Abstract
Resveratrol, a naturally occurring phytoalexin, is known to induce apoptosis in multiple cancer cell types, but the underlying molecular mechanisms remain unclear. Here, we show that resveratrol induced p53-independent, X-linked inhibitor of apoptosis protein (XIAP)-mediated translocation of Bax to mitochondria where it underwent oligomerization to initiate apoptosis. Resveratrol treatment promoted interaction between Bax and XIAP in the cytosol and on mitochondria, suggesting that XIAP plays a critical role in the activation and translocation of Bax to mitochondria. This process did not involve p53 but required accumulation of Bim and t-Bid on mitochondria. Bax primarily underwent homo-oligomerization on mitochondria and played a major role in release of cytochrome c to the cytosol. Bak, another key protein that regulates the mitochondrial membrane permeabilization, did not interact with p53 but continued to associate with Bcl-xL. Thus, the proapoptotic function of Bak remained suppressed during resveratrol-induced apoptosis. Caspase-9 silencing inhibited resveratrol-induced caspase activation, whereas caspase-8 knockdown did not affect caspase activity, suggesting that resveratrol induces caspase-9-dependent apoptosis. Together, our findings characterize the molecular mechanisms of resveratrol-induced caspase activation and subsequent apoptosis in cancer cells.
Keywords: Apoptosis, Cytochrome c, Mitochondria, p53, Resveratrol, XIAP, Bak, Bax
Introduction
Anticancer agents induce cell death in cancer and normal cells via mechanisms including apoptosis and autophagy (1–4). Therefore, there is a need for alternative anticancer agents that can promote cancer cell death while avoiding killing of normal, non-cancerous cells. Resveratrol (trans-3,5,4′-trihydroxystilbene) is a naturally occurring polyphenolic phytoalexin found at high levels in the skin of grapes and in red wine. It is also present in peanuts and other plant products. Resveratrol has been shown to possess an apoptosis-dependent anticancer activity and minimal toxicity to normal cells (5–11). How resveratrol induces apoptosis or cancer cell death is not clearly known, but available evidence indicates that resveratrol induces p53-dependent signaling, which leads to cell cycle arrest and apoptosis induction (10, 12, 13). Additionally, resveratrol targets mitochondria to induce cytochrome c release and thereby triggers caspase-dependent apoptotic cell death in multiple types of cancer cells (14–18). How resveratrol induces cytochrome c release and caspase activation to execute apoptosis remains unclear.
Caspases are activated by proteolytic processing and are broadly divided into initiator caspases (e.g. procaspase-8 and -9) and executioner caspases (such as procaspase-3 and -7) (19–22). During apoptosis, the released cytochrome c from mitochondria triggers caspase-9 activation, whereas ligation of death receptors on the plasma membrane activates caspase-8. Active caspase-8 generated upon death receptor ligation requires Bid-mediated cytochrome c release to execute apoptotic cell death in epithelial cancer cells (22–25). Proapoptotic BH3-only proteins such as activated Bid/Bim translocate to mitochondria to initiate Bax and/or Bak activation, leading to channel formation on the outer mitochondrial membrane (OMM)3 and permeabilization of mitochondria (22, 26, 27). Bak localizes on the OMM, whereas Bax mostly exists in the cytosol. Although resveratrol has been shown to modulate the levels of Bax/Bak or other Bcl-2 family proteins (14, 28), it is unclear whether and how resveratrol activates Bax/Bak to permeabilize the mitochondrial membrane.
Here, we demonstrate that resveratrol induced caspase-dependent apoptosis by targeting mitochondria. Our findings indicate that resveratrol may induce Bax oligomerization in the cytosol. Bax activation and its translocation to mitochondria seemed to be regulated by XIAP; however, p53 did not directly participate in the activation of Bax/Bak. Bax recruitment to and its oligomerization on mitochondria were associated with cytochrome c release, caspase activation, and apoptosis. These findings suggest that resveratrol may be a novel inducer of Bax-mediated caspase activation and apoptosis in cancer cells, which normally lack p53 activity or harbor mutant p53.
EXPERIMENTAL PROCEDURES
Cells and Reagents
Colon cancer cells (HCT116, HCT116-Bax-KO, and HCT-p53-KO) were kindly provided by Dr. B. Vogelstein (29, 30) and cultured in McCoy's 5A medium supplemented with 10% FBS. Prostate cancer (PC3 and LNCaP), breast cancer (MDA-MB231, MCF-7, and MDA-MB435), immortalized normal human fibroblast (GM701), Jurkat wild type (WT), Jurkat caspase-8−/−, mouse embryonic fibroblast WT, and mouse embryonic fibroblast Apaf-1−/− cells were obtained from the ATCC or from various investigators and were subcultured as described previously (31–36). The primary antibodies against cytochrome c (monoclonal antibody (mAb)), Apaf-1 and Bax (rabbit polyclonal antibody (Rb pAb)), Bid, caspase-8, XIAP, and Bcl-xL were purchased from BD Pharmingen. Bax N terminus (Rb pAb; Upstate); Bak (Rb pAb; Santa Cruz Biotechnology); p53 (Santa Cruz Biotechnology); cytochrome c oxidase subunit II (Mito Sciences); heat shock protein 60 (Hsp60) (Millipore); Hsp70 (Stressgen); Bak N terminus (Rb pAb; Upstate); VDAC-1 and Bim (Calbiochem); caspase-3 (Rb pAb; Biomol); caspase-9 (Cell Signaling Technology); and poly(ADP-ribose) polymerase, lactate dehydrogenase, and actin (mAb; ICN) were obtained from the indicated suppliers. Secondary antibodies and ECL reagents were acquired from GE Healthcare. Alexa Fluor 594- or 488-conjugated goat anti-mouse or -rabbit IgG (heavy + light) and mitochondrial dye (i.e. MitoTracker Orange CMTMRos) were purchased from Molecular Probes (Invitrogen). The fluorogenic caspase substrates DEVD-AFC, LEHD-AFC, general caspase inhibitor Z-VAD-fluoromethyl ketone, and cross-linkers were bought from Enzo Life Sciences. All other chemicals were purchased from Sigma unless specified otherwise.
Subcellular Fractionation and Western Blotting
The preparation of whole cell lysates and mitochondrial and cytosolic fractions and Western blotting were preformed as detailed previously (35).
Quantification of Apoptosis and Caspase Activity Measurement
Apoptotic cells were counted based on live cell staining with DAPI to label apoptotic nuclei (35). In addition, both live and dead cells were counted using trypan blue dye. DEVDase and LEHDase activities were measured as described previously (35).
Establishment of Cancer Cells Stably Expressing Caspase-9 or Caspase-8 siRNA Using shRNA Lentiviral Vectors
Green fluorescence protein (GFP)-tagged short hairpin RNAs (shRNAs) specific to caspase-9 and caspase-8 and negative control shRNA were cloned into the pGIPZ (Open Biosystems) lentiviral vector to generate lentiviral particles. The shRNA sequences were as follows: caspase-8, 5′-GACTTCAGCAGAAATCTTT-3′; and caspase-9, 5′-CCAGGCAGCTGATCATAGA-3′. Lentiviral particles specific for caspase-9, caspase-8, and control shRNAs were obtained from the Roswell Park Cancer Institute shRNA core resource and were directly utilized to infect cells at a multiplicity of infection of 3. After 48 h, puromycin (1 μg/ml) was added to the medium to select caspase-8 or caspase-9 knockdown cells (37).
Immunofluorescence
Cells grown on coverslips were treated with resveratrol and 15 min prior to the end of treatment were incubated live with either DAPI alone (to label nuclei) or MitoTracker Orange (CMTMRos) and DAPI (to label mitochondria and nuclei, respectively). Cells were then fixed, permeabilized, and immunolabeled for cytochrome c (32, 35).
Chemical Cross-linking and Oligomerization Assays
Freshly harvested cells or freshly purified mitochondria or cytosol (50 μg) was suspended in 45 μl of HIM buffer (200 mm mannitol, 70 mm sucrose, 10 mm HEPES-KOH, 1 mm EGTA, pH 7.5) followed by addition of freshly prepared bismaleimidohexane (BMH) or ethylene glycol bis(succinimidylsuccinate) to a final concentration of 2 mm and incubated at room temperature for 30 min. Mitochondria were then mixed with protein sample buffer and subjected to Western blotting (32).
Gel Filtration Analysis
Various types of cancer cells were treated with resveratrol or vehicle (DMSO) and washed twice in cold phosphate-buffered saline. Cytosolic and mitochondrial fractions were purified as described previously (31, 32, 34) and loaded onto a Superdex 200 HR10/30 column (GE Healthcare). Proteins were eluted at 0.5 ml/min, and fractions (0.5 ml) were collected for Western analyses (38).
Immunoprecipitation
Purified mitochondrial lysates or cytosols were precleared with mouse or rabbit (depending on the primary antibodies used) IgG-conjugated agarose beads and incubated with primary antibodies against Bax, Bak, p53, XIAP, or Rb IgG (as control) followed by addition of rabbit or mouse IgG beads. Finally, the beads were washed thoroughly and analyzed by the TrueBlot (eBioscience) Western blotting system (34).
Cell-free Reconstitution Experiments
Purified mitochondria were incubated with cytosol or homogenizing buffer in a total reaction mixture of 50 μl at 37 °C for 60 min followed by Western blotting to detect XIAP translocation (36).
Statistical Analysis
Results are presented as mean ± S.D. of data from at least three independent experiments. Statistical analysis was performed by analysis of variance using SigmaStat. Significant changes (p < 0.01) are represented by *.
RESULTS
Resveratrol Induces Caspase-dependent Apoptosis
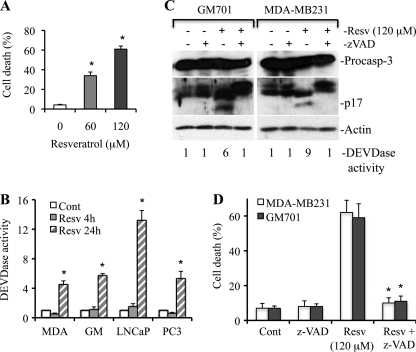
Resveratrol induces apoptotic cell death in multiple types of cancer cells, but the molecular mechanism is still unclear. To understand the involvement of apoptosis in resveratrol-induced cell death, we treated MDA-MB231, LNCaP, PC3, and GM701 cells with increasing doses (20–120 μm) of resveratrol and observed a dose-dependent apoptosis and caspase-3 activation (Fig. 1A and data not shown). Caspase-3 is a 34-kDa protein and is processed during apoptosis to generate p20, p19, and p17 fragments (39, 40). To investigate whether cleaved caspase fragments are functionally active, a substrate cleavage assay (i.e. DEVDase assay), which represents caspase-3/7 activity, was performed after treating cells with resveratrol for 4- and 24-h time periods. As shown in Fig. 1B, resveratrol induced 4-, 6-, 13-, and 5-fold increases in caspase-3 activity as compared with DMSO in MDA-MD231, GM701, LNCaP, and PC3 cells, respectively, at the 24-h time period. To further demonstrate that apoptosis induced by resveratrol is caspase-dependent, we pretreated MDA-MB231 and GM701 cells with a pan-caspase inhibitor (Z-VAD) and observed that resveratrol-induced caspase-3 processing (i.e. p17), and DEVDase activity was inhibited by Z-VAD (Fig. 1C). Similarly, resveratrol-induced cell death was inhibited in the presence of caspase inhibitor (Fig. 1D). Altogether, these findings demonstrate that resveratrol induces caspase-dependent apoptosis.
FIGURE 1.
Resveratrol induces caspase-dependent apoptosis. A, MDA-MB231 cells were treated with resveratrol (Resv) for 36 h, and the percentage of cell death (mean ± S.D. from four independent experiments) was determined using trypan blue dye. *, p < 0.01. B, MDA-MB231, GM701 (GM), LNCaP, and PC3 cells were treated with resveratrol for the indicated time intervals, and equal amounts of protein (50 μg) were subjected to caspase-3 activity measurements. Caspase activities are presented as values (mean ± S.D. from at least three independent experiments) relative to those in controls. *, p < 0.01. C, GM701 and MDA-MB231 cells were pretreated with the pan-caspase inhibitor Z-VAD (50 μm) for 1 h and then treated with resveratrol for 36 h. At the end of treatment, equal amounts of protein were used for Western blotting and DEVDase activity assay. The number in DEVDase activity represents -fold change against untreated control (for example, lane 1 is control for GM701 cells and lane 5 is control for MDA-MB231 cells). Data are the representative of four independent experiments. D, MDA-MB231 and GM701 cells were pretreated with the pan-caspase inhibitor Z-VAD (50 μm) for 1 h and then treated with resveratrol for 36 h. At the end of treatment, the percentage of cell death was determined, and data are represented as mean ± S.D. from four independent experiments. *, p < 0.01 as compared with resveratrol-treated cells. Cont, control; Resv, resveratrol; MDA, MDA-MB231; Procasp-3, procaspase-3. p17 is a cleaved caspase-3 fragment.
Resveratrol Treatment Leads to Cytochrome c Release from Mitochondria
To explore the molecular mechanism of resveratrol-induced caspase-dependent apoptosis, we purified mitochondrial and cytosolic fractions from untreated or resveratrol-treated MDA-MB231 cells followed by Western blotting to detect cytochrome c release from mitochondria, a critical step to trigger Apaf-1-dependent caspase activation. As shown in Fig. 2A, resveratrol treatment induced a low level of cytochrome c release in the cytosol as early as 12 h in MDA-MB231 cells prior to caspase activation, which happened around 24 h after resveratrol treatment. The levels of mitochondrial cytochrome c did not decrease, supporting previous findings that treatment of cells with apoptotic inducers up-regulates mitochondrial respiratory chain proteins such as cytochrome c (35, 41–44). We did not detect cytochrome c oxidase subunit II (a marker for mitochondria) in the cytosol and lactate dehydrogenase (a marker for cytosol) in mitochondria, indicating that cytosolic fractions were not contaminated with mitochondrial proteins and vice versa. The released cytochrome c was followed by caspase activation as caspase-3 was processed to p20/17 in the cytosol and mitochondria at 24 h and onward after resveratrol treatment (Fig. 2A). The substrate cleavage assay (i.e. DEVDase assay) also showed an increase in caspase-3 activity at 24 h onward upon resveratrol treatment (Fig. 1B and data not shown). Similarly, resveratrol induced cytochrome c release in MDA-MB435, HCT116, and LNCaP cells (see Figs. 4F and 7A and data not shown).
FIGURE 2.
Resveratrol induces cytochrome c release from mitochondria into cytosol and is accompanied by fragmentation of nucleus. A, MDA-MB231 cells were treated with resveratrol (Resv; 120 μm) for the indicated time intervals. At the end of the treatment, cytosolic and mitochondrial fractions were isolated, and equal amounts of protein were subjected to Western blotting for the detection of the indicated molecules. Cyt. c, cytochrome c; Procasp-3, procaspase-3; COX II, cytochrome c oxidase subunit II; LDH, lactate dehydrogenase. p20/17 represents cleaved products of caspase-3. Actin and Hsp60 serve as loading controls. B, cells were treated with resveratrol (Resv; 120 μm) for 24 h. At the end of the treatment, live cells were labeled with DAPI (panels a, c, e, and g) and MitoTracker Orange to detect the nucleus and mitochondria (data not shown), respectively. Cells were then immunolabeled for cytochrome c (Cyt. c; panels b, d, f, and h). Representative micrographs are shown, and magnification bars represent 20 μm. Consistent with the Western blot analysis, the diffuse staining for cytochrome c in individual cells reveals that it was released from mitochondria. Apoptotic cells show fragmented/shiny nucleus with DAPI staining in the panels c and g. Data are representative of at least three independent experiments.
FIGURE 4.
Bax undergoes homo-oligomerization on mitochondria, and Bax oligomerization on mitochondria is accompanied by cytochrome c release. A and B, MDA-MB231 (A) cells were treated with resveratrol (Resv; 120 μm) for the indicated time intervals, and HCT116 WT or HCT116 Bax−/− cells (B) were treated with resveratrol (50 μm). Purified mitochondria were cross-linked with BMH and subjected to Western blotting for Bax to detect its oligomers. Hsp60 serves as a loading control. C and D, MDA-MB231 and HCT116 cells were treated with resveratrol (50 μm) for the indicated time periods. At the end of treatments, cells were cross-linked with BMH or ethylene glycol bis(succinimidylsuccinate) (EGS) followed by Western blotting for Bax or Hsp60. E, mitochondrial lysates isolated from MDA-MB231 cells treated with resveratrol (120 μm for 36 h) or from unstimulated cells were subjected to immunoprecipitation followed by Western blotting for the indicated molecules. F, MDA-MB231 and MDA-MB435 cells were treated with resveratrol (50 μm) followed by mitochondrial cross-linking and cytochrome c release assayed by Western blotting. MDA, MDA-MB231; MB435, MDA-MB435; Cyt. c, cytochrome c; LDH, lactate dehydrogenase. The asterisks in A, B, and F indicate a nonspecific band. Data are representative of at least three independent experiments.
FIGURE 7.
Bax plays prominent role to induce cytochrome c release upon resveratrol treatment. A, HCT116 WT, Bax−/−, and p53−/− cells were treated with resveratrol (120 μm for 12 or 48 h). Cytosolic and mitochondrial fractions were subjected to Western blotting to detect cytochrome c release. Resv, resveratrol; Cyt. c, cytochrome c; COX II, cytochrome c oxidase subunit II. Actin and Hsp60 serve as loading controls. Data are representative of three independent experiments. B, MDA-MB231, MDA-MB435, and MCF-7 cells were pretreated for 2 h with PFT-α (30 μm) or PFT-μ (10 μm) followed by resveratrol treatment (50 μm for 24 h). At the end of treatment, the percentage of cell death was quantified. Data are mean ± S.D. (n = 3). PFT-a, PFT-α; PFT-m, PFT-μ.
Cytochrome c is encoded by the nuclear genome and is synthesized in the cytosol. This newly synthesized cytochrome c (i.e. apocytochrome c) then translocates to mitochondria where a heme moiety is attached to generate holocytochrome c (i.e. mitochondrial cytochrome c), which participates in the electron transport chain (45–47). Once released from mitochondria, holocytochrome c triggers apoptosome-dependent caspase activation (20, 35). To further demonstrate that the cytosolic increase in cytochrome c levels was not due to the up-regulation of newly synthesized cytochrome c (i.e. apocytochrome c), we performed immunolabeling to detect holocytochrome c. As shown in Fig. 2B, cytochrome c was mostly detected in mitochondria of control cells in MDA-MB231 and GM701 cells. Resveratrol treatment led to diffuse cytochrome c staining, suggesting that cytochrome c was released in response to resveratrol treatment. As expected, the released cytochrome c induced apoptosis as evidenced by the fragmentation of the nucleus (Fig. 2B, panels c and g). Altogether, these findings demonstrate that resveratrol treatment triggers cytochrome c release in multiple types of cells.
Resveratrol Treatment Causes Loss of Mitochondrial Membrane Potential
To understand the mechanism underlying cytochrome c release from mitochondria in response to resveratrol, we first evaluated the membrane potential of mitochondria by labeling MDA-MB231, LNCaP, and GM701 cells with MitoTracker Orange, which labels mitochondria in a membrane potential-dependent manner. Our data demonstrated that MitoTracker was accumulated in mitochondria of control cells. Upon resveratrol treatment, MitoTracker was not observed in mitochondria, suggesting the loss of membrane potential during apoptosis (Fig. 3A). These findings led us to conclude that resveratrol induces mitochondrial dysfunction, which would account for the observed release of cytochrome c from mitochondria.
FIGURE 3.
Resveratrol causes loss of mitochondrial membrane potential and induces Bax translocation to mitochondria and up-regulates Bim and t-Bid on mitochondria. A, MDA-MB231, LNCaP, and GM701 cells were treated without (Control; panels a, c, and e) or with resveratrol (Resv; 120 μm for 24 h) (panels b, d, and f) and just prior to fixation were labeled live with MitoTracker Orange, which accumulates in mitochondria in a membrane potential-dependent manner. The images were captured by microscope, representative micrographs are shown, and magnification bars represent 20 μm. B, MDA-MB231 cells were treated with resveratrol (Resv; 120 μm) for the indicated time intervals. Cytosolic and mitochondrial fractions were purified followed by Western blotting for the indicated molecules by loading equal amounts of protein. COX II, cytochrome c oxidase subunit II. Actin and Hsp60 serve as loading controls. Data are representative of three independent experiments.
Resveratrol Induces Bax Translocation to Mitochondria and Accumulation of Bim/t-Bid on Mitochondria
Because Bax and/or Bak has been shown to form channels on the OMM during apoptosis (48, 49), we investigated the subcellular localization of Bax/Bak by Western blotting using cytosolic and mitochondrial fractions. We found that during resveratrol-induced apoptosis, Bax translocated to mitochondria, whereas the level of Bak was not altered (Fig. 3B). Bax oligomerization on mitochondria is known to require the presence of activated proapoptotic proteins such as Bim and/or Bid (i.e. t-Bid) on mitochondria (48, 49). Indeed, we observed accumulation of Bim and t-Bid on mitochondria upon resveratrol treatment. Additionally, the level of antiapoptotic protein Bcl-xL was not modulated on mitochondria upon resveratrol treatment (Fig. 3B). These findings suggest that resveratrol induces Bim and t-Bid accumulation on mitochondria, contributing to the activation and oligomerization of Bax and/or Bak and leading to the OMM permeabilization.
Bax Oligomerizes on Mitochondria upon Resveratrol Treatment
Translocation of Bax to mitochondria is followed by its oligomerization on the mitochondrial membrane (48, 49). To investigate whether Bax undergoes oligomerization to form channels on mitochondria, freshly prepared mitochondria from untreated or resveratrol-treated MDA-MB231 cells were incubated with BMH, a noncleavable, membrane-permeable homobifunctional maleimide that covalently and irreversibly cross-links sulfhydryl groups. We observed Bax oligomers on mitochondria (Fig. 4A). Similarly, BMH cross-linking of mitochondria isolated from HCT116 WT cells treated with resveratrol also demonstrated Bax oligomerization, whereas Bax oligomers were not detected in HCT116 Bax−/− cells (Fig. 4B).
Bax has been shown to be expressed as multiple isoforms with molecular masses ranging from 19 to 24 kDa (50–52). Additionally, Bax is cleaved during apoptosis to generate p18, which has also been shown to oligomerize with full-length Bax (53). We have also detected multiple bands using Bax antibody by Western blotting upon 24 h of resveratrol treatment (Fig. 4, A and B). These findings support that multiple Bax oligomers could be detected by Western analysis, which is consistent with previous findings (54, 55).
To further demonstrate that resveratrol induces Bax oligomerization to execute apoptosis, resveratrol-treated or unstimulated MDA-MB231 and HCT116 cells were directly cross-linked with BMH, and samples were analyzed by Western blotting. As shown in Fig. 4, C and D, the expected Bax oligomers (e.g. dimers and multimers) were detected in resveratrol-treated cells. To further confirm that resveratrol induces Bax oligomerization, we cross-linked unstimulated or resveratrol-treated MDA-MB231 and HCT116 cells using another cross-linker, ethylene glycol bis(succinimidylsuccinate). We observed Bax oligomerization mostly as multimers upon resveratrol treatment (Fig. 4, C and D). Altogether, our results demonstrate that resveratrol induces Bax oligomerization to permeabilize the OMM.
Bax oligomerization during resveratrol-induced apoptosis was further demonstrated by gel filtration analysis using MDA-MB231 cells. Bax was mostly eluted as monomers in mitochondria obtained from untreated cells (Fig. 5A, fractions 22–25). Resveratrol treatment resulted in the elution of Bax (Fig. 5A, fractions 7–11, indicated by arrows) in higher molecular mass protein complexes (∼400–700 kDa) in mitochondria.
FIGURE 5.
Bax undergoes oligomerization on mitochondria; Bax translocation and activation of Bax/Bak do not involve p53. A and B, mitochondrial (A) and cytosolic (B) fractions isolated from MDA-MB231 cells treated with resveratrol (120 μm for 36 h) or unstimulated were fractionated on a Superdex 200 column. Fractions (0.5 ml) were collected, and a portion (20 μl) of fractions 6–28 was analyzed by Western blotting for Bax, Bak, and p53. Arrows in A indicate Bax oligomers. C, cytosol isolated from MDA-MB231 treated with resveratrol (120 μm for 36 h) was subjected to immunoprecipitation with p53 or Bax followed by Western blotting for the indicated proteins. D and E, MDA-MB231 (D) and MDA-MB435 (E) cells were pretreated with PFT-α (30 μm) followed by resveratrol (50 μm) treatment. Cells were cross-linked and subjected to Western blotting to detect Bax oligomerization. Resv, resveratrol. Data are representative of at least three independent experiments.
To determine whether Bak also undergoes oligomerization, mitochondrial lysates were subjected to gel filtration analysis. Bak was eluted (Fig. 5A, fractions 12–17) as higher molecular mass protein complexes (∼158–440 kDa) in both untreated and resveratrol-treated MDA-MB231 cells. This suggests that Bak is already a part of larger protein complexes, which is consistent with our previous findings (32).
Bax Forms Homo-oligomers, and Bak Continues to Associate with Bcl-xL upon Resveratrol Treatment
To understand whether Bax hetero-oligomerizes with Bak to form a Bax-Bak channel, co-immunoprecipitation (co-IP) of Bax was performed in freshly isolated mitochondria of MDA-MB231 cells treated with resveratrol (120 μm for 36 h). We observed very low levels of Bak by Western blotting, suggesting that Bax primarily undergoes homo-oligomerization to form Bax channels (Fig. 4E, lane 5). Reciprocal IP with Bak in mitochondria of resveratrol-treated MDA-MB231 cells also did not significantly precipitate Bax (Fig. 4E, lane 6). Interestingly, Bcl-xL was detected upon co-IP using Bak but not Bax, suggesting that proapoptotic functions of Bak may be inhibited by Bcl-xL (Fig. 4E, lanes 5 and 6). As expected, both Bax and Bak did not interact with each other under unstimulated conditions (Fig. 4E, lanes 1 and 2). Co-IP experiments indicated Bak sequestration by Bcl-xL in unstimulated cells (Fig. 4E, lane 2). These findings suggest that resveratrol induces the formation of Bax channels; however, a very small amount of Bax and Bak may also hetero-oligomerize to form a Bax-Bak channel on mitochondria.
Because Bak continues to associate with Bcl-xL during resveratrol-induced apoptosis on mitochondria, we examined whether Bax oligomerization is required for cytochrome c release from mitochondria. To accomplish this, we treated MDA-MB231 and MDA-MB435 cells with a lower dose (50 μm) of resveratrol. We observed that at 18-h treatment a low level of cytochrome c release in MDA-MB231 cells was accompanied by slight Bax oligomerization (Fig. 4F). In MDA-MB435 cells, Bax oligomerization was observed with concomitant release of cytochrome c upon treatment with a lower dose (50 μm) of resveratrol (Fig. 4F).
Bax Translocation to and Its Oligomerization on Mitochondria Do Not Involve p53
Having established that Bax translocates to mitochondria, we next asked how this is accomplished. Does Bax translocation to mitochondria require association with some cytosolic or mitochondrial proteins such as p53 (56) in addition to Bim or Bid? To investigate this possibility, we examined Bax co-elution with p53 by gel filtration analysis. As shown in Fig. 5B, p53 (fractions 17–21) and Bax (fractions 22–26) did not co-elute in the fractionated cytosols obtained from unstimulated or resveratrol-treated MDA-MB231 cells. To further determine the interaction of p53 with Bax, we performed co-IP with p53 using cytosol obtained from MDA-MB231 cells treated with resveratrol and observed that Bax was not precipitated with p53 co-IP (Fig. 5C, lane 3). We performed reciprocal IP using Bax antibody, which detects only activated Bax, and as expected, a very little amount of Bax was precipitated without significant pulldown of p53 despite abundant amounts of p53 in the cytosol (Fig. 5C, lane 2).
To understand whether mitochondrially localized Bax exists in p53-containing complexes, we performed gel filtration analysis using mitochondrial lysates obtained from untreated or resveratrol-treated MDA-MB231 cells. We observed that Bax was oligomerized and eluted in higher molecular protein complexes (Fig. 5A, fractions 7–11, indicated by arrows), which did not co-elute with p53 (Fig. 5A, fractions 11–16). These findings were further supported by co-IP experiments utilizing mitochondria isolated from untreated or resveratrol-treated MDA-MB231 cells. As shown in Fig. 4E (lanes 3 and 7), we did not observe Bax interaction with p53. These findings suggest that Bax and p53 interaction may not be essential for Bax recruitment and oligomerization on mitochondria in response to resveratrol treatment.
To further demonstrate that Bax oligomerization is not modulated by p53, we treated MDA-MB231 cells with pifithrin (PFT), a pharmacological inhibitor of p53, followed by cross-linking. Cyclic PFT-α inhibits transactivation of p53 (57), whereas PFT-μ inhibits translocation of p53 to mitochondria (58). We observed that resveratrol-induced Bax oligomerization was not modulated by pretreatment of MDA-MB231 cells with PFT-α (Fig. 5D). Similarly, resveratrol-induced Bax oligomerization was not altered by pretreatment of MDA-MB435 cells with PFT-α or PFT-μ (Fig. 5E and data not shown).
Bak Does Not Interact with p53 on Mitochondria during Resveratrol-induced Apoptosis
To investigate whether an association between p53 and Bak plays a role in channel formation or whether p53 activates Bak (59), gel filtration analysis using mitochondrial lysates was performed. We observed that Bak and p53 were co-eluted upon fractionation of mitochondria obtained from untreated and resveratrol-treated MDA-MB231 cells (Fig. 5A, fractions 13–17). To examine whether p53 interacts with Bak to form p53-mediated Bak channels on mitochondria, we performed a co-IP experiment using either Bak or p53. We observed that p53 did not interact with Bak, suggesting that p53 may not play a critical role in Bak oligomerization and channel formation to induce cytochrome c release (Fig. 4E, lanes 2, 3, 6, and 7). Similarly, gel filtration and IP analyses demonstrated that p53 did not interact with Bak during resveratrol-induced apoptosis in HCT116 and LNCaP cells (data not shown). Altogether, our findings demonstrate that p53 does not interact with Bak to permeabilize the mitochondrial membrane.
Resveratrol Induces Bax Oligomerization in Cytosol
BH3-only proteins such as Bid play an important role in the activation and recruitment of Bax on the OMM. How Bax translocates to mitochondria during apoptosis induction is not completely defined. However, current evidence suggests that the integration of cleaved Bid (t-Bid) on mitochondria recruits monomeric Bax, which undergoes oligomerization on the OMM to form the Bax channels (60–62). To investigate how Bax translocates to mitochondria during resveratrol-induced apoptosis, we cross-linked the cytosol obtained from untreated or resveratrol-treated MDA-MB231 cells. Surprisingly, we observed Bax oligomers in the cytosol as early as 12 h after resveratrol treatment (Fig. 6A). Gel filtration analyses demonstrated Bax elution in fractions 22–26, which represent the 14–43-kDa molecular mass range, suggesting that a small amount of Bax may exist as dimers in the unstimulated cytosols. Upon resveratrol treatment, Bax was eluted in fractions 22–25, suggesting that some amount of Bax might have been dimerized (Fig. 5B). Similar to MDA-MB231 cells, increased Bax oligomers were also observed in the cytosol of HCT116 cells treated with resveratrol (Fig. 6B). Altogether, our findings suggest that resveratrol induces Bax activation in the cytosol and Bax translocates to the mitochondrial membrane to undergo a higher order of oligomerization to form the Bax channels.
FIGURE 6.
Resveratrol induces Bax activation/oligomerization in cytosol, and XIAP interacts with Bax to regulate its activation and translocation to mitochondria. A and B, equal amounts of cytosols obtained from MDA-MB231 cells (A) treated with resveratrol (120 μm for 0, 12, 24, and 36 h) or from HCT116 cells (B) treated with resveratrol (50 μm for 0, 12, and 24 h) were cross-linked with BMH followed by Western blotting for Bax. C, purified mitochondria from resveratrol-treated (50 μm for 24 h) or untreated cells were subjected to Western blotting to detect XIAP and Bax translocation to mitochondria. D, purified cytosol and mitochondria obtained from resveratrol-treated or untreated MDA-MB231 cells were reconstituted to examine XIAP translocation to mitochondria. E and F, IP was performed using mitochondria (E) or cytosol (F) obtained from MDA-MB231 cells treated with resveratrol (50 μm for 18 h) followed by Western blotting for the indicated proteins. MDA, MDA-MB231; MB435, MDA-MB435; HCT, HCT116 WT; Resv, resveratrol; Cont, control; Cyto, cytosol; Mito, mitochondria. The asterisks in A and B indicate a nonspecific band. Data are representative of at least three independent experiments.
XIAP Regulates Bax Activation and Translocation to Mitochondria
Because Bax does not interact with p53, we asked how Bax is activated in the cytosol and how it is translocated to mitochondria. Recent findings demonstrate that XIAP translocates to mitochondria and promotes Bax-dependent permeabilization of the mitochondrial membrane (63). To determine whether XIAP translocates to mitochondria during resveratrol-induced apoptosis, we determined the levels of XIAP by Western blotting upon resveratrol treatment. As shown in Fig. 6C, XIAP translocation to mitochondria was also accompanied by a higher amount of Bax on mitochondria. To further demonstrate that XIAP may associate with Bax to promote Bax translocation to mitochondria during resveratrol-induced apoptosis, we performed reconstitution experiments using purified mitochondria and cytosol. We observed XIAP translocation to mitochondria when resveratrol-treated cytosol and mitochondria were used in the reconstitution experiments (Fig. 6D). To determine XIAP interaction with Bax, we performed co-IP experiments using Bax or XIAP in mitochondria from MDA-MB231 cells treated with resveratrol (50 μm for 24 h). Our findings clearly demonstrated that XIAP interacts with Bax as XIAP was co-precipitated with Bax IP (Fig. 6E, lane 1). However, reciprocal IP using XIAP could not precipitate Bax (Fig. 6E, lane 2), suggesting that Bax might stably associate with XIAP. This Bax-XIAP complex could not be precipitated by XIAP antibody. If XIAP interacts with Bax in mitochondria, it is possible that XIAP may associate with Bax in the cytosol to help translocate Bax to mitochondria. Indeed, we observed XIAP precipitation by Bax IP in the cytosol (Fig. 6F, lane 1) obtained from resveratrol (50 μm for 24 h)-treated MDA-MB231 cells, suggesting that XIAP associates with Bax to promote its translocation to mitochondria. Additionally, similar to a higher dose of resveratrol (i.e. in Figs. 4E and 5C), a low dose of resveratrol further demonstrated that p53 does not interact with Bax in the cytosol or in mitochondria (Fig. 6, E and F).
Bax Plays a Prominent Role in Resveratrol-induced Cytochrome c Release and Caspase Activation
Because Bak is sequestered by VDAC-2 (32, 64), leading to the inhibition of its proapoptotic function, Bax translocation and oligomerization might play a dominant role to induce cytochrome c release. Additionally, Bak continued to associate with Bcl-xL on mitochondria during resveratrol-induced apoptosis (Fig. 4E). We used MDA-MB231, HCT116, and LNCaP cells to study the molecular mechanisms of cytochrome c release from mitochondria and observed similar results with all three types of cells. Therefore, to study the importance of Bax or p53 on the cytochrome c release, we utilized HCT116 isogenic cell lines deficient for Bax or p53. We treated HCT116 WT and Bax-deficient HCT116 cells with resveratrol and observed that cytochrome c release was significantly inhibited in Bax-deficient cells (Fig. 7A, lanes 3 and 6). Because our data support that p53 does not directly associate with Bax or Bak, the proapoptotic functions of p53 should be inhibited. Thus, we evaluated whether p53 plays a role in the outer membrane permeabilization. Cytosolic and mitochondrial fractions obtained from p53-deficient cells treated with resveratrol or DMSO were subjected to Western blotting. We observed that p53 deficiency did not inhibit cytochrome c release (Fig. 7A, lane 9), whereas cytochrome c release was blocked in Bax-deficient cells, suggesting that Bax plays a critical role in resveratrol-induced cytochrome c release and thus apoptosis in a p53-independent manner.
If p53 does not regulate cytochrome c release from mitochondria in multiple types of cancer cells upon resveratrol treatment, then inhibition of p53 function using pharmacologic inhibitors should not affect the levels of resveratrol-induced apoptosis. Indeed, we observed that pretreatment of MDA-MB231, MDA-MB435, and MCF-7 cells with PFT-α or PFT-μ did not alter resveratrol-induced apoptosis (Fig. 7B).
Resveratrol Induces Caspase-9-mediated Apoptotic Cell Death
The experiments described above clearly suggest that resveratrol treatment leads to Bax-dependent cytochrome c release from mitochondria, which activates caspase-9 with subsequent activation of caspase-3 and execution of apoptosis. We first determined whether caspase-9 is required for resveratrol-induced apoptosis. To accomplish this, we silenced caspase-9 in LNCaP (expressing wild type p53) cells using shRNA (Fig. 8A). These caspase-9-silenced stable cells were then treated with resveratrol for 24 h. We observed that silencing of caspase-9 inhibited cell death in LNCaP cells (data not shown). To further validate that activation of caspase-9 is required for resveratrol-induced cell death, we treated caspase-9-silenced cells with resveratrol to measure caspase-3 (DEVDase) activity. As shown in Fig. 8B, resveratrol-induced caspase-3 activity in control LNCaP cells, whereas in caspase-9-silenced LNCaP cells caspase-3 activity was inhibited. In contrast, caspase-8-silenced LNCaP cells (Fig. 8B) showed enhanced caspase-3 activity similar to control LNCaP cells upon resveratrol treatment. Similarly, resveratrol-induced caspase activation was inhibited in caspase-9-silenced MDA-MB231 cells (expressing mutant p53) but not in caspase-8-silenced MDA-MB231 cells (Fig. 8, C and D). Altogether, these findings demonstrate that resveratrol induces the intrinsic pathway to trigger caspase-9-dependent cell death in cancer cells.
FIGURE 8.
Activation of caspase-9 is initiating event in resveratrol-induced apoptosis. A, LNCaP cells were infected with caspase-9, caspase-8, or control shRNA lentiviral particles at a multiplicity of infection of 3. Stable cells were used for Western blotting to detect caspase-9 or caspase-8 silencing. B, caspase-8- and caspase-9-silenced or control (non-targeting) cells were treated with resveratrol (120 μm for 24 h). C, caspase-8 or caspase-9 was silenced in MDA-MB231 cells. D, cells were treated with resveratrol (50 μm for 24 h). At the end of treatment, caspase-3 activity was determined. Resv, resveratrol; Casp-9, caspase-9; Casp-8, caspase-8. Actin and Hsp60 serve as loading controls. Data are mean ± S.D. (n = 3). *, p < 0.01 as compared with resveratrol-treated control cells.
DISCUSSION
A majority of epithelial cancer cells become resistant to known anticancer agents, making it necessary to apply such drugs at higher doses or to combine different anticancer drugs. These approaches are often associated with severe side effects in cancer patients. Therefore, anticancer agents that are minimally toxic to normal cells are highly sought for use in cancer therapy. Resveratrol has been shown to selectively kill cancer cells or possess anticancer properties, but how resveratrol induces apoptotic cell death is not clearly defined. Here, we show that the mitochondrion is a critical target organelle for resveratrol-induced apoptosis in epithelial cancer cells. The induction of apoptosis was initiated by caspase-9 activation. The salient features of resveratrol-induced apoptosis are as follows. Resveratrol induced mitochondrial dysfunction, leading to the loss of mitochondrial membrane potential and cytochrome c release. The release of cytochrome c from mitochondria was dependent on Bax. Translocation and oligomerization of Bax on mitochondria were not facilitated by p53. Similarly, p53 did not interact with Bak on mitochondria during resveratrol-induced apoptosis. Most importantly, resveratrol induced Bax oligomerization in the cytosol, and XIAP interacted with Bax in the cytosol and on mitochondria, suggesting that XIAP regulates Bax-mediated release of cytochrome c from mitochondria. The oligomerization of Bax on mitochondria coincided with the release of mitochondrial cytochrome c, which triggered caspase-9-dependent apoptosis. These findings suggest that resveratrol activates Bax in a p53-independent mechanism. Furthermore, Bax recruitment and a high level of oligomerization to form Bax channels on the OMM are mediated by Bim/Bid-dependent mechanisms to induce cytochrome c release, caspase activation, and apoptosis.
We have demonstrated that resveratrol induces caspase-dependent apoptosis as evidenced by the presence of catalytically active caspase-3. Caspase activity and cell death were inhibited by pretreatment of cells with a pan-caspase inhibitor (i.e. Z-VAD), further demonstrating caspase-dependent apoptosis upon resveratrol treatment. Because we did not observe a release of apoptosis-inducing factor or endonuclease G from the mitochondrial compartment,4 caspase-independent cell death does not seem to play a critical role in initiating cancer cell death upon resveratrol treatment. However, at later stages of apoptosis, the release of mitochondrial contents including apoptosis-inducing factor and endonuclease G could amplify the process of apoptotic cell death.
How is the caspase cascade initiated? We have provided the comprehensive evidence that resveratrol induces caspase-9-dependent but caspase-8-independent apoptosis. This is supported by the fact that in the absence of Apaf-1 (by utilizing Apaf-1−/− mouse embryonic fibroblasts; data not shown) or caspase-9 (silencing of caspase-9) caspase-3 activation was inhibited, whereas this was not the case in caspase-8-deficient Jurkat cells (data not shown) or caspase-8-silenced LNCaP and MDA-MB231 cells. Similarly, Western blot analysis clearly demonstrated caspase-9 activation but not caspase-8 activation in MDA-MB231 cells, suggesting that caspase-9 is the initiator caspase for caspase-dependent apoptosis in response to resveratrol in cancer cells (data not shown). Our results are consistent with previous findings that resveratrol induces mitochondrion-dependent but death receptor-independent apoptosis (65, 66). Because resveratrol primarily targets mitochondria to induce apoptosis, resveratrol could be used as a synergistic agent to enhance death receptor-mediated cancer cell death. Indeed, elegant works of other colleagues have demonstrated that resveratrol enhances TNF-related apoptosis-inducing ligand (TRAIL)-induced apoptosis in cancer cells (67–70).
Because activation of caspase-9 requires cytochrome c release from mitochondria, resveratrol should permeabilize mitochondria to induce cytochrome c release. Indeed, we observed robust cytochrome c release from mitochondria into the cytosol upon resveratrol treatment in multiple types of cells. How is cytochrome c released? The release of cytochrome c from mitochondria requires multiple mechanisms such as Bax/Bak activation, a loss of mitochondrial membrane potential, and permeability transition pores (19, 26, 49). Our data demonstrate that Bax activation and translocation to mitochondria seem to play a prominent role in cytochrome c release. Active Bax on mitochondria underwent oligomerization to form primarily Bax homo-oligomers. Although Bak existed in higher molecular protein complexes on mitochondria, Bax-deficient HCT116 cells (which still contain Bak) did not release cytochrome c from mitochondria. These data support previous observations that Bak is sequestered by VDAC-2 in the presence of Bax. Bak does not play a significant role in inducing cytochrome c release even if Bak is activated or oligomerized as activated Bak remains in a complex with VDAC-2 (32, 64), and Bak continued to associate with Bcl-xL (Fig. 4E). We did not observe VDAC-1 homo-oligomers, suggesting that resveratrol does not induce the formation of VDAC-only channels (data not shown). The permeability transition pore mechanism requires rupturing of the OMM, leading to the release of proteins localized in the intermembrane space. However, we observed that apoptosis-inducing factor and endonuclease G were not released from mitochondria, suggesting that the permeability transition pore may not be involved in resveratrol-induced cytochrome c release and apoptosis.
How is Bax translocated to mitochondria, and how is it activated? We observed a novel mechanism of Bax activation. For example, Bax is normally activated in a t-Bid-dependent manner to translocate to and oligomerize on mitochondria (22, 23, 48, 49, 60, 61). We observed the presence of full-length Bid and t-Bid on mitochondria, suggesting that resveratrol induces t-Bid accumulation on mitochondria. Another direct activator of Bax could be activation/up-regulation of Bim (48, 49), which accumulates on mitochondria to participate in the activation/oligomerization of Bax, leading to the formation of Bax-dependent channels on mitochondria. More importantly, we have demonstrated that XIAP interacts with Bax in the cytosol and on mitochondria, suggesting that XIAP regulates activation/translocation of Bax to mitochondria. XIAP generally performs prosurvival functions by inhibiting caspase activation (71); however, a recent study demonstrates that XIAP translocates to mitochondria and promotes Bax-dependent permeabilization of the mitochondrial membrane (63). Our findings further demonstrate the proapoptotic function of XIAP in resveratrol-induced apoptosis.
Is Bax only activated on the OMM? We provide the first evidence that Bax could be activated in the cytosol upon resveratrol treatment. How this could be achieved is still under investigation, but available data support that Bax could be activated by XIAP or by BH3-only proteins (Bim and Bid) in the cytosol. We observed that XIAP associated with Bax in the cytosol and on mitochondria, and translocation of XIAP to mitochondria was accompanied by increased level of Bax. These findings suggest that XIAP may regulate activation/translocation of Bax to mitochondria to promote Bax-dependent permeabilization of the mitochondrial membrane. Because only transient interaction between BH3-only proteins and Bax is required for Bax activation (72), it is quite possible that small amounts of activated Bim or t-Bid or any other BH3-only proteins in the cytosolic compartment may transiently interact with the cytosolic Bax, leading to its activation in the cytosol. Activated cytosolic Bax then undergoes oligomerization, mostly dimerization, and translocates to mitochondria to undergo a higher order of oligomerization in a t-Bid/Bim-dependent mechanism or through association with XIAP to form Bax channels. How transient interaction of Bim or Bid with Bax induces Bax oligomerization is under investigation, but dimerization of Bax in the cytosol may be explained by a recent model by Andrews and co-workers (60, 61). For example, the BH3 domain of Bax activated by Bid or Bim can interact with another activated Bax in the cytosol to form a Bax dimer, which may then be recruited by mitochondrially localized Bid or Bim. Subsequently, the recruited Bax dimer may undergo a higher order of oligomerization to form Bax channels on the OMM in a t-Bid-dependent manner. Because intracellular glutathione depletion promotes Bax translocation to mitochondria (73, 74), it is possible that glutathione might interfere with Bax activation and oligomerization in the cytosol and its subsequent translocation to mitochondria. During resveratrol-induced apoptosis, depletion of glutathione may facilitate Bax oligomerization in the cytosol, leading to Bax channel formation on the mitochondrial membrane.
Bax could also be activated by cytosolic p53 (56), but gel filtration and immunoprecipitation analysis demonstrated that Bax does not associate with p53 upon resveratrol treatment, suggesting that p53 is not critical for resveratrol-induced outer membrane permeabilization. These findings are consistent with the earlier report that resveratrol induces p53-independent apoptosis in cancer cells (65, 67, 73–76). Although Bak can be activated by p53 (59), we did not observe p53 involvement in Bak activation. It has also been reported that p53 could also suppress the antiapoptotic function of Bcl-2/Bcl-xL/Mcl-1 (77–79) and thus may cause an increase in cytochrome c release and apoptosis. Because the level of Bcl-2 is very low in MDA-MB231 cells, we tested whether levels of Bcl-xL were modified during resveratrol-induced cell death. We observed that the level of Bcl-xL was not modified during resveratrol-induced apoptosis (Fig. 3B). Additionally, we did not observe Bcl-xL interaction with p53 on mitochondria, further supporting that p53 does not play a critical role in cytochrome c release upon resveratrol treatment. We are further investigating whether resveratrol modifies the antiapoptotic function of Bcl-2/Bcl-xL/Mcl-1 through p53, but available evidence indicates that resveratrol induces caspase activation and apoptosis both in LNCaP (expressing wild type p53) and MDA-MB231 (expressing mutant p53) cells. Bax-deficient cells showed significantly reduced levels of cytochrome c release, whereas p53-deficient cells showed equal or even slightly higher levels of cytochrome c release upon resveratrol treatment (Fig. 7A). These findings support that p53 does not interact with Bax/Bak and that p53-Bax or p53-Bak interaction may not be critical for resveratrol-induced cytochrome c release in cancer cells. It has been reported that wild type p53 as well as mutant p53 can promote Bax activation on mitochondria (80, 81), whereas other studies suggest that p53 translocation to mitochondria does not induce Bax activation and apoptosis (82, 83). Expression of mutant p53 in cancer cells confers selective advantage and resistance to apoptosis (84–86). Our findings suggest that resveratrol induces cytochrome c release, caspase activation, and apoptosis in cancer cells such as in MDA-MB231 and MDA-MB435 cells that harbor mutant p53.
Altogether, our findings provide comprehensive evidence that resveratrol induces Bax-dependent but p53-independent apoptosis in epithelial cancer cells, and further analysis on how resveratrol induces Bax activation may lead to the foundation of resveratrol-based anticancer agents. It is generally believed that for chemoprevention or cancer therapy doses of resveratrol used to induce apoptotic cell death may not easily be achieved under physiological conditions through regular diets. However, various lines of evidence indicate that in vitro doses that have been shown to induce apoptotic cell death could be achieved in cancer cells through higher intake of resveratrol (87–91). For example, resveratrol at daily doses of up to 5 g for 29 days is not toxic to humans, and daily doses of 0.5 or 1 g produce levels of resveratrol in tumor cells that are sufficient to elicit anticancer effects such as induction of apoptosis in cancer cells (88). Additionally, further research on resveratrol modifications may increase the bioavailability of resveratrol in cancer tissues. Because we have demonstrated that resveratrol induces Bax activation in the cytosol and that XIAP interacts with Bax, further analysis on how to enhance Bax oligomerization on mitochondria in a p53-independent manner may provide a novel approach to enhance apoptotic cell death with lower doses of resveratrol.
Acknowledgments
We thank Drs. B. Vogelstein and Terry Beerman for providing reagents. We are thankful to Drs. Jennifer Black, Dean Tang, and Terry Beerman for critical reading of the manuscript. V. P. was a Master student at the University at Buffalo. We also thank Dr. Adrian Black for help with analysis of micrographs. We apologize to those colleagues whose publications could not be cited due to space constraints.
This work was supported, in whole or in part, by National Institutes of Health K01 Award CA123142 (to D. C.) and NCI Center Support Grant CA016056 (to the Roswell Park Cancer Institute).
V. Prabhu, R. Gogada, and D. Chandra, unpublished data.
- OMM
- outer mitochondrial membrane
- XIAP
- X-linked inhibitor of apoptosis protein
- Rb
- rabbit
- pAb
- polyclonal antibody
- AFC
- 7-amino-4-trifluoromethylcoumarin
- Z
- benzyloxycarbonyl
- BMH
- bismaleimidohexane
- IP
- immunoprecipitation
- PFT
- pifithrin
- VDAC
- voltage-dependent anion channel
- Hsp
- heat shock protein.
REFERENCES
- 1. Mathew R., White E. (2011) Curr. Opin. Genet. Dev. 21, 113–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zois C. E., Koukourakis M. I. (2009) Autophagy 5, 442–450 [DOI] [PubMed] [Google Scholar]
- 3. Blagosklonny M. V. (2002) Int. J. Cancer 98, 161–166 [DOI] [PubMed] [Google Scholar]
- 4. Lowe S. W., Lin A. W. (2000) Carcinogenesis 21, 485–495 [DOI] [PubMed] [Google Scholar]
- 5. Delmas D., Rébé C., Lacour S., Filomenko R., Athias A., Gambert P., Cherkaoui-Malki M., Jannin B., Dubrez-Daloz L., Latruffe N., Solary E. (2003) J. Biol. Chem. 278, 41482–41490 [DOI] [PubMed] [Google Scholar]
- 6. Fulda S., Debatin K. M. (2004) Cancer Res. 64, 337–346 [DOI] [PubMed] [Google Scholar]
- 7. Gill C., Walsh S. E., Morrissey C., Fitzpatrick J. M., Watson R. W. (2007) Prostate 67, 1641–1653 [DOI] [PubMed] [Google Scholar]
- 8. Niles R. M., McFarland M., Weimer M. B., Redkar A., Fu Y. M., Meadows G. G. (2003) Cancer Lett. 190, 157–163 [DOI] [PubMed] [Google Scholar]
- 9. Pervaiz S. (2003) FASEB J. 17, 1975–1985 [DOI] [PubMed] [Google Scholar]
- 10. She Q. B., Bode A. M., Ma W. Y., Chen N. Y., Dong Z. (2001) Cancer Res. 61, 1604–1610 [PubMed] [Google Scholar]
- 11. Estrov Z., Shishodia S., Faderl S., Harris D., Van Q., Kantarjian H. M., Talpaz M., Aggarwal B. B. (2003) Blood 102, 987–995 [DOI] [PubMed] [Google Scholar]
- 12. Joe A. K., Liu H., Suzui M., Vural M. E., Xiao D., Weinstein I. B. (2002) Clin. Cancer Res. 8, 893–903 [PubMed] [Google Scholar]
- 13. Hsieh T. C., Juan G., Darzynkiewicz Z., Wu J. M. (1999) Cancer Res. 59, 2596–2601 [PubMed] [Google Scholar]
- 14. Mohan J., Gandhi A. A., Bhavya B. C., Rashmi R., Karunagaran D., Indu R., Santhoshkumar T. R. (2006) J. Biol. Chem. 281, 17599–17611 [DOI] [PubMed] [Google Scholar]
- 15. Sareen D., Darjatmoko S. R., Albert D. M., Polans A. S. (2007) Mol. Pharmacol. 72, 1466–1475 [DOI] [PubMed] [Google Scholar]
- 16. Tinhofer I., Bernhard D., Senfter M., Anether G., Loeffler M., Kroemer G., Kofler R., Csordas A., Greil R. (2001) FASEB J. 15, 1613–1615 [DOI] [PubMed] [Google Scholar]
- 17. van Ginkel P. R., Sareen D., Subramanian L., Walker Q., Darjatmoko S. R., Lindstrom M. J., Kulkarni A., Albert D. M., Polans A. S. (2007) Clin. Cancer Res. 13, 5162–5169 [DOI] [PubMed] [Google Scholar]
- 18. Huang T. T., Lin H. C., Chen C. C., Lu C. C., Wei C. F., Wu T. S., Liu F. G., Lai H. C. (2011) J. Cell. Physiol. 226, 720–728 [DOI] [PubMed] [Google Scholar]
- 19. Boatright K. M., Renatus M., Scott F. L., Sperandio S., Shin H., Pedersen I. M., Ricci J. E., Edris W. A., Sutherlin D. P., Green D. R., Salvesen G. S. (2003) Mol. Cell 11, 529–541 [DOI] [PubMed] [Google Scholar]
- 20. Jiang X., Wang X. (2004) Annu. Rev. Biochem. 73, 87–106 [DOI] [PubMed] [Google Scholar]
- 21. Salvesen G. S., Dixit V. M. (1997) Cell 91, 443–446 [DOI] [PubMed] [Google Scholar]
- 22. Shi Y. (2004) Cell 117, 855–858 [DOI] [PubMed] [Google Scholar]
- 23. Ashkenazi A., Dixit V. M. (1998) Science 281, 1305–1308 [DOI] [PubMed] [Google Scholar]
- 24. Carrington P. E., Sandu C., Wei Y., Hill J. M., Morisawa G., Huang T., Gavathiotis E., Wei Y., Werner M. H. (2006) Mol. Cell 22, 599–610 [DOI] [PubMed] [Google Scholar]
- 25. Scaffidi C., Fulda S., Srinivasan A., Friesen C., Li F., Tomaselli K. J., Debatin K. M., Krammer P. H., Peter M. E. (1998) EMBO J. 17, 1675–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Garrido C., Galluzzi L., Brunet M., Puig P. E., Didelot C., Kroemer G. (2006) Cell Death Differ. 13, 1423–1433 [DOI] [PubMed] [Google Scholar]
- 27. Kuwana T., Mackey M. R., Perkins G., Ellisman M. H., Latterich M., Schneiter R., Green D. R., Newmeyer D. D. (2002) Cell 111, 331–342 [DOI] [PubMed] [Google Scholar]
- 28. Aziz M. H., Nihal M., Fu V. X., Jarrard D. F., Ahmad N. (2006) Mol. Cancer Ther. 5, 1335–1341 [DOI] [PubMed] [Google Scholar]
- 29. Zhang L., Yu J., Park B. H., Kinzler K. W., Vogelstein B. (2000) Science 290, 989–992 [DOI] [PubMed] [Google Scholar]
- 30. Bunz F., Dutriaux A., Lengauer C., Waldman T., Zhou S., Brown J. P., Sedivy J. M., Kinzler K. W., Vogelstein B. (1998) Science 282, 1497–1501 [DOI] [PubMed] [Google Scholar]
- 31. Chandra D., Bratton S. B., Person M. D., Tian Y., Martin A. G., Ayres M., Fearnhead H. O., Gandhi V., Tang D. G. (2006) Cell 125, 1333–1346 [DOI] [PubMed] [Google Scholar]
- 32. Chandra D., Choy G., Daniel P. T., Tang D. G. (2005) J. Biol. Chem. 280, 19051–19061 [DOI] [PubMed] [Google Scholar]
- 33. Chandra D., Choy G., Deng X., Bhatia B., Daniel P., Tang D. G. (2004) Mol. Cell. Biol. 24, 6592–6607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chandra D., Choy G., Tang D. G. (2007) J. Biol. Chem. 282, 31289–31301 [DOI] [PubMed] [Google Scholar]
- 35. Chandra D., Liu J. W., Tang D. G. (2002) J. Biol. Chem. 277, 50842–50854 [DOI] [PubMed] [Google Scholar]
- 36. Chandra D., Tang D. G. (2003) J. Biol. Chem. 278, 17408–17420 [DOI] [PubMed] [Google Scholar]
- 37. Jeter C. R., Badeaux M., Choy G., Chandra D., Patrawala L., Liu C., Calhoun-Davis T., Zaehres H., Daley G. Q., Tang D. G. (2009) Stem Cells 27, 993–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Malladi S., Challa-Malladi M., Fearnhead H. O., Bratton S. B. (2009) EMBO J. 28, 1916–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fernandes-Alnemri T., Armstrong R. C., Krebs J., Srinivasula S. M., Wang L., Bullrich F., Fritz L. C., Trapani J. A., Tomaselli K. J., Litwack G., Alnemri E. S. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 7464–7469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nicholson D. W., Ali A., Thornberry N. A., Vaillancourt J. P., Ding C. K., Gallant M., Gareau Y., Griffin P. R., Labelle M., Lazebnik Y. A., et al. (1995) Nature 376, 37–43 [DOI] [PubMed] [Google Scholar]
- 41. Heerdt B. G., Houston M. A., Augenlicht L. H. (1997) Cell Growth Differ. 8, 523–532 [PubMed] [Google Scholar]
- 42. Joshi B., Li L., Taffe B. G., Zhu Z., Wahl S., Tian H., Ben-Josef E., Taylor J. D., Porter A. T., Tang D. G. (1999) Cancer Res. 59, 4343–4355 [PubMed] [Google Scholar]
- 43. Sánchez-Alcázar J. A., Ault J. G., Khodjakov A., Schneider E. (2000) Cell Death Differ. 7, 1090–1100 [DOI] [PubMed] [Google Scholar]
- 44. Sánchez-Alcázar J. A., Khodjakov A., Schneider E. (2001) Cancer Res. 61, 1038–1044 [PubMed] [Google Scholar]
- 45. Poyton R. O., McEwen J. E. (1996) Annu. Rev. Biochem. 65, 563–607 [DOI] [PubMed] [Google Scholar]
- 46. Stuart R. A., Neupert W. (1990) Biochimie 72, 115–121 [DOI] [PubMed] [Google Scholar]
- 47. Stuart R. A., Nicholson D. W., Neupert W. (1990) Cell 60, 31–43 [DOI] [PubMed] [Google Scholar]
- 48. Danial N. N. (2007) Clin. Cancer Res. 13, 7254–7263 [DOI] [PubMed] [Google Scholar]
- 49. Brunelle J. K., Letai A. (2009) J. Cell Sci. 122, 437–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Oltvai Z. N., Milliman C. L., Korsmeyer S. J. (1993) Cell 74, 609–619 [DOI] [PubMed] [Google Scholar]
- 51. Cartron P. F., Oliver L., Martin S., Moreau C., LeCabellec M. T., Jezequel P., Meflah K., Vallette F. M. (2002) Hum. Mol. Genet. 11, 675–687 [DOI] [PubMed] [Google Scholar]
- 52. Antonsson B., Montessuit S., Sanchez B., Martinou J. C. (2001) J. Biol. Chem. 276, 11615–11623 [DOI] [PubMed] [Google Scholar]
- 53. Cao X., Deng X., May W. S. (2003) Blood 102, 2605–2614 [DOI] [PubMed] [Google Scholar]
- 54. Zhang H., Kim J. K., Edwards C. A., Xu Z., Taichman R., Wang C. Y. (2005) Nat. Cell Biol. 7, 909–915 [DOI] [PubMed] [Google Scholar]
- 55. Wolff S., Erster S., Palacios G., Moll U. M. (2008) Cell Res. 18, 733–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Chipuk J. E., Kuwana T., Bouchier-Hayes L., Droin N. M., Newmeyer D. D., Schuler M., Green D. R. (2004) Science 303, 1010–1014 [DOI] [PubMed] [Google Scholar]
- 57. Komarov P. G., Komarova E. A., Kondratov R. V., Christov-Tselkov K., Coon J. S., Chernov M. V., Gudkov A. V. (1999) Science 285, 1733–1737 [DOI] [PubMed] [Google Scholar]
- 58. Strom E., Sathe S., Komarov P. G., Chernova O. B., Pavlovska I., Shyshynova I., Bosykh D. A., Burdelya L. G., Macklis R. M., Skaliter R., Komarova E. A., Gudkov A. V. (2006) Nat. Chem. Biol. 2, 474–479 [DOI] [PubMed] [Google Scholar]
- 59. Pietsch E. C., Perchiniak E., Canutescu A. A., Wang G., Dunbrack R. L., Murphy M. E. (2008) J. Biol. Chem. 283, 21294–21304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang Z., Zhu W., Lapolla S. M., Miao Y., Shao Y., Falcone M., Boreham D., McFarlane N., Ding J., Johnson A. E., Zhang X. C., Andrews D. W., Lin J. (2010) J. Biol. Chem. 285, 17614–17627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Bogner C., Leber B., Andrews D. W. (2010) Curr. Opin. Cell Biol. 22, 845–851 [DOI] [PubMed] [Google Scholar]
- 62. Kim H., Tu H. C., Ren D., Takeuchi O., Jeffers J. R., Zambetti G. P., Hsieh J. J., Cheng E. H. (2009) Mol. Cell 36, 487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Owens T. W., Foster F. M., Valentijn A., Gilmore A. P., Streuli C. H. (2010) J. Biol. Chem. 285, 1081–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Cheng E. H., Sheiko T. V., Fisher J. K., Craigen W. J., Korsmeyer S. J. (2003) Science 301, 513–517 [DOI] [PubMed] [Google Scholar]
- 65. Mahyar-Roemer M., Katsen A., Mestres P., Roemer K. (2001) Int. J. Cancer 94, 615–622 [DOI] [PubMed] [Google Scholar]
- 66. Dörrie J., Gerauer H., Wachter Y., Zunino S. J. (2001) Cancer Res. 61, 4731–4739 [PubMed] [Google Scholar]
- 67. Pavet V., Beyrath J., Pardin C., Morizot A., Lechner M. C., Briand J. P., Wendland M., Maison W., Fournel S., Micheau O., Guichard G., Gronemeyer H. (2010) Cancer Res. 70, 1101–1110 [DOI] [PubMed] [Google Scholar]
- 68. Mader I., Wabitsch M., Debatin K. M., Fischer-Posovszky P., Fulda S. (2010) FASEB J. 24, 1997–2009 [DOI] [PubMed] [Google Scholar]
- 69. Rushworth S. A., Micheau O. (2009) Br. J. Pharmacol. 157, 1186–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ganapathy S., Chen Q., Singh K. P., Shankar S., Srivastava R. K. (2010) PLoS One 5, e15627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Shi Y. (2002) Mol. Cell 9, 459–470 [DOI] [PubMed] [Google Scholar]
- 72. Perez D., White E. (2000) Mol. Cell 6, 53–63 [PubMed] [Google Scholar]
- 73. Guha P., Dey A., Sen R., Chatterjee M., Chattopadhyay S., Bandyopadhyay S. K. (2011) J. Pharmacol. Exp. Ther. 336, 206–214 [DOI] [PubMed] [Google Scholar]
- 74. Honda T., Coppola S., Ghibelli L., Cho S. H., Kagawa S., Spurgers K. B., Brisbay S. M., Roth J. A., Meyn R. E., Fang B., McDonnell T. J. (2004) Cancer Gene. Ther. 11, 249–255 [DOI] [PubMed] [Google Scholar]
- 75. Chow S. E., Wang J. S., Chuang S. F., Chang Y. L., Chu W. K., Chen W. S., Chen Y. W. (2010) Cancer Gene. Ther. 17, 872–882 [DOI] [PubMed] [Google Scholar]
- 76. Kim M. Y., Trudel L. J., Wogan G. N. (2009) Anticancer Res. 29, 3733–3740 [PubMed] [Google Scholar]
- 77. Mihara M., Erster S., Zaika A., Petrenko O., Chittenden T., Pancoska P., Moll U. M. (2003) Mol. Cell 11, 577–590 [DOI] [PubMed] [Google Scholar]
- 78. Chipuk J. E., Bouchier-Hayes L., Kuwana T., Newmeyer D. D., Green D. R. (2005) Science 309, 1732–1735 [DOI] [PubMed] [Google Scholar]
- 79. Leu J. I., Dumont P., Hafey M., Murphy M. E., George D. L. (2004) Nat. Cell Biol. 6, 443–450 [DOI] [PubMed] [Google Scholar]
- 80. Yamaguchi H., Woods N. T., Piluso L. G., Lee H. H., Chen J., Bhalla K. N., Monteiro A., Liu X., Hung M. C., Wang H. G. (2009) J. Biol. Chem. 284, 11171–11183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Yamaguchi H., Chen J., Bhalla K., Wang H. G. (2004) J. Biol. Chem. 279, 39431–39437 [DOI] [PubMed] [Google Scholar]
- 82. Mahyar-Roemer M., Fritzsche C., Wagner S., Laue M., Roemer K. (2004) Oncogene 23, 6226–6236 [DOI] [PubMed] [Google Scholar]
- 83. Essmann F., Pohlmann S., Gillissen B., Daniel P. T., Schulze-Osthoff K., Jänicke R. U. (2005) J. Biol. Chem. 280, 37169–37177 [DOI] [PubMed] [Google Scholar]
- 84. Di X., Gennings C., Bear H. D., Graham L. J., Sheth C. M., White K. L., Jr., Gewirtz D. A. (2010) Breast Cancer Res. Treat. 124, 349–360 [DOI] [PubMed] [Google Scholar]
- 85. Forrester K., Lupold S. E., Ott V. L., Chay C. H., Band V., Wang X. W., Harris C. C. (1995) Oncogene 10, 2103–2111 [PubMed] [Google Scholar]
- 86. Oren M., Rotter V. (2010) Cold Spring Harb. Perspect. Biol. 2, a001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Bishayee A. (2009) Cancer Prev. Res. 2, 409–418 [DOI] [PubMed] [Google Scholar]
- 88. Patel K. R., Brown V. A., Jones D. J., Britton R. G., Hemingway D., Miller A. S., West K. P., Booth T. D., Perloff M., Crowell J. A., Brenner D. E., Steward W. P., Gescher A. J., Brown K. (2010) Cancer Res. 70, 7392–7399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Chow H. H., Garland L. L., Hsu C. H., Vining D. R., Chew W. M., Miller J. A., Perloff M., Crowell J. A., Alberts D. S. (2010) Cancer Prev. Res. 3, 1168–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Baur J. A., Sinclair D. A. (2006) Nat. Rev. Drug Discov. 5, 493–506 [DOI] [PubMed] [Google Scholar]
- 91. Boocock D. J., Faust G. E., Patel K. R., Schinas A. M., Brown V. A., Ducharme M. P., Booth T. D., Crowell J. A., Perloff M., Gescher A. J., Steward W. P., Brenner D. E. (2007) Cancer Epidemiol. Biomarkers Prev. 16, 1246–1252 [DOI] [PubMed] [Google Scholar]