Abstract
N-Acetylglucosamine (GlcNAc) stimulates important signaling pathways in a wide range of organisms. In the human fungal pathogen Candida albicans, GlcNAc stimulates hyphal cell morphogenesis, virulence genes, and the genes needed to catabolize GlcNAc. Previous studies on the GlcNAc transporter (NGT1) indicated that GlcNAc has to be internalized to induce signaling. Therefore, the role of GlcNAc catabolism was examined by deleting the genes required to phosphorylate, deacetylate, and deaminate GlcNAc to convert it to fructose-6-PO4 (HXK1, NAG1, and DAC1). As expected, the mutants failed to utilize GlcNAc. Surprisingly, GlcNAc inhibited the growth of the nag1Δ and dac1Δ mutants in the presence of other sugars, suggesting that excess GlcNAc-6-PO4 is deleterious. Interestingly, both hxk1Δ and an hxk1Δ nag1Δ dac1Δ triple mutant could be efficiently stimulated by GlcNAc to form hyphae. These mutants could also be stimulated to express GlcNAc-regulated genes. Because GlcNAc must be phosphorylated by Hxk1 to be catabolized, and also for it to enter the anabolic pathways that form chitin, N-linked glycosylation, and glycosylphosphatidylinositol anchors, the mutant phenotypes indicate that GlcNAc metabolism is not needed to induce signaling in C. albicans. Thus, these studies in C. albicans reveal a novel role for GlcNAc in cell signaling that may also regulate critical pathways in other organisms.
Keywords: Cell Differentiation, Gene Regulation, Signal Transduction, Yeast Genetics, Yeast Metabolism, Candida albicans, Hyphae, N-Acetylglucosamine
Introduction
N-Acetylglucosamine (GlcNAc) is an interesting molecule because it plays important roles in both cell structure and cell signaling. GlcNAc contributes significantly to cell-surface structure in a very wide range of organisms; it is a key component of the bacterial cell wall peptidoglycan layer, fungal cell wall chitin, and the extracellular matrix glycosaminoglycans in animal cells. Eukaryotic cells also use GlcNAc to modify cell-surface proteins by N-linked glycosylation and in the formation of glycosylphosphatidylinositol anchors. Additionally, GlcNAc is widely used as a signaling molecule. For example, bacterial cells respond to extracellular GlcNAc by altering the production of CURLI fibers that function in biofilm formation (1), and some yeast species are induced by GlcNAc to switch from budding to a filamentous hyphal morphology that promotes invasive growth (2, 3). Animal cells respond to increased GlcNAc by promoting the post-translational attachment of GlcNAc to intracellular proteins, including the transcription factors c-Myc, p53, and NF-κB that play key roles in cellular regulation (4). This type of O-linked GlcNAc attachment has been implicated in a range of human diseases, including cancer and diabetes (5).
GlcNAc signaling is under investigation in the human fungal pathogen Candida albicans because it is a potent inducer of hyphal growth, whereas other sugars are not (2). GlcNAc induces two sets of responses that have significance for understanding the mechanisms of pathogenesis. One pathway stimulates C. albicans to switch from budding to hyphal growth and to induce expression of virulence genes. These responses are activated by stimulation of adenylyl cyclase and increased cAMP signaling (2, 6–8). Activation of cAMP signaling by GlcNAc also promotes C. albicans cells to undergo an epigenetic switch from the white phase morphology to a distinct state known as the opaque phase that is thought to be better suited for mucosal infections (9). This is consistent with the expected presence of GlcNAc in the gastrointestinal tract mucosa as the result of cell wall remodeling by the endogenous bacterial population. GlcNAc also activates a pathway that is independent of cAMP to induce the expression of the genes needed to catabolize GlcNAc (8, 10, 11). The signaling pathways induced by GlcNAc in C. albicans are not yet defined well. One reason for this is that the commonly studied model yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe lack the genes needed to catabolize GlcNAc (12). Also, post-translational modification of proteins by O-GlcNAc attachment is not known to occur in C. albicans as it does in humans and other metazoans. Thus, C. albicans must use distinct GlcNAc signaling strategies whose identification may help to identify novel signaling pathways in metazoans and other organisms.
Previous studies indicated that GlcNAc must enter C. albicans cells to induce signaling. Transport of GlcNAc into C. albicans is mediated by a recently discovered plasma membrane transporter, Ngt1, which represents the first eukaryotic GlcNAc transporter to be identified (12). Ngt1 appears to specifically transport GlcNAc, and not other related sugars. Mutant cells lacking Ngt1 (ngt1) showed greatly reduced GlcNAc uptake and required about 1,000-fold higher levels of GlcNAc to induce signaling (12). This is similar to the results for galactose-induced signaling in S. cerevisiae, which is activated by intracellular galactose binding to the cytoplasmic Gal3 protein that then shuttles to the nucleus to regulate the Gal4 transcriptional regulator (13, 14). It is not known how GlcNAc is sensed and whether signaling requires its catabolism, which would release acetate and ammonia in the cell. Therefore, in this study we examined how GlcNAc signaling was affected in cells lacking the proteins that catabolize GlcNAc. The genes that mediate GlcNAc catabolism have been identified previously and are known to encode Hxk1, a kinase that phosphorylates GlcNAc to create GlcNAc-6-PO4, Dac1, a deacetylase that converts it to glucosamine-6-PO4, and Nag1, a deaminase that converts it to fructose-6-PO4 that can then be used in glycolysis (10, 11, 15). These experiments also test whether signaling requires the exogenous GlcNAc to enter the anabolic pathways that form chitin, N-linked glycosylation, and glycosylphosphatidylinositol anchors on proteins (16). Phosphorylation of exogenous GlcNAc by Hxk1 is needed to create GlcNAc-6-PO4, which is then converted to UDP-GlcNAc for use as a substrate in the anabolic pathways (16). The results demonstrate that GlcNAc metabolism is not required for induction of hyphal growth or expression of the catabolic genes, thereby identifying a novel role for GlcNAc in cell signaling.
EXPERIMENTAL PROCEDURES
Strains and Media
The genotypes of the C. albicans strains used in this study are listed in Table 1. C. albicans cells were propagated on rich YPD medium or on synthetic medium (17). The medium was supplemented with 80 mg/liter uridine to permit growth of ura3 mutants. The ability of cells to grow on different sugars was assayed by spotting dilutions of cells onto solid media agar plates followed by incubation at 37 °C.
TABLE 1.
C. albicans strains used in this study
| Strain | Genotype |
|---|---|
| BWP17 | ura3Δ::limm434/ura3Δ::limm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| DIC185 | ura3Δ::limm434/URA3 his1::hisG/HIS1 arg4::hisG/ARG4 |
| AG673 | NAG1/nag1::ARG4 ura3Δ::limm434/ura3Δ::limm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| AG676 | nag1::HIS1/nag1::ARG4 ura3Δ::limm434/ura3Δ::limm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| AG734 | nag1::HIS1/nag1::ARG4 URA3/ura3Δ::limm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| AG767 | nag1::HIS1/nag1::ARG4 NAG1::URA3 ura3Δ::limm434/ura3Δ::limm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| AG648 | DAC1/dac1::ARG4 ura3Δ::limm434/ura3Δ::limm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| AG712 | dac1::HIS1/dac1::ARG4 ura3Δ::limm434/ura3Δ::limm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| AG732 | dac1::HIS1/dac1::ARG4 URA3/ura3Δ::limm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| AG762 | dac1::HIS1/dac1::ARG4 DAC1::URA3 ura3Δ::limm434/ura3Δ::limm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| AG697 | HXk1/hxk1::ARG4 ura3Δ::limm434/ura3Δ::limm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| AG689 | hxk1::URA3/hxk1::ARG4 ura3Δ::limm434/ura3Δ::limm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| AG736 | hxk1::URA3/hxk1::ARG4 ura3Δ::limm434/ura3Δ::limm434 HIS1/his1::hisG arg4::hisG/arg4::hisG |
| SN772 | hxk1::URA3/hxk1::ARG4 HXK1::HIS1 ura3Δ::limm434/ura3Δ::limm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| AG695 | Heterozygous h-d strain; [hxk1Δ nag1Δ dac1Δ]::ARG4 /HXK1 NAG1 DAC1 his1::hisG/his1::hisG ura3Δ::limm434/ura3Δ::limm434 arg4::hisG/arg4::hisG |
| AG692 | Homozygous h-d strain; [hxk1 nag1 dac1]::ARG4/[ hxk1Δ nag1Δ dac1Δ]::URA3 his1::hisG/his1::hisG ura3Δ::limm434/ura3Δ::limm434 arg4::hisG/arg4::hisG |
| AG738 | His+ homozygous h-d strain; [hxk1 nag1 dac1]::ARG4/[ hxk1 nag1Δ dac1Δ]::URA3 HIS1/his1::hisG ura3Δ::limm434/ura3Δ::limm434 arg4::hisG/arg4::hisG |
| SN778 | Complemented h-d strain; [hxk1Δ nag1Δ dac1Δ]::ARG4/[ hxk1Δ nag1Δ dac1Δ]::URA3 [HXK1 NAG1 DAC1]::HIS1 his1::hisG /his1::hisG ura3Δ::limm434/ura3Δ::limm434 arg4::hisG/arg4::hisG |
| SN787 | NGT1-GFPγ::HIS1 ura3Δ::limm434/ura3Δ::limm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| SN791 | hxk1Δ strain AG689 plus NGT1-GFPγ::HIS1 |
| SN794 | hxk1Δ nag1Δ dac1Δ strain AG692 plus NGT1-GFPγ::HIS1 |
| 180 | cph1::HIS1/cph1::LEU2 his1/his1 leu2/leu2 arg4/arg4 |
| CJN305 | cph1::UAU/cph1::URA3 his1::hisG/his1::hisG leu2/leu2 arg4/arg4 |
| 1112 | mkc1::HIS1/mkc1::LEU2 his1/his1 leu2/leu2 arg4/arg4 |
| 544 | cek1::HIS1/cek1::LEU2 his1/his1 leu2/leu2 arg4/arg4 |
| 060 | efg1::HIS1/efg1::LEU2 his1/his1 leu2/leu2 arg4/arg4 |
| SN250 | LEU2/leu2 HIS1/his1 arg4/arg4 |
Homozygous dac1Δ/dac1Δ and nag1Δ/nag1Δ deletion mutants that removed the entire open reading frames were constructed in C. albicans strain BWP17 (arg4Δ his1Δ ura3Δ) using methods described previously (18). In brief, PCR primers containing ∼70 bp of sequence homologous to the sequences flanking the open reading frame of NAG1 or DAC1 were used to amplify the ARG4 and the HIS1 selectable marker genes. Integration of these deletion cassettes at the appropriate sites to delete the corresponding copies of the NAG1 or DAC1 genes was verified by PCR using combinations of primers that flanked the integration and also primers that annealed within the introduced cassettes. Complementation of the nag1Δ and dac1Δ strains was performed by introducing a plasmid carrying one wild-type copy of NAG1 or DAC1 into the genome. These plasmids were constructed by PCR amplification of the genomic DNA from 1,000 bp upstream of the initiator ATG to 300 bases downstream of the terminator codon of NAG1 or DAC1. These DNA fragments were then inserted between the SacI and SacII restriction sites of the URA3 plasmid pDDB57 (19). The resulting DAC1 plasmid was linearized in the promoter region by digestion with PstI and integrated into the dac1Δ/dac1Δ strain AG732 using URA3 selection to create complemented strains AG762. Similarly, the NAG1 plasmid was linearized by digestion with NcoI and then integrated into a nag1Δ/nag1Δ strain to create the complemented strain AG767. To create prototrophic versions of the dac1Δ/dac1Δ and nag1Δ/nag1Δ mutants, they were transformed with a URA3-containing DNA fragment to restore URA3 at its native locus. The URA3 fragments were liberated from plasmid pBSK-URA3 (20) by digestion with restriction enzymes PstI and NotI.
For reasons that are unclear, homozygous deletions mutants lacking HXK1 or lacking the whole set of contiguous genes HXK1, DAC1, and NAG1 could not be isolated using selection with ARG4 and HIS1. Therefore, a similar strategy was used in which the corresponding genes were deleted with ARG4 and URA3 to create the hxk1Δ mutant AG736 and the h-d mutant (hxk1Δ nag1Δ dac1Δ) AG738. Complementing plasmids were then created using a derivative of pDDB57 in which URA3 was replaced with HIS1. The complementing plasmids were linearized by digestion with SphI and integrated into the corresponding deletion mutants using selection for HIS1 to create strains SN772 and SN778. Prototrophic versions of the mutants were created by transformation with a DNA fragment containing HIS1.
C. albicans strains carrying NGT1-GFP strain were created by homologous recombination of GFP sequences into the 3′ end of the NGT1 open reading frame using previously described methods (21). PCR primers containing ∼70 bp of sequence homologous to the 3′ end of the NGT1 open reading frame were used to amplify a cassette containing a more photostable version of enhanced GFP (CaGFPγ) and a URA3 selectable marker (21). The resulting Ura+ colonies from the transformation into C. albicans were then screened for GFP-positive cells by fluorescence microscopy and confirmed by PCR.
Microscopy
The ability of the wild-type control strain DIC185 and mutant cells to form hyphae was carried out with cells grown overnight to early log phase in synthetic medium with galactose. The cells were then resuspended in synthetic medium containing 50 mm galactose plus or minus the indicated concentration of GlcNAc and grown for 2 h at 37 °C and then photographed using differential interference contrast microscopy. Induction of Ngt1-GFP was detected in cells that were grown overnight to log phase in synthetic medium containing dextrose, washed, and then resuspended in medium containing dextrose or GlcNAc as indicated. Ngt1-GFP was detected by fluorescence microscopy, and light microscope images were taken with differential interference contrast optics. Images were captured using an Olympus BH2 microscope equipped with a Zeiss AxioCam digital camera. Fluorescent cell signals were quantified using AxioVision software.
Analysis of mRNA Levels Using Real Time Quantitative RT-PCR (qRT-PCR)4
Cells were grown overnight to log phase, washed, and then incubated in synthetic medium containing 50 mm dextrose or GlcNAc for 2 h, and the cell pellets were then frozen at −80 °C. RNA was extracted from a pellet of about 3 × 108 cells using a RiboPure-Yeast RNA isolation kit (Ambion). RNA samples were tested by PCR to confirm the absence of DNA contamination. cDNA was then synthesized from total RNA using an oligo(dT) primer (Invitrogen) and Superscript III reverse transcriptase (Invitrogen). The cDNA samples were treated with RNase A (New England Biolabs) and purified using a PCR cleanup column (Qiagen). The cDNA samples were then used as templates for qRT-PCR in a Mastercycler EP Realplex2 (Eppendorf). The reaction mix included 2× iQ SYBR Green Supermix (Bio-Rad), 1 μl of first-strand cDNA reaction mixture, and 0.1 μm of primers per 10-μl reaction. The amplification program was initiated with a 95 °C denaturation step for 5 min, followed by 40 cycles of 95 °C for 15 s, 55 °C for 15 s, and 72 °C for 30 s. Raw data were analyzed using Mastercycler Realplex2 software analysis module (ΔΔCT method) to determine the relative differences in gene expression, which were normalized to the level of ACT1 mRNA. The specificity of the reaction products was assessed by melting curve analysis at the end of the qPCR program. The results reported represent the average of at least three independent assays each done in triplicate. Where indicated, the results include the analysis of two independent RNA preparations from independently derived mutant strains. The following primers used for qPCR analysis were designed using Primer3 software and ordered from Invitrogen: ACT1-F 5′-TCCAGAAGCTTTGTTCAGACCAGC-3′ and ACT1-R 5′-TGCATACGTTCAGCAATACCTGGG-3′; NGT1-F 5′-TCGTGCCAAAATTGGTTGGGCT-3′ and NGT1-R 5′-TGGACATGGCTCCCAATACCCA-3′; HXK1-F 5′-TGTGAGCTCTCGTTGTTTGG-3′ and HXK1-R 5′-TCAATTCCGGCAAGTACCTC-3′; NAG1-F 5′-GAAGCGGGATCATCAAGAAA-3′ and NAG1-R 5′-TGGCAATTTCGTCTGAGTTG-3′; DAC1-F 5′-CGTTGCCGCAATGTGTGCGT-3′ and DAC1-R 5′-GACGACTGGACTTCACGTCCCA-3′; GIG1-F 5′-GCAAACCCCACCCACTTCACCA-3′ and GIG1-R 5′-TGTTTGCTGTCGTGATCGAGCA-3′; GAL10-F 5′-AGGAGCAAACAACTTGCATGGTGG-3′ and GAL10-R 5′-GCTTCAAGCTCACCTGGGAACC-3′; and PMA1-F 5′-TTGCCAAGATTGTGGGGTAT-3′ and PMA1-R 5′-GACCAGAATGGACCTTGAGC-3′.
RESULTS
Deletion of HXK1, NAG1, and DAC1
Previous studies identified the C. albicans HXK1, NAG1, and DAC1 genes that encode the enzymes needed to catabolize GlcNAc (10, 11, 22). Although mutants lacking these genes were constructed previously, there are now concerns regarding the approaches used for recycling of the URA-selectable marker to delete both copies of each gene in the diploid C. albicans genome and also for the use of control strains carrying a reintroduced copy of the deleted gene (23, 24). Therefore, as described under “Experimental Procedures,” we constructed new deletion mutants using the auxotrophic strain BWP17 (arg4Δ his1Δ ura3Δ) (18). In addition to creating individual hxk1Δ, nag1Δ, and dac1Δ mutants, we also made a triple mutant. All three genes could be deleted in one step because they are clustered on chromosome 6. For simplicity, the triple hxk1Δ nag1Δ dac1Δ mutant will be referred to as the h-d mutant as it removes the gene cluster from HXK1 to DAC1.
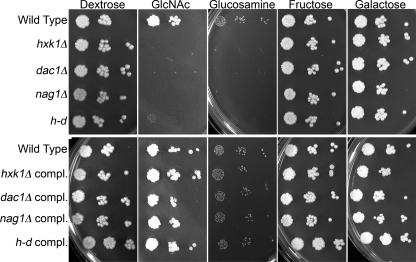
The homozygous deletion mutants were examined for their ability to grow on different carbon sources by spotting dilutions of cells onto solid agar medium (Fig. 1). As expected, all of the mutants failed to grow on medium containing GlcNAc as the sole carbon and energy source. There was no growth on GlcNAc for the mutants even after extended periods of time (1 month). Any minor amount of growth observed on GlcNAc medium was similar to that seen for cells grown on a medium lacking any added sugar (data not shown). Control studies showed that the mutants grew well on media containing dextrose, fructose, and the unrelated sugar galactose but failed to grow on glucosamine medium. It is not clear why the GlcNAc deacetylase mutant (dac1Δ) did not grow on glucosamine. However, growth on GlcNAc and glucosamine media was restored in the complemented strains in which one copy of the wild-type gene was reintroduced (Fig. 1, lower panel).
FIGURE 1.
Growth of mutant strains on media containing different sugars. Dilutions of a wild-type control and the indicated mutants strains were spotted onto the solid agar media plates containing 50 mm of the indicated sugar and then incubated at 37 °C for 2 days. The strains used in the upper panel are the wild-type control (DIC185), hxk1Δ (AG736), dac1Δ (AG732), nag1Δ (AG734), and a triple hxk1Δ dac1Δ nag1Δ mutant referred to as the h-d strain (AG738). The complemented (compl.) strains in the lower panel, which carry a wild-type copy of the corresponding gene integrated into the genome of the mutant cells, included hxk1Δ + HXK1 (SN772), dac1Δ + DAC1 (AG762), nag1Δ + NAG1 (AG767), and h-d + HXK1 NAG1 DAC1 (SN778) strains.
GlcNAc Inhibits Growth of nag1Δ and dac1Δ Mutants
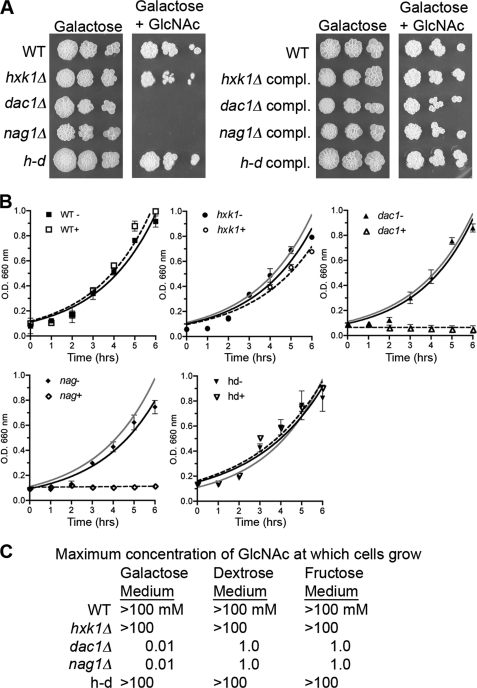
The response of the mutants to GlcNAc was tested by growing cells in galactose medium and then adding GlcNAc as an inducer. Galactose was selected to provide a source of carbon and energy because it is an unrelated sugar that does not repress induction of GlcNAc catabolic genes, as does dextrose (8, 12). Surprisingly, GlcNAc inhibited the growth of the nag1Δ and dac1Δ mutants on solid agar plates containing galactose (Fig. 2A). Growth rate studies in liquid showed that addition of 10 mm GlcNAc rapidly blocked growth of the nag1Δ and dac1Δ mutants even though 50 mm galactose was present as a nutrient source (Fig. 2B). These studies also revealed a very slight delay in the growth of the hxk1Δ mutant but no effect on the growth of the h-d mutant. The observation that the hxk1Δ and h-d mutants grow well in the presence of GlcNAc indicates that it must be converted to GlcNAc-6-PO4 to cause the strong inhibitory effects.
FIGURE 2.
GlcNAc inhibits growth of nag1Δ and dac1Δ mutants. A, dilutions of the indicated strains were spotted onto medium containing 50 mm galactose plus or minus 10 mm GlcNAc and then incubated for 2 days at 37 °C. B, short term growth assays for cells incubated at 37 °C in medium containing 50 mm galactose plus (+) or minus (−) 10 mm GlcNAc as indicated. For comparison, the wild-type growth curve is shown as a gray line in the mutant growth curves. C, maximum concentration of GlcNAc at which cells can grow in synthetic medium that also contains 50 mm galactose, 50 mm dextrose, or 50 mm fructose. Cells were tested for growth in the presence of the indicated sugar plus 10-fold different concentrations of GlcNAc from 10 μm to 100 mm. Strains are as described in the legend to Fig. 1. compl., complemented.
Dose-response assays demonstrated that 0.1 mm GlcNAc inhibited the growth of the nag1Δ and dac1Δ mutants in the presence of 50 mm galactose (Fig. 2C). This inhibitory effect was not specific to galactose medium. GlcNAc also inhibited the growth of the nag1Δ and dac1Δ mutants in dextrose or fructose media (Fig. 2C), although 100-fold higher concentrations of GlcNAc were required to inhibit growth. These results are consistent with dextrose or fructose having a stronger ability to repress the expression of the GlcNAc transporter (12).
Even though their growth was blocked, the nag1Δ and dac1Δ mutants maintained a very high degree of viability after 24 h of incubation in galactose plus GlcNAc, indicating GlcNAc inhibited their growth but did not kill them (data not shown). Prolonged incubation of the cells in liquid cultures revealed that the nag1Δ mutant eventually recovered after 3 days and grew to high density. This growth was not due to a spontaneous mutation, as the cells were still inhibited when inoculated into fresh medium. In contrast, the nag1Δ and dac1Δ mutants did not show obvious growth after extended incubation on solid medium agar plates containing galactose plus GlcNAc (Fig. 2A and data not shown).
Similar inhibitory effects of GlcNAc have also been reported for mutant bacteria lacking the orthologs of NAG1 or DAC1 (25). In bacteria, the inhibitory effects of GlcNAc could be rapidly reversed by adding uridine to the medium (26). However, addition of uridine did not quickly rescue growth of C. albicans nag1Δ and dac1Δ mutants as it did in bacteria. Uridine did not rescue growth of cells on agar plates, even after extended incubation. Addition of uridine also did not significantly improve the growth of the nag1Δ mutant in liquid culture, but it did enable a C. albicans dac1Δ mutant to resume growth in liquid after a lag of about 3 days. Thus, the effects of uridine on growth of the C. albicans nag1Δ and dac1Δ mutants are distinct from the effects on the analogous E. coli mutants.
GlcNAc Catabolism Is Not Needed to Induce Hyphal Growth
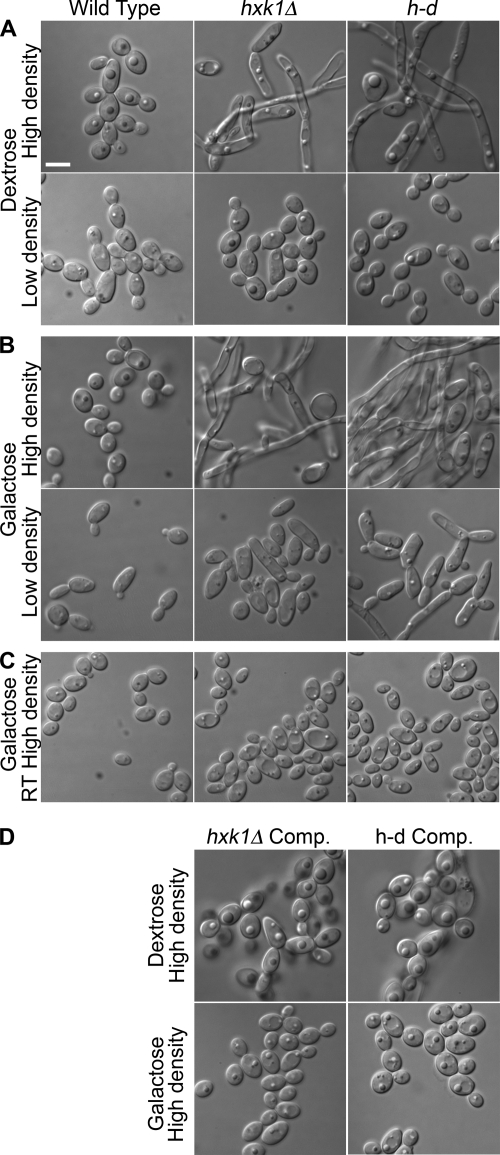
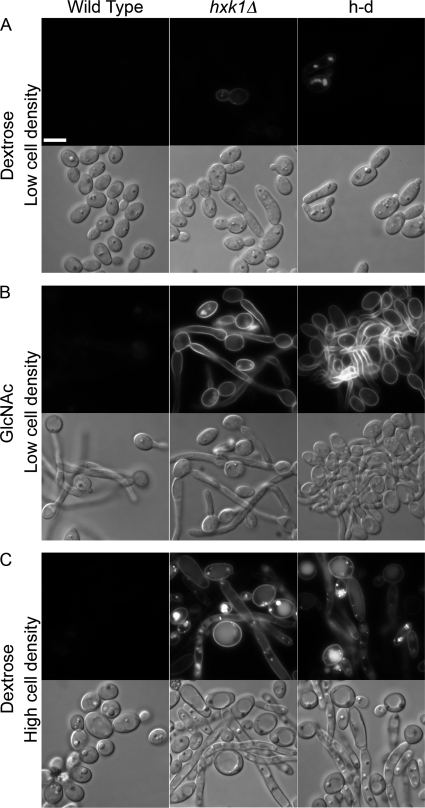
The hxk1Δ and h-d mutants grew well in the presence of galactose plus GlcNAc, so they were tested for the ability to be induced by GlcNAc to form hyphae. Interestingly, control studies showed that the hxk1Δ and the h-d mutants displayed a high basal level of filamentous growth in the absence of GlcNAc (Fig. 3). Deletion of HXK1 was reported previously to cause filamentous growth as part of a study aimed at defining the mechanisms of epithelial cell colonization (27). Analysis of mutant cells grown in dextrose medium revealed that the filamentous growth was most pronounced in cell cultures grown to high density, whereas the cells maintained at low density were grew primarily as buds (Fig. 3A). Similar results were observed for cells grown in galactose, except that there was a higher degree of abnormal-looking elongated cells in the low density cultures (Fig. 3B), consistent with the expected increase in expression of the GlcNAc transporter in galactose medium. The high level of filamentous growth was specific for the hxk1Δ and h-d mutations, as it was not seen in the complemented strains (Fig. 3D). Also, it appears to be due to activation of the hyphal pathway because the cells formed normal looking buds at room temperature, which prevents activation of the hyphal pathway (Fig. 3C). These data suggest that the failure of the hxk1Δ and h-d mutants to catabolize GlcNAc released during cell wall remodeling and septation results in it accumulating in the medium and stimulating morphological changes of cells grown to high density. This is supported by the observation that the hxk1Δ and h-d mutants also show elevated levels of Ngt1-GFP at high cell density, consistent with the accumulation of GlcNAc in the medium (see below).
FIGURE 3.
Mutant cell morphology varies with cell density in the absence of GlcNAc. Different dilutions of the wild-type control and mutant strains were grown overnight at 37 °C in synthetic medium containing either dextrose (A) or galactose (B). Light microscopic images were then captured for cells that were maintained in early log phase (labeled Low density, ≤106 cells/ml) or grown to saturation (labeled High density, ∼108 cells/ml). C, cells grown to high density in galactose medium at room temperature. D, complemented (Comp.) strains in which a copy of the wild-type gene was reintroduced into the mutant cells. Strains are as described in the legend to Fig. 1. Bar, 5 μm.
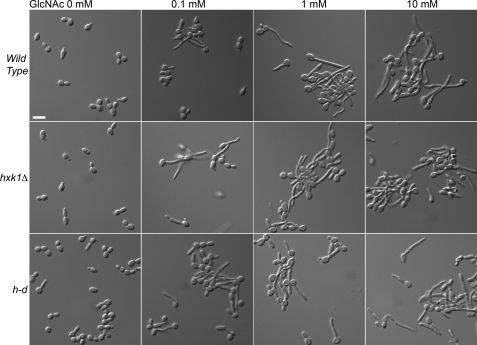
The wild-type and mutants were then tested for capacity to be induced by GlcNAc to form hyphae (Fig. 4). Interestingly, the hxk1Δ and h-d mutants grown in galactose medium could be induced by GlcNAc to form long filamentous cells with parallel walls, indicative of true hyphae and not just pseudohyphal cells. Dose-response assays showed that the mutants could be induced efficiently, even at low doses of GlcNAc (e.g. 0.1 mm). Thus, GlcNAc catabolism is not needed for induction of hyphal growth.
FIGURE 4.
GlcNAc induction of hyphal growth in the hxk1Δ and h-d mutants. Cells were grown in synthetic medium containing 50 mm galactose plus the indicated concentration of GlcNAc for 2 h at 37 °C. The hxk1Δ and h-d strains were stimulated by GlcNAc at all concentrations tested to form elongated hyphae similar to the wild type. Strains used were the wild-type control (DIC185), hxk1Δ (AG736), and h-d (AG738) mutant. Bar, 10 μm.
A previous study reported that the hxk1Δ, nag1Δ, and dac1Δ mutants were defective in forming hyphae in response to serum (11). However, we found no defects in the ability of these mutants or the h-d mutant to be induced by serum (data not shown). Another study reported a defect in GlcNAc-induced hyphal formation for a mutant in which NAG1, the promoter for DAC1, and a portion of HXK1 were deleted (28). However, it appears that this mutant was primarily defective in forming new growth on GlcNAc medium, perhaps because of an inhibitory effect or the absence of a suitable carbon and energy source.
GlcNAc Catabolism Is Not Needed to Induce Expression of the NGT1, HXK1, NAG1, and DAC1 Genes
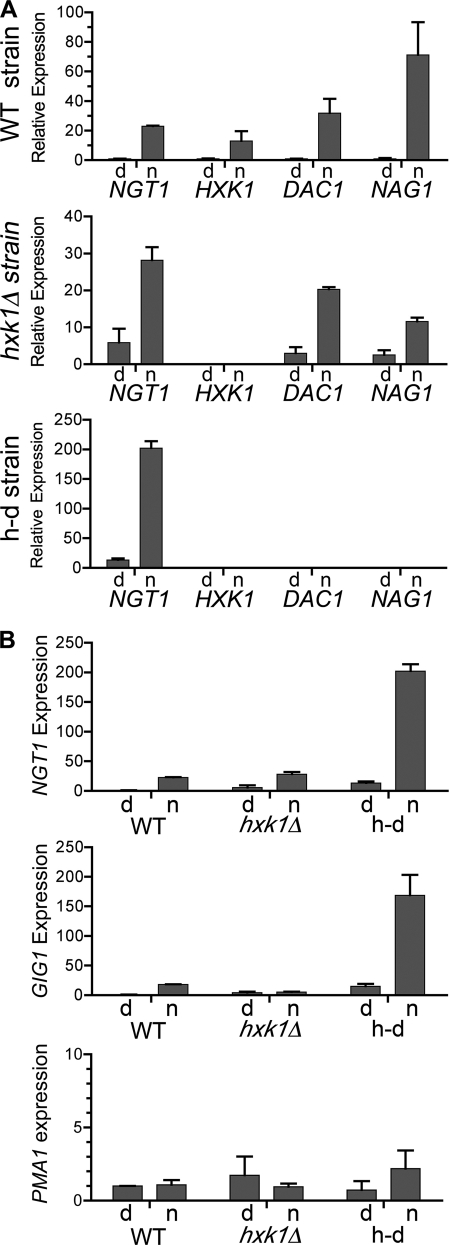
The ability of GlcNAc to activate the separate signaling pathway that induces expression of the catabolic genes was analyzed using qPCR. In dextrose medium, wild-type cells showed a low level of expression of the GlcNAc transporter NGT1 that was induced about 23-fold when cells were switched to GlcNAc medium (Fig. 5A). The basal level of NGT1 expression was higher in both the hxk1Δ and h-d mutants, analogous to what was observed for hyphal growth. NGT1 was induced by GlcNAc in the hxk1Δ mutant to about the same level as the wild type, but because of the higher basal level the relative induction was 4.8-fold. Interestingly, despite the higher basal level of NGT1 expression in the h-d mutant, GlcNAc still induced NGT1 expression 15.4-fold. Thus, the overall GlcNAc-induced level of NGT1 expression was about 8.8-fold higher in the h-d mutant than in the wild type. Similar results were observed for the expression of the NAG1 and DAC1 genes in the hxk1Δ mutant. As expected, we failed to detect expression of HXK1 in the hxk1Δ mutant or expression of the HXK1, NAG1 or DAC1 genes in the h-d mutant (Fig. 5A).
FIGURE 5.
GlcNAc induces the catabolic genes in the mutant cells. A, expression of the GlcNAc transporter (NGT1), GlcNAc-6-PO4 deacetylase (DAC1), and glucosamine-6-PO4 deaminase (NAG1) genes. The graphs for the wild-type control, hxk1Δ, and h-d strains are indicated on the left. Cells were grown in medium containing 50 mm dextrose (d) or 50 mm N-acetylglucosamine (n) for 2 h at 37 °C. B, relative expression of NGT1, the GlcNAc-induced gene GIG1, and PMA1. The level of expression for each gene was normalized to that for the wild-type control strain grown in dextrose. Note that PMA1 is a control gene that is not regulated by GlcNAc. The mRNA levels were analyzed by qPCR as described under “Experimental Procedures.” The results represent the mean ± S.D. of four independent assays each carried out in triplicate. The data include analyses of two completely independent mutant isolates and RNA preparations for each strain.
To determine whether the elevated NGT1 expression observed in the h-d mutant was seen for other genes, we examined the expression of the GlcNAc-induced gene GIG1, which is thought to play a role in GlcNAc metabolism (8). Both the hxk1Δ and the h-d mutants showed higher basal levels of GIG1 relative to the wild type (Fig. 5B). Although GIG1 was poorly induced in the hxk1Δ mutant, it was highly induced in the h-d mutant. Similar to the relative levels of NGT1 expression after GlcNAc induction, GIG1 was expressed 7.9-fold higher in the h-d mutant than in the wild type. Additional control studies showed the expression of PMA1, the plasma membrane ATPase, was not significantly altered by GlcNAc induction in the h-d mutant (Fig. 5B). Thus, higher levels of GlcNAc-induced gene expression are seen in the h-d mutant.
Ngt1-GFP Is Induced by GlcNAc in the hxk1Δ and h-d Mutants
The results for GlcNAc-regulated gene expression were analyzed further by examining an NGT1-GFP reporter gene that was introduced into the wild-type and mutant cells. Control studies showed that wild-type cells grown in dextrose displayed no detectable Ngt1-GFP fluorescence, but Ngt1-GFP was detectable in the plasma membrane of cells grown in GlcNAc for 2 h (Fig. 6, A and B). Some GFP signal was also detected in the vacuole, presumably because of endocytosis. A high basal level of Ngt1-GFP was detected for a subset of the hxk1Δ and h-d mutant cells when grown in dextrose, consistent with the elevated expression of NGT1 detected by qPCR. Interestingly, the fraction of cells producing detectable Ngt1-GFP increased with increasing cell density for the mutants but not for the wild type. Approximately 5% of the nag1Δ and dac1Δ mutants showed elevated levels of Ngt1-GFP when grown to a low cell density (∼106 cells/ml; Fig. 6A), but this increased to essentially 100% of the cells when they were grown to saturation (∼108 cells/ml; Fig. 6C). GlcNAc was able to highly induce Ngt1-GFP in the hxk1Δ and h-d mutants, similar to the induction of the NGT1 mRNA detected by qPCR (Fig. 6B). Comparison of signal intensity in the digital images showed that Ngt1-GFP was induced to ∼4-fold higher in the hxk1Δ mutant and ∼8-fold higher in the h-d mutant relative to the level detected in the wild-type cells. Thus, the Ngt1-GFP results confirm the trends seen for the expression of GlcNAc-induced genes in the hxk1Δ and h-d mutants.
FIGURE 6.
GlcNAc induction of Ngt1-GFP in hxk1Δ and h-d mutants. Analysis of Ngt1-GFP production in cells grown overnight in dextrose medium to early log phase (low cell density) and then switched to medium containing 50 mm dextrose (A) or 50 mm GlcNAc (B) for 2 h. C, cells grown overnight to saturation (High cell density) in medium containing 50 mm dextrose. The cells were then photographed using a fluorescence microscope to detect GFP. A gene fusion between the GlcNAc transporter and the green fluorescent protein (NGT1-GFP) was introduced into cells to create a wild-type control strain (SN787), an hxk1Δ mutant (SN791), and an h-d mutant (SN794). Bar, 5 μm.
GlcNAc-induced Expression of Galactose-regulated Genes
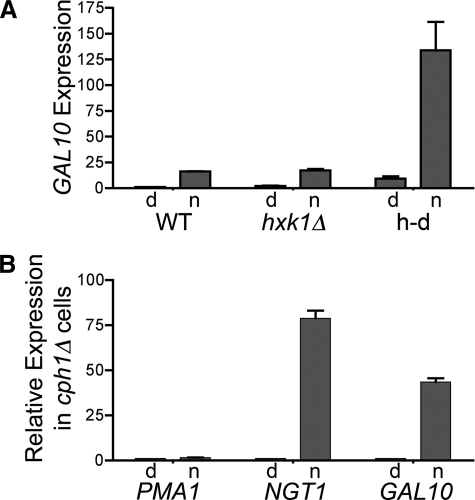
Microarray analysis of gene expression in C. albicans unexpectedly revealed that GlcNAc stimulated the expression of a subset of galactose-regulated genes, GAL1, GAL7, and GAL10 (8). GlcNAc catabolism is not needed for this induction, because qPCR analysis showed that GlcNAc induced GAL10 to similar levels in the wild-type and hxk1Δ cells, and GAL10 was hyper-induced in the h-d mutant (Fig. 7A). We previously speculated that the GAL genes are induced by activation of Cph1 in response to GlcNAc, because Cph1 is a transcription factor involved in inducing both hyphal genes and galactose-regulated genes (8, 29, 30). This prediction was also based on the fact that GAL1, -7, and -10 are the only genes in the C. albicans genome that contain typical Cph1-binding sites in their promoters (29). To test this idea, we examined the ability of GlcNAc to induce gene expression in two independently constructed Cph1 mutants (31, 32), both of which were confirmed to lack CPH1 by PCR methods. Surprisingly, GlcNAc efficiently induced GAL10 in the cph1Δ mutants (Fig. 7B). Thus, these results demonstrate that the induction of GAL10 does not require GlcNAc stimulation of Cph1. They also demonstrate that Cph1 is not needed to induce NGT1, consistent with the ability of cph1Δ mutant cells to grow well on GlcNAc medium (see below).
FIGURE 7.
GlcNAc induction of GAL10 is independent of CPH1. A, relative GAL10 mRNA levels were analyzed by qPCR for the wild-type strain (DIC185), hxk1Δ (AG736), and h-d (AG738). B, PMA1, NGT1, and GAL10 expression levels in cph1Δ mutant cells. The data represent the mean ± S.D. for RNA preparations obtained from two different cph1Δ strains (strains 180 and CJN305). Cells were grown in synthetic medium containing dextrose (d) or N-acetylglucosamine (n), and then qPCR was performed as described under “Experimental Procedures.”
Mkc1 and Cek1 Are Not Needed for GlcNAc Signaling
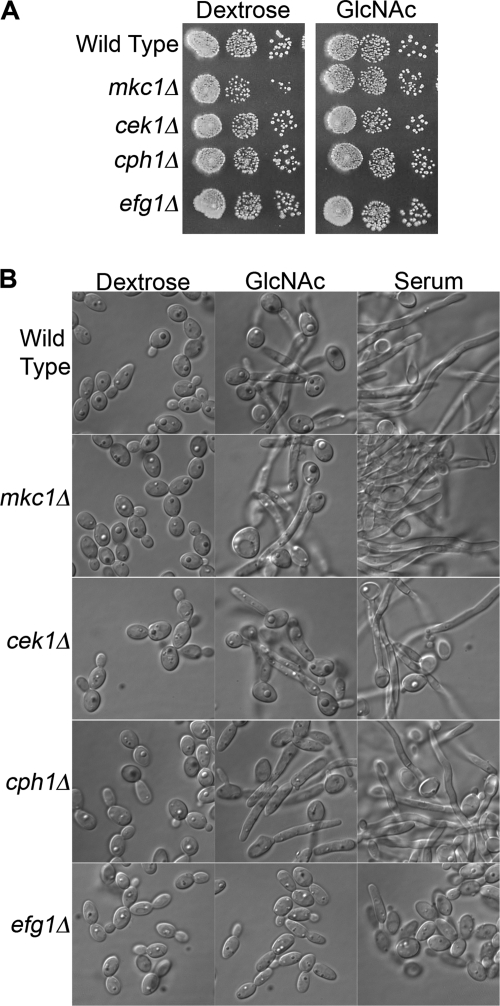
The Mkc1 and Cek1 MAPKs regulate cell-signaling pathways in C. albicans and were therefore tested for a role in GlcNAc signaling. However, mutant cells lacking these kinases grew well on GlcNAc medium and could be induced to form hyphae in response to GlcNAc (Fig. 8). A cph1Δ mutant also grew well on GlcNAc, consistent with its ability to induce GlcNAc catabolic genes (Fig. 7). However, as expected from previous studies (33), this mutant displayed a partial defect in forming hyphae in response to both GlcNAc and serum. An efg1 mutant that lacks a transcription factor activated by cAMP signaling showed a stronger defect in hyphal induction, as expected from previous studies (33), but it still grew well on GlcNAc medium. Thus, these genes do not play an essential role in GlcNAc signaling.
FIGURE 8.
Mck1, Cek1, Cph1, and Efg1 are not required for GlcNAc catabolism or GlcNAc induction of hyphal growth. A, dilutions of a wild-type control and the indicated mutants strains were spotted onto solid agar media plates containing 50 mm of the indicated sugar and then incubated at 37 °C. B, cells were grown overnight to log phase and then incubated in medium containing 50 mm dextrose, 50 mm GlcNAc, or 50 mm dextrose plus 10% serum at 37 °C for 2 h. The wild-type control (SN250), mkc1 (1112), cek1 (544), and cph1Δ (180) strains were described by Noble et al. (32), and the efg1 strain (060) was described by Homann et al. (40).
DISCUSSION
Nonphosphorylated GlcNAc Activates Cell Signaling in C. albicans
GlcNAc plays important roles in cell structure and signaling in a wide range of organisms from bacteria to man. However, the mechanisms of GlcNAc signaling are not well understood. Previous studies of the human fungal pathogen C. albicans indicated that GlcNAc must be transported into the cells to stimulate hyphal morphogenesis and gene expression (12). Therefore, mutant cells lacking the GlcNAc catabolic genes were analyzed to determine whether GlcNAc or a subsequent breakdown product was important for signaling. Interestingly, hxk1Δ cells lacking the GlcNAc kinase or the h-d mutant that lacks all three catabolic genes (hxk1Δ nag1Δ dac1Δ) could be induced efficiently to undergo hyphal morphogenesis (Fig. 4) and turn on expression of GlcNAc-regulated genes (Fig. 5). These results indicate that GlcNAc does not have to be phosphorylated and catabolized to activate signaling. The phenotypes of the hxk1Δ and h-d mutants also indicate that GlcNAc does not have to enter the anabolic pathways to induce signaling. Phosphorylation of exogenous GlcNAc by Hxk1 is required to create GlcNAc-6-PO4, which is then converted to UDP-GlcNAc for use as a substrate in the anabolic pathways that form chitin, N-linked glycosylation, and glycosylphosphatidylinositol anchors on proteins (16). Thus, the results indicate that GlcNAc metabolism is not required for it to activate signaling.
The discovery that nonphosphorylated GlcNAc from exogenous sources can activate signaling in C. albicans therefore identifies a novel role for GlcNAc in cell signaling that may be used to regulate intercellular signaling in other organisms, including humans. GlcNAc signaling in C. albicans does not require O-GlcNAc transferase as in humans, because this enzyme uses UDP-GlcNAc as a substrate for protein modification (5). This conclusion is also supported by the lack of a detectable O-GlcNAc transferase gene in C. albicans. Sensing nonphosphorylated GlcNAc would also have the advantage of allowing cells to distinguish exogenous nonphosphorylated GlcNAc that is transported into the cell from the GlcNAc-6-PO4 that is synthesized within the cell. Intracellular GlcNAc synthesis involves conversion of fructose-6-PO4 to glucosamine-6-PO4 and then to GlcNAc-6-PO4 (16). Detecting nonphosphorylated GlcNAc is also expected to provide increased sensitivity for sensing extracellular GlcNAc, because there is a high rate of flux of GlcNAc-6-PO4 through the anabolic pathways in cells to keep up with the demands for the structural roles of GlcNAc. The ability of C. albicans to sense nonphosphorylated GlcNAc is also supported by the observation that glucosamine does not induce hyphal morphogenesis. Glucosamine is expected to be phosphorylated to glucosamine-6-PO4 and then converted to GlcNAc-6-PO4 or catabolized to fructose-6-PO4 (16).
Other sugar signaling pathways also sense nonphosphorylated sugars. For example, S. cerevisiae Gal3, an enzymatically inactive paralog of the galactose kinase Gal1, binds nonphosphorylated galactose and then migrates to the nucleus to stimulate Gal4 to induce gene expression by removing the inhibitory Gal80 factor (13). Similarly, the S. cerevisiae glucose kinase Hxk2 binds glucose and then effects changes in transcription in the nucleus (34). Although these sugar kinases have been directly implicated in gene regulation stimulated by other carbohydrates, analysis of the hxk1Δ and h-d mutants indicates that the GlcNAc kinase Hxk1 is not needed for induction of GlcNAc-regulated genes in C. albicans (Fig. 5).
Inhibitory Effects of GlcNAc on nag1Δ and dac1Δ Mutants
The nag1Δ and dac1Δ mutants quickly ceased growth in the presence of GlcNAc, even when another sugar was available for nutrition (Fig. 2). The inability of these mutants to deacetylate and deaminate GlcNAc-6-PO4 suggests that the resulting excess of GlcNAc-6-PO4 is toxic to cells. Similar deleterious effects that are attributable to excess GlcNAc-6-PO4 have been observed in other mutant organisms. For example, GlcNAc inhibited growth of S. cerevisiae cells that express C. albicans HXK1 but not cells that co-expressed HXK1 with NAG1 and DAC1 (15). GlcNAc also inhibited the growth of mutant E. coli cells that lack GlcNAc deacetylase or deaminase activity (25). Growth was quickly restored by adding exogenous uridine (26), suggesting that conversion of excess GlcNAc-6-PO4 to UDP-GlcNAc may have depleted uridine levels. In contrast, extra uridine did not quickly rescue the C. albicans nag1Δ and dac1Δ mutants from inhibition by GlcNAc (Fig. 2). Another example of this type of inhibition was reported for Leishmania, in which the GlcNAc sensitivity of a gnd mutant lacking glucosamine deaminase was attributed to depletion of ATP (35). The inhibitory effect of GlcNAc on the Leishmania gnd mutant appears to be distinct from that seen for the C. albicans mutants because it could be overcome by adding alternative carbon sources, such as glycerol.
It is interesting to speculate that the inhibitory effects of GlcNAc on nag1Δ and dac1Δ mutants could have played a role in loss of the GlcNAc catabolic genes in S. cerevisiae, S. pombe, and related species. The HXK1, NAG1, and DAC1 genes are in a cluster termed the NAG regulon in C. albicans that is also conserved in other species (36). This clustering of genes might facilitate their simultaneous loss in the case of an adverse mutation of NAG1 or DAC1.
Activation of GAL Genes
GlcNAc induction of a subset of the galactose-regulated genes was unexpected, because there is no obvious overlap between galactose and GlcNAc catabolism (8). Furthermore, GlcNAc catabolism was not required for GAL10 to be induced in the hxk1Δ and h-d mutants (Fig. 7), and galactose does not induce GlcNAc catabolic genes (29, 37). We previously proposed that GlcNAc induction of a subset of GAL genes might be due to activation of Cph1 (8), a transcription factor that is implicated in the regulation of both hyphal- and galactose-stimulated genes (29, 30). The GAL1, GAL7, and GAL10 genes that were induced by GlcNAc were reported to be the only C. albicans genes that contain a putative Cph1 recognition motif in their upstream regulatory regions (29). Other galactose-stimulated genes whose promoters lack this recognition site were not induced by GlcNAc (8, 29). Despite this suggestive evidence, two independently constructed cph1Δ mutants could be stimulated by GlcNAc to induce GAL10 efficiently (Fig. 7), demonstrating that Cph1 is not required for this effect. Nonetheless, the induction of GAL1, GAL7, and GAL10 by GlcNAc may be physiologically significant as it has been suggested that GAL10 functions in cell wall integrity even in the absence of galactose (38).
GlcNAc and Virulence
Previous studies observed that C. albicans hxk1Δ, dac1Δ, and nag1Δ mutants were defective for pathogenesis in a mouse model of systemic infection and concluded that GlcNAc catabolism was important for virulence (11, 28). However, the relationship between GlcNAc catabolism and virulence is unclear. One reason is that, unfortunately, the gene deletion methods used in these studies are now known to require additional controls before they can be reliably used to assess virulence (23, 24). Furthermore, the new phenotypes uncovered for the GlcNAc catabolic mutants could affect pathogenesis independently of the ability to use GlcNAc as a nutrient source. For example, the hxk1Δ and h-d mutants display a higher basal level of filamentous growth (Fig. 3) that could decrease virulence similar to the C. albicans mutants that are locked into the hyphal mode of growth (39). Also, the strong inhibitory effects of GlcNAc on the proliferation of the nag1Δ and dac1Δ mutants could impact growth in vivo in some niches (Fig. 2). It is unclear whether C. albicans cells in a systemic infection typically encounter much GlcNAc, because serum is low in GlcNAc and high in glucose. However, GlcNAc catabolism is likely to be important for commensal growth of C. albicans on mucosal surfaces. The extracellular matrix glycosaminoglycans that line the mucosa are rich in GlcNAc. GlcNAc is also expected to be present in the gastrointestinal tract because of remodeling of the bacterial peptidoglycan. Thus, GlcNAc plays a multifaceted role in cell signaling and structure of C. albicans during commensal growth and pathogenesis.
Acknowledgments
We thank the members of our laboratory for their helpful advice and comments on the manuscript. We also thank Dr. Aaron Mitchell for providing strains and the Fungal Genetics Stock Center for providing strains deposited by Dr. Suzanne Noble.
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 GM087368 from USPHS (to J. B. K.).
- qRT-PCR
- quantitative RT-PCR.
REFERENCES
- 1. Barnhart M. M., Lynem J., Chapman M. R. (2006) J. Bacteriol. 188, 5212–5219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Simonetti N., Strippoli V., Cassone A. (1974) Nature 250, 344–346 [DOI] [PubMed] [Google Scholar]
- 3. Pérez-Campo F. M., Domínguez A. (2001) Curr. Microbiol. 43, 429–433 [DOI] [PubMed] [Google Scholar]
- 4. Hart G. W., Housley M. P., Slawson C. (2007) Nature 446, 1017–1022 [DOI] [PubMed] [Google Scholar]
- 5. Slawson C., Copeland R. J., Hart G. W. (2010) Trends Biochem. Sci. 35, 547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Castilla R., Passeron S., Cantore M. L. (1998) Cell. Signal. 10, 713–719 [DOI] [PubMed] [Google Scholar]
- 7. Leberer E., Harcus D., Dignard D., Johnson L., Ushinsky S., Thomas D. Y., Schröppel K. (2001) Mol. Microbiol. 42, 673–687 [DOI] [PubMed] [Google Scholar]
- 8. Gunasekera A., Alvarez F. J., Douglas L. M., Wang H. X., Rosebrock A. P., Konopka J. B. (2010) Eukaryot. Cell 9, 1476–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang G., Yi S., Sahni N., Daniels K. J., Srikantha T., Soll D. R. (2010) PLoS Pathog. 6, e1000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kumar M. J., Jamaluddin M. S., Natarajan K., Kaur D., Datta A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 14218–14223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yamada-Okabe T., Sakamori Y., Mio T., Yamada-Okabe H. (2001) Eur. J. Biochem. 268, 2498–2505 [DOI] [PubMed] [Google Scholar]
- 12. Alvarez F. J., Konopka J. B. (2007) Mol. Biol. Cell 18, 965–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sellick C. A., Reece R. J. (2005) Trends Biochem. Sci. 30, 405–412 [DOI] [PubMed] [Google Scholar]
- 14. Douglas H. C., Condie F. (1954) J. Bacteriol. 68, 662–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wendland J., Schaub Y., Walther A. (2009) Appl. Environ. Microbiol. 75, 5840–5845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Milewski S., Gabriel I., Olchowy J. (2006) Yeast 23, 1–14 [DOI] [PubMed] [Google Scholar]
- 17. Sherman F. (1991) Methods Enzymol. 194, 3–21 [DOI] [PubMed] [Google Scholar]
- 18. Wilson R. B., Davis D., Mitchell A. P. (1999) J. Bacteriol. 181, 1868–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wilson R. B., Davis D., Enloe B. M., Mitchell A. P. (2000) Yeast 16, 65–70 [DOI] [PubMed] [Google Scholar]
- 20. Park H., Myers C. L., Sheppard D. C., Phan Q. T., Sanchez A. A., Edwards J., Filler S. G. (2005) Cell. Microbiol. 7, 499–510 [DOI] [PubMed] [Google Scholar]
- 21. Zhang C., Konopka J. B. (2010) Eukaryot. Cell 9, 224–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Natarajan K., Datta A. (1993) J. Biol. Chem. 268, 9206–92014 [PubMed] [Google Scholar]
- 23. Lay J., Henry L. K., Clifford J., Koltin Y., Bulawa C. E., Becker J. M. (1998) Infect. Immun. 66, 5301–5306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen X., Magee B. B., Dawson D., Magee P. T., Kumamoto C. A. (2004) Mol. Microbiol. 51, 551–565 [DOI] [PubMed] [Google Scholar]
- 25. White R. J. (1968) Biochem. J. 106, 847–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bernheim N. J., Dobrogosz W. J. (1970) J. Bacteriol. 101, 384–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wendland J., Hellwig D., Walther A., Sickinger S., Shadkchan Y., Martin R., Bauer J., Osherov N., Tretiakov A., Saluz H. P. (2006) J. Basic Microbiol. 46, 513–523 [DOI] [PubMed] [Google Scholar]
- 28. Singh P., Ghosh S., Datta A. (2001) Infect. Immun. 69, 7898–7903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martchenko M., Levitin A., Hogues H., Nantel A., Whiteway M. (2007) Curr. Biol. 17, 1007–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu H., Köhler J., Fink G. R. (1994) Science 266, 1723–1726 [DOI] [PubMed] [Google Scholar]
- 31. Nobile C. J., Mitchell A. P. (2005) Curr. Biol. 15, 1150–1155 [DOI] [PubMed] [Google Scholar]
- 32. Noble S. M., French S., Kohn L. A., Chen V., Johnson A. D. (2010) Nat. Genet. 42, 590–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lo H. J., Köhler J. R., DiDomenico B., Loebenberg D., Cacciapuoti A., Fink G. R. (1997) Cell 90, 939–949 [DOI] [PubMed] [Google Scholar]
- 34. Gancedo J. M. (2008) FEMS Microbiol. Rev. 32, 673–704 [DOI] [PubMed] [Google Scholar]
- 35. Naderer T., Heng J., McConville M. J. (2010) PLoS Pathog. 6, e1001245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fitzpatrick D. A., O'Gaora P., Byrne K. P., Butler G. (2010) BMC Genomics 11, 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brown V., Sabina J., Johnston M. (2009) Curr. Biol. 19, 436–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Singh V., Satheesh S. V., Raghavendra M. L., Sadhale P. P. (2007) Fungal. Genet. Biol. 44, 563–574 [DOI] [PubMed] [Google Scholar]
- 39. Whiteway M., Oberholzer U. (2004) Curr. Opin. Microbiol. 7, 350–357 [DOI] [PubMed] [Google Scholar]
- 40. Homann O. R., Dea J., Noble S. M., Johnson A. D. (2009) PLoS Genet. 5, e1000783. [DOI] [PMC free article] [PubMed] [Google Scholar]