033–3044. doi: 10.1021/jm0512903. [DOI] [PubMed] [Google Scholar]; b Mahindroo N, Liou JP, Chang JY, Hsieh HP. Expert Opin. Ther. Pat. 2006;16:647–691. [Google Scholar]; c Jordan MA, Wilson L. Nat. Rev. Cancer. 2004;4:253–265. doi: 10.1038/nrc1317. [DOI] [PubMed] [Google Scholar]; d Chaudhary A, Pandeya SN, Kumar P, Sharma PP, Gupta S, Soni N, Verma KK, Bhardwaj G. Mini Rev. Med. Chem. 2007;7:1186–1205. doi: 10.2174/138955707782795647. [DOI] [PubMed] [Google Scholar]; e Hsieh HP, Liou JP, Mahindroo N. Curr. Pharm. Des. 2005;11:1655–1677. doi: 10.2174/1381612053764751. [DOI] [PubMed] [Google Scholar]
- 10.a Hatanaka T, Fujita K, Ohsumi K, Nakagawa R, Fukuda Y, Nihei Y, Suga Y, Akiyama Y, Tsuji T. Bioorg. Med. Chem. Lett. 1998;8:3371–3374. doi: 10.1016/s0960-894x(98)00622-2. [DOI] [PubMed] [Google Scholar]; b Ohsumi K, Nakagawa R, Fukuda Y, Hatamaka T, Morinaga Y, Nihei Y, Ohishi K, Suga Y, Akiyama Y, Tsuji T. J. Med. Chem. 1998;41:3022–3032. doi: 10.1021/jm980101w. [DOI] [PubMed] [Google Scholar]
- 11.a Delmonte A, Sessa C. Exp. Opin. Invest. Drugs. 2009;18:1541–1548. doi: 10.1517/13543780903213697. [DOI] [PubMed] [Google Scholar]; b Siemann DW, Chaplin DJ, Walike P. Expert Opin. Invest. Drugs. 2009;18:189–197. doi: 10.1517/13543780802691068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.a Hori K, Saito S. Br. J. Cancer. 2003;89:1334–1344. doi: 10.1038/sj.bjc.6601261. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Hori K, Saito S. Br. J. Cancer. 2004;90:549–53. doi: 10.1038/sj.bjc.6601582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romagnoli R, Baraldi PG, Sarkar T, Carrion MD, Cruz-Lopez O, Lopez-Cara C, Tolomeo M, Grimaudo S, Di Cristina A, Pipitone MR, Balzarini J, Gambari R, Ilaria L, Saletti R, Brancale A, Hamel E. Bioorg. Med. Chem. 2008;16:8419–8426. doi: 10.1016/j.bmc.2008.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gaukroger K, Hadfield JA, Lawrence NJ, Nlan S, McGown AT. Org. Biomol. Chem. 2003;1:3033–3037. doi: 10.1039/b306878a. [DOI] [PubMed] [Google Scholar]
- 15.Beria I, Baraldi PG, Cozzi P, Caldarelli M, Geroni C, Marchini S, Mongelli N, Romagnoli R. J. Med. Chem. 2004;47:2611–2623. doi: 10.1021/jm031051k. [DOI] [PubMed] [Google Scholar]
- 16.Romagnoli R, Baraldi PG, Cruz-Lopez O, Lopez-Cara C, Preti D. Mini-Rev. Med. Chem. 2009;9:81–94. doi: 10.2174/138955709787001640. [DOI] [PubMed] [Google Scholar]
- 17.a Nam NH, Kim Y, Yu YJ, Hong DH, Kim HM, Ahn BZ. Bioorg. Med. Chem. 2003;11:1021–1029. doi: 10.1016/s0968-0896(02)00514-x. [DOI] [PubMed] [Google Scholar]; b Pettit GR, Anderson CR, Herald DL, Jung MK, Lee DJ, Hamel E, Pettit RK. J. Med. Chem. 2003;46:525–531. doi: 10.1021/jm020204l. [DOI] [PubMed] [Google Scholar]
- 18.2-Hydroxy-3-nitrobenzaldehyde (6e), 2-hydroxy-5-nitrobenzaldehyde (6b), and 1-(2-hydroxy-5-nitrophenyl)ethanone (6c) are commercially available and were used as received; 2-hydroxy-6-nitrobenzaldehyde (6a) was prepared by following the procedure reported here: Miranda LP, Meutermans WDF, Smythe ML, Alewood PF. J. Org. Chem. 2000;65:5460–5468. doi: 10.1021/jo991340+.. For the preparation of 6d, see: MacMillan KS, Nguyen T, Hwang I, Boger DL. J. Am. Chem. Soc. 2009;131:1187–1194. doi: 10.1021/ja808108q.. For the preparation of 1-(2-hydroxy-5-methoxy-3-nitrophenyl)ethanone (6 f), see Cushman M, Zhu H, Geahlen RL, Kraker AJ. J. Med. Chem. 1994;37:3353–3362. doi: 10.1021/jm00046a020.. For the synthesis of 2-hydroxy-4-methoxy-5-nitrobenzaldehyde (6g), see: Guru M, Rao S, Srikantia C, Iyengar MS. J. Chem. Soc. 1924:556–560. For the preparation of 1-(2-hydroxy-4-methoxy-5-nitrophenyl)ethanone (6h), see: Banks H. J. Am. Chem. Soc. 1938;60:1370.. For the synthesis of 2-hydroxy-3-methoxy-5-nitrobenzaldehyde (6i), see: Kowalewska M, Kwiecień H. Tetrahedron. 2008;64:5085–5090.. For the synthesis of 1-(2-hydroxy-3-methoxy-5-nitrophenyl)ethanone (6j), see: Horton W, Spence JT. J. Am. Chem. Soc. 1958;80:2453–2456.
- 19.a Hamel E. Cell Biochem. Biophys. 2003;38:1–21. doi: 10.1385/CBB:38:1:1. [DOI] [PubMed] [Google Scholar]; b Verdier-Pinard P, Lai J-Y, Yoo H-D, Yu J, Marquez B, Nagle DG, Nambu M, White JD, Falck JR, Gerwick WH, Day BW, Hamel E. Mol. Pharmacol. 1998;53:62–76. doi: 10.1124/mol.53.1.62. [DOI] [PubMed] [Google Scholar]
- 20.Rello S, Stockert J, Moreno V, Gamez A, Pacheco M, Juarranz A, Canete M, Villanueva A. Apoptosis. 2005;10:201–208. doi: 10.1007/s10495-005-6075-6. [DOI] [PubMed] [Google Scholar]
- 21.Steller H. Science. 1995;267:1445–1449. doi: 10.1126/science.7878463. [DOI] [PubMed] [Google Scholar]
- 22.a Nhan TQ, Liles WC, Scwartz SM. Am. J. Pathol. 2006;169:729–737. doi: 10.2353/ajpath.2006.060105. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Graf D, Bode JG, Haussinger D. Arch. Biochem. Biophys. 2007;462:162–170. doi: 10.1016/j.abb.2007.03.029. [DOI] [PubMed] [Google Scholar]
- 23.a Susin SA, Lorenzo HK, Zamzami N, Marzo I, Brenner C, Larochette N, Prevost MC, Alzari PM, Kroemer G. J. Exp. Med. 1999;189:381–394. doi: 10.1084/jem.189.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Denault J-B, Salvesen GS. Chem. Rev. 2002;102:4489–4499. doi: 10.1021/cr010183n. [DOI] [PubMed] [Google Scholar]
- 24.Soldani C, Scovassi AI. Apoptosis. 2002;7:321–328. doi: 10.1023/a:1016119328968. [DOI] [PubMed] [Google Scholar]
- 25.Ratnam K, Low JA. Clin. Cancer Res. 2007;13:1383–1388. doi: 10.1158/1078-0432.CCR-06-2260. [DOI] [PubMed] [Google Scholar]
- 26.Knudson CM, Korsmeyer SJ. Nature Genet. 1997;16:358–363. doi: 10.1038/ng0897-358. [DOI] [PubMed] [Google Scholar]
- 27.a Jiang X, Wang X. Annu. Rev. Biochem. 2004;73:87–106. doi: 10.1146/annurev.biochem.73.011303.073706. [DOI] [PubMed] [Google Scholar]; b Kluck RM, Bossy-Wetzel E, Green DR, Newmeyer DD. Science. 1997;275:1132–1136. doi: 10.1126/science.275.5303.1132. [DOI] [PubMed] [Google Scholar]
- 28.Vincent L, Kermani P, Young LM, Cheng J, Zhang F, Shido K, Lam G, Bompais-Vincent H, Zhu Z, Hicklin DJ, Bohlen P, Chaplin DJ, May C, Rafii S. J. Clin. Invest. 2005;115:2992–3006. doi: 10.1172/JCI24586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.a Cuccia DJ, Bevilacqua F, Durkin AJ, Tromberg B. J. Opt. Lett. 2005;30:1354–1356. doi: 10.1364/ol.30.001354. [DOI] [PubMed] [Google Scholar]; b Cuccia DJ, Bevilacqua F, Durkin AJ, Ayers FR, Tromberg BJ. J. Biomed. Opt. 2009;14:024012. doi: 10.1117/1.3088140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brindle K. Nature Rev. Cancer. 2008;8:94–107. doi: 10.1038/nrc2289. [DOI] [PubMed] [Google Scholar]
- 31.Hamel E, Lin CM. Biochemistry. 1984;23:4173–4184. doi: 10.1021/bi00313a026. [DOI] [PubMed] [Google Scholar]
- 32.Rubio S, Quintana J, Eiroa JL, Triana J, Estévez F. Carcinogenesis. 2007;28:2105–2013. doi: 10.1093/carcin/bgm131. [DOI] [PubMed] [Google Scholar]
- 33.Bradford MM. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 34.Erickson TA, Mazhar A, Cuccia A, Durkin J, Tunnel JW. J. Biom. Opt. 2010;15:036013. doi: 10.1117/1.3431728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ravelli RBG, Gigant B, Curmi PA, Jourdain I, Lachkar S, Sobel A, Knossow M. Nature. 2004;428:198–202. doi: 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
- 36.Molecular Operating Environment (MOE 2010.10) Chemical Computing Group Inc.; Montreal, QC (Canada): [July 6, 2011]. http://www.chemcomp.com. [Google Scholar]
- 37.Korb O, Stützle T, Exner TE. PLANTS: Application of Ant Colony Optimization to Structure-Based Drug Design. In: Dorigo M, Gambardella LM, Birattari M, Martinoli A, Poli R, St tzle T, editors. Ant Colony Optimization and Swarm Intelligence, 5th International Workshop, ANTS 2006; Brussels (Belgium). September 4–7, 2006; Berlin: Springer; 2006. pp. 247–258. LNCS 4150. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.