Abstract
CYP24A1 is a mitochondrial cytochrome P450 (CYP) that catabolizes 1α,25-dihydroxyvitamin D3 (1α,25-(OH)2D3) to different products: calcitroic acid or 1α,25-(OH)2D3-26,23-lactone via multistep pathways commencing with C24 and C23 hydroxylation, respectively. Despite the ability of CYP24A1 to catabolize a wide range of 25-hydroxylated analogs including 25-hydroxyvitamin D3, the enzyme is unable to metabolize the synthetic prodrug, 1α-hydroxyvitamin D3 (1α-OH-D3), presumably because it lacks a C25-hydroxyl. In the current study we show that a single V391L amino acid substitution in the β3a-strand of human CYP24A1 converts this enzyme from a catabolic 1α,25-(OH)2D3-24-hydroxylase into an anabolic 1α-OH-D3-25-hydroxylase, thereby forming the hormone, 1α,25-(OH)2D3. Furthermore, because the mutant enzyme retains its basal ability to catabolize 1α,25-(OH)2D3 via C24 hydroxylation, it can also make calcitroic acid. Previous work has shown that an A326G mutation is responsible for the regioselectivity differences observed between human (primarily C24-hydroxylating) and opossum (C23-hydroxylating) CYP24A1. When the V391L and A326G mutations were combined (V391L/A326G), the mutant enzyme continued to form 1α,25-(OH)2D3 from 1α-OH-D3, but this initial product was diverted via the C23 hydroxylation pathway into the 26,23-lactone. The relative position of Val-391 in the β3a-strand of a homology model and the crystal structure of rat CYP24A1 is consistent with hydrophobic contact of Val-391 and the substrate side chain near C21. We interpret that the substrate specificity of V391L-modified human CYP24A1 toward 1α-OH-D3 is enabled by an altered contact with the substrate side chain that optimally positions C25 of the 1α-OH-D3 above the heme for hydroxylation.
Keywords: Drug Metabolism, Metabolism, Mutant, Protein Structure, Vitamin D, CYP24A1, Substrate Docking, Vitamin D Analog
Introduction
The activation of vitamin D3 (1) is accomplished by two sequential cytochrome P450-mediated steps (2) to give the hormonal form, 1α,25-dihydroxyvitamin D3 (1α,25-(OH)2D3),2 which transactivates vitamin D-dependent genes through a nuclear vitamin D receptor (3). The first step in activation of the vitamin D molecule is hepatic 25-hydroxylation, mediated by the microsomal enzyme, CYP2R1 (4, 5), although other P450s, such as the mitochondrial CYP27A1 (6) and the microsomal CYP3A4, may also play a role (7). The second step in activation, 1α-hydroxylation, is carried out by CYP27B1 and occurs mainly in the kidney and to a lesser extent in extra-renal tissues (8, 9). Synthetic 1α-hydroxylated prodrug forms of vitamin D are also activated in the liver and bypass the kidney entirely (10). In vitro, hepatic CYP27A1 primarily 25-hydroxylates 1α-OH-D3 but also yields lesser amounts of 24- and 26-hydroxylated products (6). Surprisingly, the 25-hydroxylation of 1α-OH-D2 by CYP27A1 is minimal, and the only efficient 25-hydroxylation of the vitamin D2 side chain is mediated by CYP2R1. The vitamin D catabolic enzyme, CYP24A1, exhibits minimal side-chain metabolic activities toward the 1α-OH-D3 prodrug but instead demonstrates significant 24-hydroxylase activity toward the 1α-OH-D2 prodrug (11), yielding a metabolite, 1α,24-(OH)2D2, with potent anti-proliferative activity and reduced calcemic activity. On the other hand, there is little evidence that CYP27A1, CYP27B1, or CYP2R1 can degrade vitamin D compounds to inactive products in vivo. Thus, the catabolic function appears to be the exclusive domain of CYP24A1 in the kidney and other vitamin D target cells.
The catabolism of 1α,25-(OH)2D3 by CYP24A1 occurs through multiple, sequential hydroxylation/oxidation reactions targeting specific carbon atoms in the vitamin D side chain. These multicatalytic activities of CYP24A1 (12–15) separate into two regioselective hydroxylation/oxidation pathways involving an initial attack at C24 to yield a side-chain truncated product, calcitroic acid (14, 15), or at C23 to give 1α,25-(OH)2D3-26,23-lactone (16). Several laboratories have shown that the proportion of these catabolic products is species-dependent with calcitroic acid production predominating in rodents (17), 26,23-lactone predominating in opossum and guinea pig (18, 19), and a mixture of the two products being formed in humans (2). Previously, we have shown that substrate contact with a species-specific residue in the I-helix adjacent to the heme group of the cytochrome P450 exerts considerable control over pathway selection, with Ala-326 directing substrate into the C24 pathway in human and Gly-326 conducting 23-hydroxylation in opossum (20). As a result, we have shown that catabolic pathway selection can be engineered into hCYP24A1. Other substrate contact residues having a lesser influence on regioselectivity in rat include Thr-416 (Met-416; human) and Ile-500 (Thr-500; human) (17).
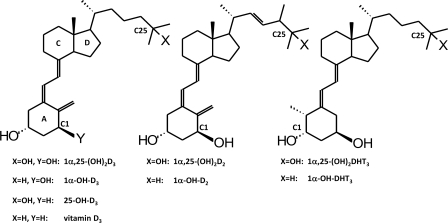
These regioselective-determining residues and other contact residues in the active site of CYP24A1 allow precise positioning the vitamin D side chain over a heme catalytic center by a combination of ionic and hydrophobic interactions. Thus, the failure of the enzyme to metabolize certain substrates may result from an inability to correctly position the side chain rather than the inability to bind the substrate. These two mechanisms of substrate specificity help the enzyme discriminate between a potential substrate and products or non-substrates. Thus, the objectives of this study were to 1) examine if selected amino acid residues in human CYP24A1 enzyme affect substrate specificity and the regioselectivity of side-chain metabolism by studying the catalytic properties of wild-type and mutant forms of hCYP24A1 and 2) examine if wild-type and mutant forms of CYP24A1 metabolize selected vitamin D analogs in which the ring structure or side chain had been altered differently. Both an A-ring analog dihydrotachysterol (DHT3) and vitamin D2 and D3 compounds with and without 25-hydroxyl group were used (Fig. 1).
FIGURE 1.
Structures of vitamin D analogs used as substrates in the current study.
We report here the generation of de novo 25-hydroxylase activity toward 1α-OH-D3 and increases in 26-hydroxylase activity toward the 1α-OH-D2 by making the V391L modification in hCYP24A1, whereas the subsequent catabolism of the 1α,25-(OH)2D3 product from 1α-OH-D3 can be engineered to proceed to either calcitroic acid or 1α,25-(OH)2D3-26,23-lactone by a A326G substitution.
EXPERIMENTAL PROCEDURES
Preparation of Plasmid Constructs
Full-length hCYP24A1, containing its mitochondrial targeting sequence described previously (21), was subcloned as an NheI-XhoI fragment into pcDNA5/FRT (Invitrogen), which was modified to include the C-terminal V5-His epitope from pcDNA3.1 V5-HisB. The V391L mutation was introduced into wild-type hCYP24A1 and A326G constructs (19, 20) using QuikChange (Stratagene Corp., La Jolla, CA) according to the manufacturer's protocol and an oligonucleotide pair based on 5′ GAG GCT TAC GCC GGG TGT ACC ATT TAC AAC TCG G (Cortec, Kingston, ON, Canada). Full-length hCYP27A1 (8) was subcloned as an NheI-XhoI fragment into the pcDNA5/FRT vector described above. Mutations were confirmed by sequencing (Cortec), and the plasmids used for transfection were purified using Qiagen Plasmid Maxi kit (Qiagen Inc., Mississauga, ON, Canada).
Preparation of Stably Transfected Cell Lines
The Flp-In transfection system (Invitrogen) (22) was used to stably transfect hCYP24A1 and hCYP27A1 constructs into Chinese Hamster lung fibroblast cells (V79-4; ATCC CCL-93). The vector pFRT/lacZeo, containing an Flp recombinase target site was transfected into V79-4 cells using Lipofectamine and Plus reagents (Invitrogen). The transfected cells were selected in 375 μg/ml zeocin, and the zeocin-resistant clones were subjected to screening by Southern blotting and β-galactosidase assays to determine the number of integration sites and estimate the transcriptional activity at the site(s) of integration. Clones containing a single copy of the Flp recombinase target site possessing a relatively high level of β-galactosidase activity were selected as the host cell line. The host cell line was co-transfected with pOG44 encoding the Flp recombinase, and single plasmid constructs containing hCYP24A1, hCYP24A1 mutants A326G, V391L, or V391L/A326G, or hCYP27A1 using a 9:1 ratio of pOG44 and CYP construct. Cells exhibiting site-specific recombination of the expression plasmid were selected in medium containing 93 μg/ml hydromycin B.
Western Blotting
Approximately 2.4 × 106 cells were lysed on ice in lysis buffer (23) containing protease inhibitor mixture (Sigma). A 100-μg amount of protein from each lysate was separated by 10% SDS-PAGE and transferred to a nitrocellulose membrane. V5 epitope-tagged CYP24A1 proteins were detected by blocking (1 h) and incubating the membrane overnight in blocking buffer containing a 1:5000 dilution of mouse monoclonal anti-V5 horseradish peroxidase (HRP) conjugate. GAPDH was detected by incubating with 1:4000 primary monoclonal anti-GAPDH antibody overnight followed by a 1-h incubation with secondary anti-mouse HRP conjugate (GE Healthcare). Proteins were detected using ECL Western Lightning reagents (GE Healthcare). All blocking and incubation steps were carried out in Tris-buffered saline supplemented with Tween 20 and 5% nonfat dry milk powder.
Analysis of CYP24A1 Activity
Transfected cell lines were maintained in a humidified atmosphere of 5% CO2 in air using a Dulbecco's Modified Eagle's Medium (Invitrogen) supplemented with 10% (v/v) fetal bovine serum (HyClone, Logan, UT), 1% antibiotic/antimycotic (Invitrogen), 2 g/liter glucose, and 93 μg/ml hydromycin B (Invitrogen). Cells were plated and maintained at 2.4 × 106 cells/P60 cell culture dishes and allowed to recover overnight. Cells were incubated for 36 h in 2 ml of medium without glucose, where FCS was substituted with 1% w/v bovine serum albumin, supplemented with 9 μm 1α,25-(OH)2D3, 1α,25-(OH)2D2, 1α,25-(OH)2DHT3, 25-OH-D3, 1α-OH-D3, 1α-OH-D2, 1α-OH-DHT3, or vitamin D3 (Fig. 1). Media and cells were lipid-extracted and analyzed by HPLC as previously described (20, 21).
Species Ortholog and Rotamer Frequencies
Multisequence alignments consisting of fragments of 53 species orthologs of CYP24A1 were retrieved from GenBankTM, Ensembl and Pre!Ensembl databases and aligned using Genedoc according to observed secondary structure. The fragmentary nature of the sequences gave 45–50 residues at any given position from which frequencies of the most predominant amino acid were calculated as percents. Four levels of conservation were computed and are depicted in supplemental Figs. S1A. rCYP24A1 crystal structures (PDB codes 3k9v and 3k9y) were used to identify side-chain dihedral angle P1 rotamers as m-, p-, and t-type (24) and residues displaying rotamer variation (supplemental Figs. S1B). Side-chain replacements in the crystal and model structures were performed using backbone-independent rotamer selection if they were in accordance with rotamers cataloged in 25 selected P450 crystal structures.
Homology Modeling and Structure Alignment
The construction of the hCYP24A1 model was described previously (20, 21). The model and the A and B chains of the rat (r) CYP24A1 were structurally aligned using a triangulation method based on the heme methyl carbons (20). The methyl carbons in each structure were used to geometrically calculate three anchor points (CMA′, CMB′, and CMD′) at a distance of 30 Å from the proximal face of the heme. All atoms in the CHAPS B chain (including the waters), Cymol-5-containing chains A ad B and model were triangulated off corresponding anchors, and the distances were used to numerically translate coordinates onto the CHAPS chain A structure using Mathematica (Wolfram Research, Champaign, IL). The resulting superimposed structures were used to overlap the cavity mesh surfaces seen in supplemental Figs. S1C. The root mean squared deviations between the model, CHAPS chain A, and Cymal-5 chain A were calculated as the square root of the average square of the distances between α-carbons in the aligned structures (supplemental Figs. S2).
Substrate Docking
1α-OH-D2 and 1α-OH-D3 were docked into the model before the release of the crystal structures of rCYP24A1. Regioselective substrate orientations corresponding to C23-C26 hydroxylation were geometry-minimized and subjected to constrained molecular dynamics in a water-free structure using the Discover module in Insight II (Accelrys, San Diego, CA) by triangulating the corresponding atom directly off the heme methyls as previously (20) and the hydroxylation target hydrogens (CMA = 6.00; CMB = 6.87; CMD = 6.27) corresponding to C23-proS and C24-proR hydroxylation. The substrate binding cavity in the model was directed along a pw2a trajectory substrate access channel (25). The substrate A-ring conformation featured axial 3β- and equatorial 1α-hydroxyls with the substrate C18 methyl facing the β-5 hairpin. A second set of regioselectively docked structures was collected with the substrate C18 methyl facing away from the β-5 hairpin. All docked substrates in the model, the CHAPS ligands (3k9v.pdb, chain B), and the Cymal-5 ligands (PDB code 3k9y, chain A) were numerically translated onto the CHAPS chain A structure by the triangulation method.
RESULTS
Alignments of CYP24A1 Orthologs from Different Species and Preliminary Analysis of the Homology Models of hCYP24A1 and Crystal Structure of rCYP24A1
An alignment of species orthologs of CYP24A1 (supplemental Figs. S3) permitted examination of conservation and mapping of residues using an homology model for hCYP24A1 with its docked 1α,25-(OH)2D3 substrate (20, 21) and the crystal structure of rCYP24A1 (26). About 43% of the residues of the entire protein are >95% conserved, especially in the secondary structures forming the core of the enzyme and the lining of the active-site cleft (supplemental Figs. S1A). However, certain key active-site residues are less-well conserved, including Ala-326 (67% alanine; 33% glycine), Leu-148 (86% conserved), and Thr-395 (80% conserved). The A′ and A helices and meander region of CYP24A1 show considerable amino acid variability, consistent with their purported membrane binding role. The four protein chains of the two crystal structures of rCYP24A1 (26) indicate that two-thirds of the amino acid rotamers, presumably set during protein folding, did not randomize during protein purification and crystallization. Rotamer variability primarily affects solvent-accessible surface residues (supplemental Figs. S1B), including several possible substrate access or contact residues (e.g. Ile-131, Leu-129, Thr-395, Thr-416) and the active site catalytic Thr-330.
The excellent alignment of the two crystal structures and homology model (20, 21, 26) allowed the active site cavities to be superimposed (supplemental Figs. S1C). From this, we postulated that Val-391 contacts the substrate side chain of 1α,25-(OH)2D3 near C21. This valine is highly conserved in 48/50 species orthologs, and among the two dozen P450 crystal structures reported to date, a hydrophobic amino acid invariably occupies this position. Consequently, we performed site-mutagenesis of Val-391 to further test its role in the enzyme.
1α,25-(OH)2D3 Metabolism by Wild-type and V391L Mutants
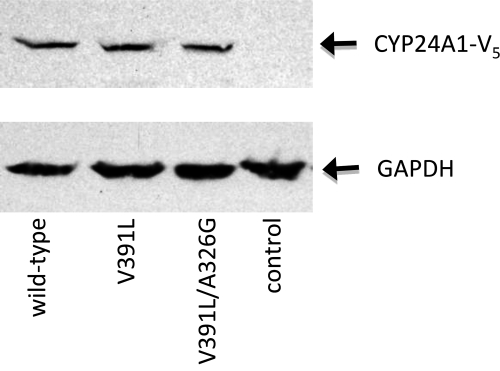
Wild-type hCYP24A1 and its V391L or V391L/A326G double mutants were stably transfected into V79-4 cells, and Western blotting confirmed that the CYP24A1 proteins were expressed at similar levels (Fig. 2). Metabolic profiles were determined for a series of vitamin D analogs including 1α,25-(OH)2D3, 1α-OH-D3, 25-OH-D3, vitamin D3, 1α,25-(OH)2D2, 1α-OH-D2, 1α,25-(OH)2DHT3, and 1α-OH-DHT3 (Fig. 1). The assay was optimized by manipulation of substrate concentration and incubation times so that enzymes produced early to middle pathway intermediates, as summarized in Table 1 and depicted chromatographically in Fig. 3.
FIGURE 2.
Analysis of expression of CYP24A1 mutants in stably transfected V79-4 cells. Relative expression levels of V5 epitope-tagged CYP24A1 in whole cell lysates were determined by Western blotting using an anti-V5 antibody. A 100-μg amount of total protein was added to each lane, and a lysate prepared from non-transfected V79-4 cells was used as a negative control. Hybridizing bands were detected at 57 and 38 kDa using the anti-V5 or anti-GAPDH antibodies, respectively. An anti-V5-hybridizing band was not detected in the non-transfected cell control.
TABLE 1.
Metabolism of vitamin D analogs by hCYP24A1 mutants
| Assay |
Activity and regioselectivity toward specific substrates |
||||||
|---|---|---|---|---|---|---|---|
| Enzyme | Parameter | 1α,25-(OH)2D3 | 25-OH-D3 | 1α-OH-D3 | 1α,25-(OH) 2D2 | 1α-OH-D2 | 1α-OH-DHT3 |
| wt CYP24A1 | Activitya | 3160 ± 100 | 6060 ± 140 | 130 ± 2 | 832 ± 25 | 1220 ± 10 | 312 ± 22 |
| % wtb | 100 | 138 | 4 | 27 | 39 | 10 | |
| C24/C23c | 18 | 21 | all C24 | ||||
| C24/C25d | 4 | 2.3 | 1.7 | ||||
| C24/C26d | 2.1 | ||||||
| A326G | Activity | 46 ± 3 | 269 ± 10 | 55 ± 2 | 386 ± 10 | 61 ± 3 | 164 ± 15 |
| % wt | 1.5 | 6 | 2 | 12 | 2 | 5.6 | |
| C24/C23 | 0 | 0 | all C24 | ||||
| C24/C25 | 3.4 | all C24 | all C24 | ||||
| C24/C26 | all C24 | ||||||
| V391L | Activity | 5160 ± 160 | 7150 ± 130 | 2630 ± 70 | 1060 ± 30 | 1450 ± 90 | 2900 ± 50 |
| % wt | 163 | 118 | 84 | 34 | 47 | 93 | |
| C24/C23 | 42 | 61 | 8.8 | all C24 | 0.2 | 0 | |
| C24/C25 | 0.17 | 3.2 | 2 | ||||
| C24/C26 | 1.0 | ||||||
| V391L/A326G | Activity | 2060 ± 10 | 3430 ± 150 | 1810 ± 120 | 1810 ± 30 | 1240 ± 10 | 2810 ± 40 |
| % wt | 65 | 57 | 58 | 58 | 40 | 90 | |
| C24/C23 | 1.2 | 0.6 | 0.7 | all C24 | 0 | ||
| C24/C25 | 0.09 | 32.7 | 1.2 | ||||
| C24/C26 | 5.9 | ||||||
| wt CYP27A1 | Activity | 0 | 2300 ± 90 | 0 | 1070 ± 30 | 2770 ± 80 | |
| % wt | 0 | 73 | 0 | 35 | 89 | ||
| C24/C23 | |||||||
| C24/C25 | 0.1 | all C24 | 0.03 | ||||
| C24/C26 | 2.1 | 0.25 | |||||
b Percentage of wild-type CYP24A1 activity toward 1α,25-(OH)2D3.
c C24/C23 regioselectivity ratio based on downstream products of 25-OH substrates. A ratio of 0 indicates that all metabolites were C23-pathway products. “all C24” indicates that all of the products detected were from the C24-pathway.
d C24/C25 ratio based upon weighted catabolites of 25-OH substrates or products.
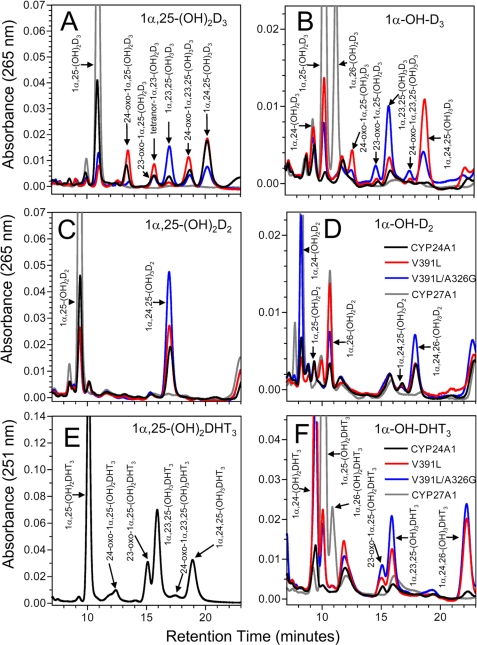
FIGURE 3.
Metabolism of vitamin D analogs and prodrugs by V391L-modified hCYP24A1. The HPLC metabolic profiles of 1α,25-(OH)2D3 and the analogs 1α,25-(OH)2D2 and 1α,25-(OH)2DHT3 (A, C, and E) are compared against those of the corresponding 1α-hydroxylated prodrugs 1α-OH-D3, 1α-OH-D2, and 1α-OH-DHT3 (B, D, and E) using wild-type hCYP24A1 and the mutants V391L and V391L/A326G. CYP27A1 is shown as a negative control for the catabolism of 25-hydroxylated substrates and as a positive control for metabolism of 1α-hydroxylated prodrugs. In panels B, D, and E, the substrates (not shown) typically had retention times of 4–6 min. The scales in panels E and F were expanded by a factor of 2 to accommodate the increased UV absorption by DHT analogs. Due to the limited availability of 1α,25-(OH)2DHT3, only wild-type hCYP24A1 was studied with this analog. The legend for the chromatograms presented in panels D and F apply to all panels in this figure. Table 1 presents a quantitative analysis of the same chromatographic data.
The metabolism of 1α,25-(OH)2D3 by hCYP24A1 (Fig. 3A) gave different intermediates characteristic of the C24 and C23 pathways with, as expected, C24 hydroxylation predominating over C23 hydroxylation with a ratio of 18 (Table 1). Total weighted activity of the V391L mutant showed an increase in total activity (163% of wild-type) and no significant change in the C24/C23 hydroxylation ratio, suggesting that it remains a better 24-hydroxylase. In contrast, the V391L/A326G double mutant shifted catabolism toward the C23 hydroxylation pathway with a C24/C23 ratio of 1.2 and a slightly lower total activity (65% of wild-type). These results suggest that although V391L is better able to position the substrate side chain for 24-hydroxylation, the A326G mutation facilitates the substrate being drawn deeper into the active site for 23-hydroxylation. Similar results were observed with the catabolism of 25-OH-D3, suggesting that lack of a 1α-hydroxyl group has minimal impact on substrate specificity and regioselectivity of hCYP24A1 (Table 1, supplemental Figs. S4).
Prodrug 1α-OH-D3 Metabolism by Wild-type and V391L Mutants
Metabolism of the prodrug 1α-OH-D3 by wild-type hCYP24A1 (Fig. 3B) was extremely low, amounting to only 4% of wild-type 1α,25-(OH)2D3 activity. The enzyme formed small amounts of 1α,24-(OH)2D3 and 1α,25-(OH)2D3, with a C24/C25 ratio of 4. This confirmed that the 1α-OH-D3 prodrug, lacking a 25-hydroxyl group, is a poor substrate for hCYP24A1 despite having a reported binding affinity for rCYP24A1 similar to that for 1α,25-(OH)2D3 (27, 28). Somewhat surprisingly, the V391L mutant exhibited a 21-fold increase in total enzymatic activity (84% of activity shown for 1α,25-(OH)2D3), producing 1α,25-(OH)2D3 as the principal product rather than 1α,24-(OH)2D3 and a major 24-fold shift in the C24/C25 ratio from 4 to 0.17. The V391L mutation thus broadens the substrate specificity of the enzyme to include the anabolic activation of 1α-OH-D3, favoring 25-hydroxylation and permitting, to a lesser extent, some 24-hydroxylation. As shown in Fig. 3B, the 1α,25-(OH)2D3 formed by V391L was subsequently catabolized via the C24 and C23 pathways to 1α,24,25-(OH)3D3 and 1α,23,25-(OH)3D3 with a C24/C23 ratio of 8.8. The V391L/A326G double mutant retained total activity (58% of wild type) and diverted the catabolism of the 1α,25-(OH)2D3 product toward 1α,23,25-(OH)3D3 and the C23 pathway (C24/C23 ratio of 0.7). Interestingly, the total activity of V391L was found to be comparable with the mitochondrial 25-hydroxylase, CYP27A1, in our expression system (Table 1).
Metabolism of Vitamin D2 Analogs by Wild-type and V391L Mutants
The pronounced 25-hydroxylase activity of the V391L mutants toward 1α-OH-D3 prompted us to examine the metabolism of analogs with a vitamin D2 side chain. Multiple rounds of 24- and 23-hydroxylation of the vitamin D2 side chain by hCYP24A1 are blocked by the presence of the 24S-methyl and the C22-C23 double bond, and thus, 1α,25-(OH)2D2 cannot be catabolized beyond 1α,24,25-(OH)3D2 in the classical C24 oxidation pathway (11). As expected, the wild-type enzyme converted 1α,25-(OH)2D2 to 1α,24,25-(OH)3D2 (Fig. 3C) (total activity of 27% of wild type with 1α,25-(OH)2D2). The V391L and V391L/A326G mutations possessed small increases in enzyme activity toward 1α,25-(OH)2D2 (34 and 58% of wild-type), but hydroxylation patterns were unaltered.
The metabolism of the prodrug 1α-OH-D2 (Fig. 3D) by hCYP24A1 produced small amounts of 1α,24-(OH)2D2, 1α,25-(OH)2D2, and 1α,26-(OH)2D2 (total activity of 39% of wild type). One or more of these metabolites were subsequently metabolized to 1α,24,25-(OH)3D2, 1α,24,26-(OH)3D2, and 1α,24,28-(OH)3D2. The V391L mutant displayed no change in total activity (47% of wild type) but significantly greater formation of 1α,26-(OH)2D2. The V391L mutation shifted the C24/C25 ratio (2.3–3.2) and the C24/C26 ratio (2.1 to 1), indicating that it became a better 26-hydroxylase. The total activity of the V391L/A326G mutant was similar to the wild-type enzyme (40%), but this was mostly directed to 24-hydroxylation with a C24/C25 ratio of 32.7 and a C24/C26 ratio of 5.9.
Metabolism of A-ring-modified Vitamin D Analogs by Wild-type and V391L Mutants
The broadened substrate specificity and altered regioselectivity of the V391L mutants toward the vitamin D3 side chain provided a unique opportunity to explore how the A-ring of vitamin D, which is 13–15 Å from the hydroxylation sites in the side chain, is bound by the enzyme and how it might influence regioselectivity. DHT3, an analog of vitamin D3, has an A-ring rotated 180° and the C19-methylene reduced to a methyl (29). Although these changes have a modest effect on the shape of the A-ring of DHT3, the ring rotation exchanges positions of 1α- and 3β-hydroxyls without changing their apparent stereochemistry. Thus, 1α,25-(OH)2DHT3 and 1α-OH-DHT3 represent useful probes to determine the effects of hydrophobic contacts between C19 and the top of the active site cavity/access channel. We also studied the metabolism of vitamin D3, lacking the A-ring 1α-hydroxyl group altogether.
1α,25-(OH)2DHT3 was catabolized into intermediates analogous to those in the C23 and C24 pathways. Wild-type CYP24A1 activity toward 1α,25-(OH)2DHT3 was 2801 ± 83 pmol/2.4 × 106 cells/36 h ±S.E., which was 90% that of 1α,25-(OH)2D3. The hydroxylation pattern for 1α,25-(OH)2DHT3 was shifted toward C23-hydroxylation with a C24/C23 ratio of 0.4 for the compared with a ratio of 18 for 1α,25-(OH)2D3 (Fig. 3E). This suggests that the altered hydrophobic contacts with the A-ring do not interfere with substrate access from penetrating the active site, thereby allowing greater 23-hydroxylation.
Similar to 1α-OH-D3, the 1α-OH-DHT3 was poorly metabolized by hCYP24A1 (10% activity), yielding a small amount of 1α,24-(OH)2DHT3 (Fig. 3F). In comparison, the V391L mutant was fully active (total activity 93% that of wild type) and made 1α,24-(OH)2DHT3 and 1α,25-(OH)2DHT3 in a C24/C25 hydroxylation ratio of 2, with some of the former product being further metabolized to 1α,24,26-(OH)3DHT3 and some of the latter metabolized to 1α,23,25-(OH)3DHT3 (C23 pathway) and 23-oxo-1α,25-(OH)2DHT3. The metabolic patterns for the V391L/A326G and V391L mutants were similar. Thus, the data for 1α-OH-DHT3 are consistent with that for 1α-OH-D3, suggesting that the V391L residue helps to orient the side chain lacking a 25-hydroxyl to be efficiently 25-hydroxylated.
Given that vitamin D3 has been previously shown to bind poorly to rCYP24A1 (28), we were somewhat surprised to observe 25-hydroxylation of vitamin D3 to 25-OH-D3 by V391L and subsequent catabolism to 24,25-(OH)2D3 and more polar products (supplemental Figs. S5). Overall activity levels appeared to be lower than that observed for 1α-OH-D3 and 1α-OH-DHT3, and no metabolism was detected for the double mutant V391L/A326G.
Modeling of Substrate Docking and Contact Residues
To assess the directionality of substrate orientation, precisely position the target carbon for hydroxylation, and orient its hydrogen for abstraction, we have previously triangulated a reactive hydroxylation “sweet-spot” using calculated distances between the heme A-, B-, and D-ring methyl carbons and the target atom in ligand-bound P450 crystal structures (21). To extend this work further here, we employed a strategy to dock vitamin D structures into the active site of our hCYP24A1 model (20, 21) and the rCYP24A1 crystal structure (26) to investigate how experimentally observed substrate specificity and regioselectivity could be influenced by selected contact residues and ligand orientation.
The docking strategy to orient any of C23-C26 carbons for hydroxylation was based on a few key assumptions. The first assumption was that the vitamin D A-ring hydroxyls are held by hydrogen bonds near Gln-82 and two adjacent water molecules seen in both CHAPS-containing rCYP24A1 crystal structures chains to interact with Gln-82 and His-108 (26). This assumption orients the vitamin D molecule into a pw2a access channel (25) and, without precluding other entry trajectories, provides a way to maximize the length of the vitamin D molecule during docking. The second assumption was that the C18 methyl perpendicular to the C- and D-rings of the vitamin D is directed toward the β-5 hairpin. This assumption is reasonable because the rCYP24A1 cavity is wide enough to accommodate any axial rotation of the substrate, and because the C17-C20 bond is free to rotate the side chain into a gauche(+), gauche(−), or anti-conformation (30), the axial rotation on the substrate should have little effect on the constrained target atom (supplemental Figs. S6). Although knowledge of the side-chain conformer might reveal all substrate contacts, this cannot be deduced from other protein-bound vitamin D crystal structures as the side chain is gauche(+) in CYP2R1 and CYP105A1, gauche(-) in vitamin D receptor, and anti- in vitamin D-binding protein. The third assumption is that, when modeling Leu-148 and Leu-391 from Met-148 and Val-391 in rCYP24A1, respectively, rotamer selection (22) can be guided by that observed in other P450 crystal structures. This assumption should be acceptable for the packing of large amino acids like leucine into structurally similar and rotamerically stable locations, and a survey of existing crystal structures shows Leu-148 adopts a t-type rotamer in CYP2R1, CYP119, and CYP125, whereas Leu-391 adopts an m-type rotamer in CYP1A2, CYP2A13, CYP2C5, CYP2C9, and CYP51. Finally, no analysis of vitamin D side chain docking would be complete without a hydrogen-bonding interaction with the 25-hydroxyl group. Such an interaction would have to support both C23- and C24-hydroxylation and be flexible enough to accommodate various pathway intermediates. In the absence of crystallographic proof, we have theorized that the heme propionate binding residue Arg-128 can perform this function if the 25-hydroxyl is able to bring a water molecule to bridge within a 4.5 Å distance. Based upon the above assumptions, the docking of the substrate 1α-OH-D3 into the active site of hCYP24A1 was successfully carried out.
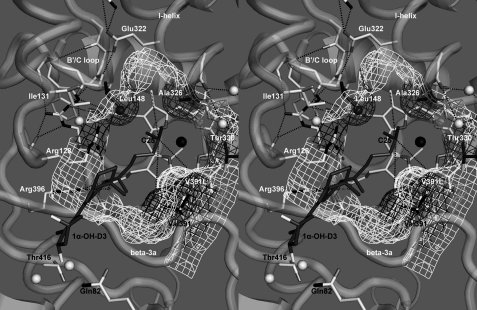
Fig. 4 depicts 1α-OH-D3 oriented for 25-hydroxylation and qualitatively shows the better contact of the mutant Leu-391 side chain with the substrate, which is consistent with the V391L mutants being more efficient 25-hydroxylases toward 1α-OH-D3. In the absence of a 25-hydroxyl, the side chain orientation process appears to be impaired, and metabolism is reduced in the wild-type enzyme. Docking showed that the D3-type side chain contacts the wild-type Val-391 (white mesh) and the mutant Leu-391 (black mesh) differently to alter regioselectivity, and the possible interpretations for the increased activity include the following. (i) Steric conflict of Val-391 with C21 (methyl) is released in the V391L mutant, increasing substrate access. (ii) The new shape of the mutant cavity reduces side chain mobility and restricts it to productive orientations. (iii) The substrate side chain is adsorbed onto Leu-391 by hydrophobic forces and held closer to the preferred regioselectivity. In contrast, a docked 1α-OH-D2 shows more steric conflict between the mutant Leu-391 side chain and the 24S-methyl when the substrate is oriented for 25-hydroxylation (supplemental Fig. S7F) than with either 24- or 26-hydroxylation (supplemental Fig. S7, G and H).
FIGURE 4.
Stereogram of 1α-OH-D3 docked within the substrate binding pocket of hCYP24A1. The mesh depicts selected substrate contact surfaces around the heme group positioned by residues in the β3a strand, I-helix, and the B′/C-loop. The surface shown highlights differences in the binding pocket between the wild-type (white mesh) and the V391L mutant (black mesh) enzymes modeled from the crystal structure of rat CYP24A1 (26). Gray mesh denotes the influence of substrate contact residues Leu-148, Ala-326, and Thr-330. The black sphere denotes the heme iron, and the white/gray spheres represent water molecules. Dotted lines represent key hydrogen bonds as well as the distances from the heme A-, B-, and D-ring methyl carbons used to triangulate the predicted position of C25 of 1α-OH-D3.
DISCUSSION
We have shown that a V391L mutation in the β3a sheet of hCYP24A1 converts this catabolic enzyme from a 1α,25-(OH)2D3-24-hydroxylase into an anabolic 1α-OH-D3-25-hydroxylase, forming 1α,25-(OH)2D3, which is subsequently degraded via a pathway of our choice depending on the amino acid at position 326 in the I-helix (Ala for C24-hydroxylation or Gly for C23-hydroxylation) (Fig. 5). Homology modeling (24) and substrate docking studies using the crystal structure of rCYP24A1 (26) revealed that Leu at position 391 results in a number of structural consequences including reduction of steric conflict with C21, restriction of side chain mobility, and provision of an alternative hydrophobic platform for the side chain, which culminate in an optimal binding orientation for efficient C25-hydroxylation of 1α-OH-D3. Given that V391L also appeared to extend the substrate specificity of CYP24A1 to include vitamin D3, which lacks a 1α-hydroxyl group necessary for strong binding affinity to rCYP24A1 (supplemental Figs. S5), we must acknowledge that improved substrate binding affinity might also contribute to relatively high total enzyme activities observed with the V391L mutants. A reconstituted enzyme system not available in our studies would be necessary to test this hypothesis. The residue Val-391 is among several residues identified in cytochrome P450s that play important roles in regioselectivity and substrate specificity (17, 20, 27, 31, 32).
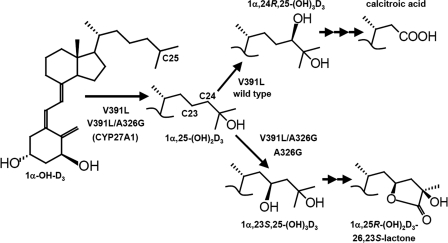
FIGURE 5.
Summary of observed metabolism by wild-type CYP24A1 and its V391L mutants. CYP24A1 catalyzes the catabolism of 1α,25-(OH)2D3 and 25-OH-D3 via C24 or C23 lactone pathways but does not significantly metabolize the prodrug, 1α-OH-D3. The mutation V391L broadened the substrate specificity to accommodate the anabolic activation of 1α-OH-D3 to produce 1α,25-(OH)2D3, which was subsequently catabolized by the C24 pathway. When combined with the A326G mutation, previously shown to direct catabolism toward the C23 lactone, the double mutant converted 1α-OH-D3 to 1α,25-(OH)2D3 and catabolized it via the C23 lactone pathway. Overall, 1α-hydroxylated prodrugs, without sterically hindering modifications on the side chain, appeared most conducive to anabolic C25-hydroxylation by V391L and, to a lesser extent, vitamin D3, which lacks a 1α-hydroxyl (not shown; supplemental Figs. S5). However, analogs investigated possessing a C25-hydroxyl group were easily catabolized by V391L whether formed in situ or given directly as substrate.
Previous work by Omdahl and co-workers (27, 28) has revealed that CYP24A1 is highly specific for vitamin D secosteroids and shows little enzyme activity toward cholesterol and bile acids and that the 1α-hydroxyl group is a critical determinant of strong binding affinity to rCYP24A1; however, this is not sufficient for catalytic activity, suggesting that a putative hydrogen bond with a 25-hydroxyl group is necessary to optimally orient the side chain above the heme for hydroxylation (26). A C24S-methyl could potentially act as a surrogate C25 hydroxyl group to stabilize the side chain of 1α-OH-D2, to which wild-type CYP24A1 exhibited greater total activity than observed with 1α-OH-D3, in agreement with our previous results (11). Furthermore, Reddy and co-workers demonstrated that the analog 1α,24R-(OH)2D3, could be C25-hydroxylated by CYP24A1 implying that a hydroxyl group at C24 can also act in lieu of a C25-hydroxyl group to stabilize the side chain (33). We have achieved stabilization of the unmodified (D3) side chain by making a subtle V391L mutation in the β3a sheet, providing the first enzyme activity-based evidence that the 1α-OH-D3 (or 1α-OH-DHT3) prodrug lacking a C25-hydroxyl group can productively bind, become activated, and then be catabolized by hCYP24A1.
The observation that the predicted position of the α-carbon of Val-391 in our homology model was within 1 Å of that observed in the crystal structure points to the growing sophistication and predictive value of the homology modeling approach. Furthermore, in the absence of a 1α,25-(OH)2D3 ligand crystallized within the active site of rCYP24A1, we used a novel average hydroxylation-susceptible carbon approach to dock the substrate and position the target carbon above the heme iron in hCYP24A1. Our approach was based upon certain assumptions that we have justified here. The substrate fit best with the anti- conformation about the C17-C20 bond because it made good contact with spatially conserved waters in the crystal structure near Gln-82 and His-108. Another important consideration for the position of the side chain, within the wild-type and mutated active site, was the selection of side chain rotamers at protein contact residues. For example, rotamer selection for residue 148 was important in this study as the substrate side chain is situated between residue 148 in the B′/C-loop and Val-391 in the β3a sheet in the active site. Thus, when modeling Leu-148 and Leu-391 in hCYP24A1 from Met-148 and Val-391 in rCYP24A1, rotamer selection was guided by observations from other crystal structures. Docking studies showed that the D3-type side chain contacts the wild-type Val-391 and the mutant Leu-391 differently (Fig. 4) and offers explanations for why the two enzymes show different regioselectivity. In contrast, a docked 1α-OH-D2 shows more steric conflict between the mutant Leu-391 side chain and the 24S-methyl when the substrate is oriented for 25-hydroxylation than with either 24- or 26-hydroxylation. This is consistent with the metabolic data showing that wild-type hCYP24A1 is a better 24-hydroxylase toward 1α-OH-D3 and that the V391L mutants are better 24- and 26-hydroxylases toward 1α-OH-D2.
Our observation that hCYP24A1 can be converted into an anabolic enzyme is an intriguing concept from the perspective of vitamin D analog design. If a 1α-hydroxylated analog could be designed to mimic the effect of V391L, the analog could potentially be activated and degraded locally in the target cell in addition to receiving activated analog on vitamin D-binding protein arising from hepatic C25-hydroxylation by CYP27A1/CYP2R1. A knock-in V391L mouse model with and without CYP27A1/CYP2R1 ablation, administered 1α-OH-D3, could theoretically test the physiological impact of CYP24A1 as the sole source of active hormone at the target cell level and evaluate the potential of an analog containing a V391L-mimicking modification. Locally produced hormone would potentially lead to increased cellular concentrations of hormone and/or and an altered half-life. In cancer cells that possess increased levels of CYP24A1, the test analog would be locally activated before becoming degraded, resulting in a longer-lived anti-proliferative/pro-differentiation signal, originally evaded by high CYP24A1 levels. This concept parallels that of the extra-renal CYP27B1 identified in breast, prostate, colon, and smooth muscle cells, which draws upon the circulating pool of 25-OH-D3 via megalin/cubilin-mediated endocytosis to become locally activated in the target cell (10). The 1α-hydroxylated test analog might possess several advantages, including a slightly lower affinity for the vitamin D binding protein than 1α,25-(OH)2D3, suggesting increased transport into target cells, circumventing the requirement of the cell to express megalin/cubilin or CYP27B1. Perhaps a 1α-hydroxylated prodrug that can be activated and degraded by wild-type CYP24A1 already exists, as it has already been shown that 1α-OH-D2 can be activated to 1α,25-(OH)2D2 and 1α,24-(OH)2D2 before being degraded by CYP24A1 (11). Taken together, this suggests that the altered regioselectivity of anabolic CYP24A1 reactions can give rise to indirect changes in the vitamin D receptor response in the target-cell environment.
Unlike with the other proteins involved in the vitamin D signal transduction pathway, CYP24A1-inactivating mutations and a CYP24A1-deficient phenotype have yet to be reported in humans. Inactivating mutations in the CYP24A1-null mouse result in hypercalcemia and nephrocalcinosis (34), suggesting that these would also be features of the human phenotype. Furthermore, a recent genome-wide association study for the determinants of serum 25-OH-D, the key vitamin D biomarker, have identified 4 major genes including CYP24A1 as well as CYP2R1, vitamin D-binding protein, and 7-dehydrocholesterol reductase (35). These data suggest that mutants in the CYP24A1 gene do exist but have yet to be specifically identified. Our work predicts that even very modest changes in key secondary structures of hCYP24A1 will result in drastic changes of its enzymatic properties, either total activity, or regioselectivity. Moreover, our developed methodology to mutate and express mutants using the Flp-In system is ideal to quickly generate and test natural polymorphisms or mutations. There is currently acute interest in unraveling the broad-ranging effects of vitamin D around the body, and our work points to the fact that vitamin D metabolism is still a fertile area to explore for clinically relevant applications.
Supplementary Material
Acknowledgments
We acknowledge Lindsay Wilson, Dr. Donald H. Maurice, Julia Kim, and Dr. Sameh Basta at Queen's University for help with Western blotting.
This study was supported by Canadian Institutes of Health Research Grants MA-9475, MMA-38116, and MMA-69106 and a research grant from Cytochroma Inc.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S7.
- 1α,25-(OH)2D3
- 1α,25-dihydroxyvitamin D3
- 1α-OH-D3
- 1α-hydroxyvitamin D3
- CYP
- cytochrome P450
- h-
- human
- DHT3
- dihydrotachysterol3
- r-
- rat.
REFERENCES
- 1. DeLuca H. F. (1974) Fed. Proc. 33, 2211–2219 [PubMed] [Google Scholar]
- 2. Prosser D. E., Jones G. (2004) Trends Biochem. Sci. 29, 664–773 [DOI] [PubMed] [Google Scholar]
- 3. Haussler M. R., Whitfield G. K., Haussler C. A., Hsieh J. C., Thompson P. D., Selznick S. H., Dominguez C. E., Jurutka P. W. (1998) J. Bone Miner. Res. 13, 325–349 [DOI] [PubMed] [Google Scholar]
- 4. Cheng J. B., Motola D. L., Mangelsdorf D. J., Russell D. W. (2003) J. Biol. Chem. 278, 38084–38093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Strushkevich N., Usanov S. A., Plotnikov A. N., Jones G., Park H. W. (2008) J. Mol. Biol. 380, 95–106 [DOI] [PubMed] [Google Scholar]
- 6. Guo Y. D., Strugnell S., Back D. W., Jones G. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 8668–8672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gupta R. P., He Y. A., Patrick K. S., Halpert J. R., Bell N. H. (2005) J. Clin. Endocrinol. Metab. 90, 1210–1219 [DOI] [PubMed] [Google Scholar]
- 8. Fraser D. R., Kodicek E. (1970) Nature 228, 764–766 [DOI] [PubMed] [Google Scholar]
- 9. Hewison M., Adams J. (2005) in Vitamin D (Feldman D., Pike J. W., Glorieux F. H. eds.) 2nd Ed., pp. 1379–1403, Elsevier Academic Press, New York [Google Scholar]
- 10. Holick M. F., Tavela T. E., Holick S. A., Schnoes H. K., DeLuca F., Gallagher B. M. (1976) J. Biol. Chem. 251, 1020–1024 [PubMed] [Google Scholar]
- 11. Masuda S., Strugnell S. A., Knutson J. C., St-Arnaud R., Jones G. (2006) Biochim. Biophys. Acta 1761, 221–234 [DOI] [PubMed] [Google Scholar]
- 12. Akiyoshi-Shibata M., Sakaki T., Ohyama Y., Noshiro M., Okuda K., Yabusaki Y. (1994) Eur. J. Biochem. 224, 335–343 [DOI] [PubMed] [Google Scholar]
- 13. Beckman M. J., Tadikonda P., Werner E., Prahl J., Yamada S., DeLuca H. F. (1996) Biochemistry 35, 8465–8472 [DOI] [PubMed] [Google Scholar]
- 14. Makin G., Lohnes D., Byford V., Ray R., Jones G. (1989) Biochem. J. 262, 173–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Reddy G. S., Tserng K. Y. (1989) Biochemistry 28, 1763–1769 [DOI] [PubMed] [Google Scholar]
- 16. Sakaki T., Sawada N., Komai K., Shiozawa S., Yamada S., Yamamoto K., Ohyama Y., Inouye K. (2000) Eur. J. Biochem. 267, 6158–6165 [DOI] [PubMed] [Google Scholar]
- 17. Hamamoto H., Kusudo T., Urushino N., Masuno H., Yamamoto K., Yamada S., Kamakura M., Ohta M., Inouye K., Sakaki T. (2006) Mol. Pharmacol. 70, 120–128 [DOI] [PubMed] [Google Scholar]
- 18. Horiuchi N., Saikatsu S., Akeno N., Abe M., Kimura S., Yamada S. (1995) Horm. Metab. Res. 27, 83–89 [DOI] [PubMed] [Google Scholar]
- 19. Pedersen J. I., Hagenfeldt Y., Björkhem I. (1988) Biochem. J. 250, 527–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Prosser D. E., Kaufmann M., O'Leary B., Byford V., Jones G. (2007) Proc. Natl. Acad. Sci. 104, 12673–12678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Masuda S., Prosser D. E., Guo Y. D., Kaufmann M., Jones G. (2007) Arch. Biochem. Biophys. 460, 177–191 [DOI] [PubMed] [Google Scholar]
- 22. O'Gorman S., Fox D. T., Wahl G. M. (1991) Science 251, 1351–1355 [DOI] [PubMed] [Google Scholar]
- 23. Gambhir V., Kim J., Siddiqui S., Taylor M., Byford V., Jones G., Petrof E. O., Basta S. (2011) Immunobiology, in press [DOI] [PubMed] [Google Scholar]
- 24. Lovell S. C., Word J. M., Richardson J. S., Richardson D. C. (2000) Proteins Struct. Funct. Genet. 40, 389–408 [PubMed] [Google Scholar]
- 25. Cojocaru V., Winn P. J., Wade R. C. (2007) Biochim. Biophys. Acta 1770, 390–401 [DOI] [PubMed] [Google Scholar]
- 26. Annalora A. J., Goodin D. B., Hong W. X., Zhang Q., Johnson E. F., Stout C. D. (2010) J. Mol. Biol. 396, 441–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Annalora A. J., Bobrovnikov-Marjon E., Serda R., Pastuszyn A., Graham S. E., Marcus C. B., Omdahl J. L. (2007) Arch. Biochem. Biophys. 460, 262–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Annalora A., Bobrovnikova-Marjon E., Serda R., Lansing L., Chiu M. L., Pastuszyn A., Iyer S., Marcus C. B., Omdahl J. L. (2004) Arch. Biochem. Biophys. 425, 133–146 [DOI] [PubMed] [Google Scholar]
- 29. Qaw F., Calverley M. J., Schroeder N. J., Trafford D. J., Makin H. L., Jones G. (1993) J. Biol. Chem. 268, 282–292 [PubMed] [Google Scholar]
- 30. Yamada S., Yamamoto K., Masuno H., Ohta M. (1998) J. Med. Chem. 41, 1467–1475 [DOI] [PubMed] [Google Scholar]
- 31. Böttner B., Schrauber H., Bernhardt R. (1996) J. Biol. Chem. 271, 8028–8033 [DOI] [PubMed] [Google Scholar]
- 32. Swart A. C., Storbeck K. H., Swart P. (2010) J. Steroid Biochem. Mol. Biol. 119, 112–120 [DOI] [PubMed] [Google Scholar]
- 33. Astecker N., Bobrovnikova E. A., Omdahl J. L., Gennaro L., Vouros P., Schuster I., Uskokovic M. R., Ishizuka S., Wang G., Reddy G. S. (2004) Arch. Biochem. Biophys. 431, 261–270 [DOI] [PubMed] [Google Scholar]
- 34. St.-Arnaud R., Arabian A., Travers R., Barletta F., Raval-Pandya M., Chapin K., Depovere J., Mathieu C., Christakos S., Demay M. B., Glorieux F. H. (2000) Endocrinology 141, 2658–2666 [DOI] [PubMed] [Google Scholar]
- 35. Wang T. J., Zhang F., Richards J. B., Kestenbaum B., van Meurs J. B., Berry D., Kiel D. P., Streeten E. A., Ohlsson C., Koller D. L., Peltonen L., Cooper J. D., O'Reilly P. F., Houston D. K., Glazer N. L., Vandenput L., Peacock M., Shi J., Rivadeneira F., McCarthy M. I., Anneli P., de Boer I. H., Mangino M., Kato B., Smyth D. J., Booth S. L., Jacques P. F., Burke G. L., Goodarzi M., Cheung C. L., Wolf M., Rice K., Goltzman D., Hidiroglou N., Ladouceur M., Wareham N. J., Hocking L. J., Hart D., Arden N. K., Cooper C., Malik S., Fraser W. D., Hartikainen A. L., Zhai G., Macdonald H. M., Forouhi N. G., Loos R. J., Reid D. M., Hakim A., Dennison E., Liu Y., Power C., Stevens H. E., Jaana L., Vasan R. S., Soranzo N., Bojunga J., Psaty B. M., Lorentzon M., Foroud T., Harris T. B., Hofman A., Jansson J. O., Cauley J. A., Uitterlinden A. G., Gibson Q., Järvelin M. R., Karasik D., Siscovick D. S., Econs M. J., Kritchevsky S. B., Florez J. C., Todd J. A., Dupuis J., Hyppönen E., Spector T. D. (2010) Lancet 376, 180–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.