Abstract
The methionine chain-elongation pathway is required for aliphatic glucosinolate biosynthesis in plants and evolved from leucine biosynthesis. In Arabidopsis thaliana, three 3-isopropylmalate dehydrogenases (AtIPMDHs) play key roles in methionine chain-elongation for the synthesis of aliphatic glucosinolates (e.g. AtIPMDH1) and leucine (e.g. AtIPMDH2 and AtIPMDH3). Here we elucidate the molecular basis underlying the metabolic specialization of these enzymes. The 2.25 Å resolution crystal structure of AtIPMDH2 was solved to provide the first detailed molecular architecture of a plant IPMDH. Modeling of 3-isopropylmalate binding in the AtIPMDH2 active site and sequence comparisons of prokaryotic and eukaryotic IPMDH suggest that substitution of one active site residue may lead to altered substrate specificity and metabolic function. Site-directed mutagenesis of Phe-137 to a leucine in AtIPMDH1 (AtIPMDH1-F137L) reduced activity toward 3-(2′-methylthio)ethylmalate by 200-fold, but enhanced catalytic efficiency with 3-isopropylmalate to levels observed with AtIPMDH2 and AtIPMDH3. Conversely, the AtIPMDH2-L134F and AtIPMDH3-L133F mutants enhanced catalytic efficiency with 3-(2′-methylthio)ethylmalate ∼100-fold and reduced activity for 3-isopropylmalate. Furthermore, the altered in vivo glucosinolate profile of an Arabidopsis ipmdh1 T-DNA knock-out mutant could be restored to wild-type levels by constructs expressing AtIPMDH1, AtIPMDH2-L134F, or AtIPMDH3-L133F, but not by AtIPMDH1-F137L. These results indicate that a single amino acid substitution results in functional divergence of IPMDH in planta to affect substrate specificity and contributes to the evolution of specialized glucosinolate biosynthesis from the ancestral leucine pathway.
Keywords: Amino Acid, Arabidopsis, Crystal Structure, Dehydrogenase, Enzyme Catalysis, Metabolism
Introduction
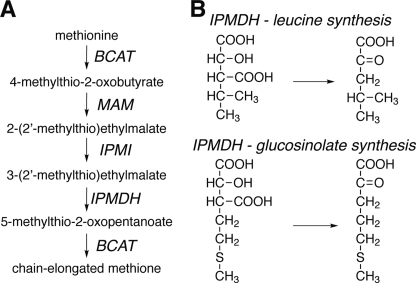
To compensate for their sessile nature, plants evolved mechanisms to cope with rapid environmental changes and challenges (1). The production of specialized metabolites is one of the important mechanisms for the survival and fitness of plants (2). The molecular diversity of these specialized compounds arises from differential modification of common backbone structures, which necessitates the evolution of homologous enzymes with varied specificities (1). In plants, glucosinolates constitute a diverse group of sulfur-containing specialized metabolites (3–4). Biosynthesis of methionine-derived glucosinolates is initiated by the sequential addition of methylene groups to produce chain-elongated methionine derivatives via an iterative three-step chain-elongation process that mimics the chemistry of leucine synthesis (Fig. 1A).
FIGURE 1.
IPMDH pathways. A, overview of the methionine chain-elongation pathway of aliphatic glucosinolate biosynthesis in A. thaliana. Note that an elongated 2-oxo acid can serve as a substrate for MAM1 and MAM3 in subsequent rounds through the pathway to yield longer side-chain products. B, IPMDH catalyze the conversion of 3-isopropylmalate to 4-methyl-2-oxovalerate in leucine synthesis and the conversion of 3-(2′-methylthio)ethylmalate to 5-methylthio-2-oxopentoate in glucosinolate synthesis.
To date, all the genes involved in the methionine chain-elongation process have been identified and characterized in Arabidopsis thaliana (5–14). The different enzymes of the methionine chain-elongation pathway for glucosinolate synthesis have evolved from leucine synthesis by gene duplication and functional specification (14–15). For example, four genes in Arabidopsis encode isopropylmalate synthases (IPMS)4 with two (IPMS1 and IPMS2) serving in leucine biosynthesis and the other two genes encoding methylthioalkylmalate (MAM) synthases (MAM1 and MAM3), which catalyze the committed step in methionine chain-elongation (5–6, 16). A recent study showed that loss of a C-terminal regulatory domain and a few amino acid exchanges can covert IPMS into MAM (14). Specialization of the Arabidopsis isopropylmalate isomerases (IPMI) for different catalytic properties occurs by changes in the oligomeric composition of these enzymes. IPMI are heterodimeric enzymes consisting of a large subunit encoded by a single gene and a small subunit encoded by one of three genes (8–9, 12). Metabolic profiling of the large subunit mutant revealed accumulation of intermediates in both the leucine pathway and the methionine chain-elongation pathway, demonstrating the dual function of this subunit in both leucine and glucosinolate biosynthesis (10). In contrast, the small subunits are specialized to either leucine biosynthesis or methionine chain-elongation (2, 10, 12). Furthermore, among the six branched-chain aminotransferases (BCATs) in Arabidopsis, BCAT4 in the cytosol is specifically involved in glucosinolate biosynthesis, whereas BCAT3 in the plastids functions in both amino acid and glucosinolate biosynthesis (7, 9). The molecular changes that tailor BCAT activity are unclear.
Previously, we showed that A. thaliana isopropylmalate dehydrogenase 1 (AtIPMDH1) catalyzes the oxidative decarboxylation step in the methionine chain-elongation of glucosinolate biosynthesis and that AtIPMDH2 and AtIPMDH3 are primarily involved in leucine biosynthesis (Fig. 1B) (11, 13). These studies highlight the functional specialization of these isoforms, but do not reveal how these activities evolved.
Here we examine the molecular basis for the functional evolution of the IPMDH family in Arabidopsis. The crystal structure of AtIPMDH2, the first determined for a plant IPMDH, reveals an active site structure similar to that of the bacterial enzymes and provides a template for modeling substrate binding in the active site. Analysis of the AtIPMDH2 structure, sequence comparisons, and site-directed mutagenesis demonstrates that a single residue difference in the active site drastically alters substrate specificity of the AtIPMDH isoforms both in vitro and in vivo. This work demonstrates the basis for functional divergence of an AtIPMDH isoform for glucosinolate biosynthesis from those of leucine biosynthesis.
EXPERIMENTAL PROCEDURES
Plants and Growth
Seeds of A. thaliana ecotype Columbia (Col-0) and SALK mutant atipmdh1 (Salk_063423C) were obtained from the Arabidopsis Biological Resource Center (ABRC). Seed germination and plant growth conditions were as previously described (11, 13).
Plasmid Construction and Plant Transformation
The full-length coding sequences of AtIPMDH1, AtIPMDH2, and AtIPMDH3 were amplified using the Platinum Pfx DNA Polymerase (Invitrogen) with appropriate primer pairs, as follows: AtIPMDH1-F, 5′-dCCATGGCGGCGTTTTTGCAA-3′; AtIMPDH1-R, 5′-dCACGTGTTAAACAGTAGCTGGAAC-3′; AtIMPDH2-F; 5′-dCCATGGCGGCGGCTCTGCAGACG-3′; AtIMPDH2-R, 5′-dCACGTGTTAAACAGAAGCTGGAACT-3′; AtIPMDH3-F, 5′-dCCATGGCGGCGTTTTTGCAAACTAA-3′; and AtIPMDH3-R, 5′-dCACGTGTTAAACAGGAACTTTGGAG-3′. PCR products were firstly cloned into pSC-B-amp/kan vector using StrataClone Blunt PCR Cloning Kit (StrataClone), and then sequenced. Correct fragments were subcloned into the AtIPMDH1pro::GUS vector (11) to generate constructs for each isoform and/or mutant under control of the AtIPMDH1 promoter. The resulting constructs were introduced into Agrobacterium tumefaciens strain C58C1 followed by transformation into atipmdh1 plants. Transgenic plants were selected for hygromycin resistance and homozygous plants used for subsequent analysis.
Glucosinolate Analysis
Rosette leaves of 4-week-old plants and mature seeds were used for glucosinolate analysis. Glucosinolates were analyzed using HPLC-mass spectrometry, as previously described (11, 13).
Protein Expression, Purification, Assays, Crystallization, and Structure Determination
Expression and purification of wild-type and mutant AtIPMDHs as histidine-tagged proteins for functional analysis was performed using nickel-affinity chromatography, as previously described (11). IPMDH assay conditions using either 3-isopropylmalate or 3-(2′-methylthio)ethylmalate as a substrate and the analysis of steady-state kinetic parameters were as previously described (11). All kinetic parameters were determined by directing fitting data to the Michaelis-Menten equation in SigmaPlot.
For crystallization of AtIPMDH2, the histidine tag was removed by thrombin digestion and the protein further purified using size-exclusion chromatography (17). Crystals of AtIPMDH2 were obtained in 5 μl hanging drops of a 1:1 mixture of protein and crystallization buffer (0.16 m ammonium sulfate, 0.08 m sodium acetate trihydrate, 20% PEG 4000, 20% glycerol) at 4 °C over a 0.7 ml reservoir. Data collection (100 K) was performed at beamline 19-ID at the Advanced Photon Source Argonne National Laboratory. Diffraction data were integrated and reduced using HKL3000 (18). The structure of AtIPMDH2 was solved by molecular replacement performed with PHASER (19) using the structure of IPMDH from Salmonella typhimurium (20) as a search model. Model building was performed in COOT (21) and all refinements were performed with PHENIX (22). Data collection and refinement statistics are reported in Table 1. The atomic coordinates and structure factors for AtIPMDH2 have been deposited in the Protein Data Bank (PDB ID code 3R8W).
TABLE 1.
Data collection and refinement statistics
| Crystal | |
|---|---|
| Space group | P21 |
| Cell dimensions | a = 76.81 Å, b = 211.0 Å, c = 76.90 Å; β = 90.15° |
| Data Collection | |
| Wavelength | 0.979 Å |
| Resolution range (highest shell) | 37.0–2.25 Å (2.29–2.25 Å) |
| Reflections (total/unique) | 433,041/114,991 |
| Completeness (highest shell) | 99.7% (100%) |
| <I/σ> (highest shell) | 31.2 (1.7) |
| Rsyma (highest shell) | 5.5% (58.4%) |
| Refinement | |
| Rcrystb/Rfreec | 17.5%/20.7% |
| No. of protein atoms | 10,885 |
| No. of water molecules | 459 |
| No. of ligand atoms | 64 |
| R.m.s. deviation, bond lengths | 0.007 Å |
| R.m.s. deviation, bond angles | 0.98° |
| Avg. B-factor - protein, water, ligand | 57.0, 60.3, 80.1 Å2 |
| Stereochemistry: most favored, allowed, generously allowed, prohibited | 97.1, 2.9, 0, 0% |
a Rsym = Σ|Ih − <Ih>|/ΣIh, where <Ih> is the average intensity over symmetry.
b Rcryst = Σ|Fo − <Fc>|/ΣFo, where summation is over the data used for refinement.
c Rfree is defined the same as Rcryst, but was calculated using 5% of data excluded from refinement.
Site-directed Mutagenesis and Mutant Protein Analysis
Site-directed mutagenesis was performed using the QuikChange PCR method (Stratagene). Bacterial expression vectors for each AtIPMDH (11, 13) were used as templates with specific oligonucleotide pairs, as follows: AtIPMDH1-F137L-F, 5′-dCTGAGACCTGAGATGGCTCTGCTTTACCTTAGAAGAGATCTC-3′; AtIPMDH1-F137L-R, 5′-dGAGATCTCTTCTAAGGTAAAGCAGAGCCATCTCAGGTCTCAG-3′; AtIPMDH2-L133F-F, 5′-dCTGAGGCCTGAGAAGGGGTTATTTCAGATTCGTGCAGCTCTC-3′; AtIPMDH2-L133F-R, 5′-dGAGAGCTGCACGAATCTGAAATAACCCCTTCTCAGGCCTCAG-3′; AtIPMDH3-L134F-F, 5′-dCTGAGACCTGAGATGGGTCTGTTTAACATTCGAAGAGA-3′; AtIPMDH3-L134F-R, 5′d-TCTCTTCGAATGTTAAACAGACCCATCTCAGGTCTCAG-3′ (mutated codon in underlined and mutation shown in bold). Mutant protein expression, purification, and assays were performed as described above for wild-type enzyme.
RESULTS
Differential Expression and AtIPMDH Metabolic Specialization
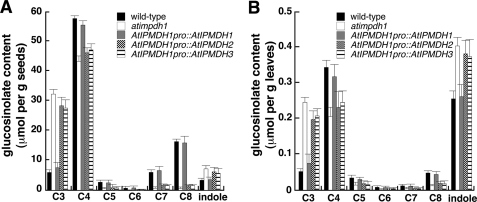
The three IPMDH genes in Arabidopsis have overlapping, yet distinct expression patterns. AtIPMDH1 (At5g14200) is highly expressed in leaves and roots; AtIPMDH2 (At1g80560) is weakly expressed throughout the plant; and AtIPMDH3 (At1g31180) is constitutively expressed at high levels in all tissues (11, 13, 23). To test the possible contribution of differential expression to the specialization of AtIPMDHs, each gene was placed under control of the native AtIPMDH1 promoter and then transformed into an atipmdh1 mutant line (11). As shown in Fig. 2, the altered glucosinolate profile of the atipmdh1 mutant could only be rescued by expression of AtIPMDH1. In addition, the atipmdh1 glucosinolate phenotype could not be rescued if expression was driven using either AtIPMDH2 or AtIPMDH3 promoter (data not shown). These results indicate that AtIPMDH1 sequence and differential expression are important in AtIPMDH specialization.
FIGURE 2.
Glucosinolate profile analysis. Glucosinolates in seeds (A) and leaves (B) from wild-type, atipmdh1 mutant, and transgenic plants harboring each AtIPMDH driven by the AtIPMDH1 promoter were analyzed. Levels of aliphatic glucosinolates with varied methylene chain length (C3-C8) are shown. All indole glucosinolates are combined into a single group. Data are mean ± S.D. (n = 3).
Structure of AtIPMDH2
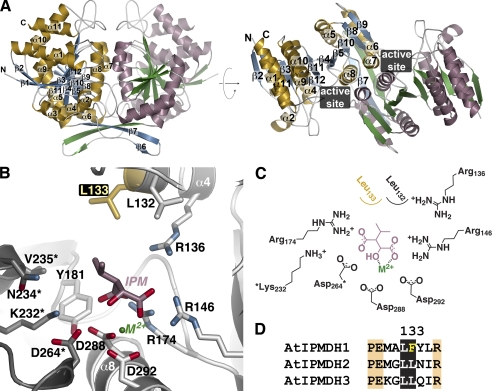
To determine the molecular architecture of a plant IPMDH, the 2.25 Å resolution x-ray crystal structure of AtIPMDH2 was solved by molecular replacement (Table 1). There were four molecules in the asymmetric unit representing two AtIPMDH2 dimers. The monomers of each dimer are related by non-crystallographic symmetry. Each AtIPMDH2 monomer consists of two domains (Fig. 3A). Domain 1 contains seven α-helices (α1–4 and α9–11) and five β-strands (β1–3 and β11–12), along with the N and C termini. Four α-helices (α5–8) and seven β-strands (β4–10) comprise domain 2. Between the two domains, β4 and β5 form the interdomain region. The second domain also serves as the dimerization interface with β6 and β7 of each monomer as part of an inter-subunit β-sheet and α7 and α8 of each monomer forming a four-helix bundle at the dimer interface. The overall structure of AtIPMDH2 is similar to those of the IPMDHs from various bacteria, including Salmonella typhimurium and Thermus thermophilus (20, 24, 25), with a root mean square deviation of 1.3–1.7 Å2 over ∼350 residues. Because the plant and bacterial IPMDHs share ∼50% sequence identity, conservation of key residues defines the active site region situated in a cleft between the two domains of each monomer (Fig. 3A).
FIGURE 3.
Structure of AtIPMDH2. A, ribbon diagrams of the AtIPMDH2 dimer. Monomer A is shown with gold α-helices and blue β-strands and monomer B is drawn with rose α-helices and green β-strands. Secondary structure features are labeled on the A monomer. The right view is rotated 90° to show the two domains of each monomer. The position of the active site cleft is indicated. B, active site view and model of 3-isopropylmalate (IPM) and divalent metal (M2+). Side chains of active site residues are shown with those from the adjacent subunit (gray) indicated by an asterisk. The positions of the substrate and metal are modeled based on the bacterial structures (20, 24, 25). The active site difference among the AtIPMDH isoforms is highlighted in gold. C, schematic of the active site model. D, sequence comparison of the region including residue 133 (AtIPMDH2 numbering).
The active site (Fig. 3B) is roughly delineated by α8 at the bottom and with α4 of one monomer and α7 of the adjacent monomer forming opposite sides of the site. Within the active site, all of the residues previously identified in structures of bacterial IPMDHs in complex with isopropylmalate and Mg2+ are also conserved in AtIPMDH2 (24, 25). Because efforts to obtain a structure of AtIPMDH2 in complex with ligands did not yield crystals, 3-isopropylmalate and Mg2+ were manually modeled into the plant enzyme based on the positions of these ligands observed in the bacterial structures (Fig. 3, B and C) (24, 25). This comparison shows that Asp264* (asterisk denotes a residue from the adjacent non-crystallographic symmetry related monomer), Asp-288, and Asp-292 are positioned to interact with a catalytically essential divalent metal (i.e. Mg2+ or Mn2+) and that a trio of arginines (Arg-136, Arg-146, and Arg-174) is poised to form charge-charge interactions with the carboxylate groups of the substrate. Residues corresponding to Leu-132, Leu-133, Tyr-181, Lys-232*, Asn-234*, and Val-235* form a largely hydrophobic region around the isopropyl group of the substrate.
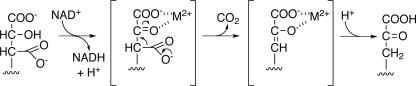
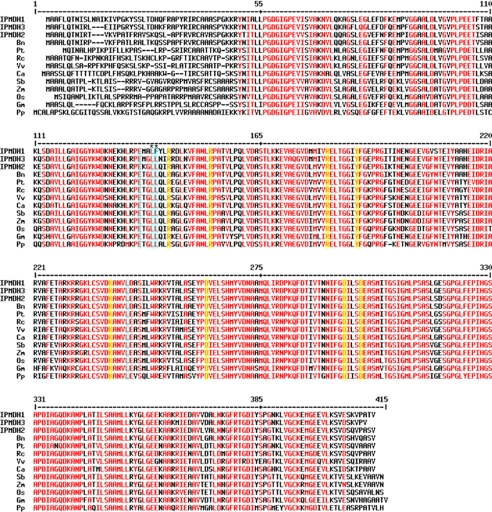
Although all of these amino acids are invariant in the bacterial and plant IPMDHs involved in leucine biosynthesis, the side-chain corresponding to Leu-133 is replaced with a phenylalanine in AtIPMDH1 (Fig. 3D), which is the isoform previously shown to be primarily involved in glucosinolate synthesis in Arabidopsis (11). Mechanistically, the conversion of 3-isopropylmalate to 4-methyl-2-oxovalerate in leucine synthesis and the conversion of 3-malate derivatives (e.g. 3-(2′-methylthio)ethylmalate) to 2-oxo acids (e.g. 5-methylthio-2-oxopentoate) in glucosinolate synthesis likely use a common metal-dependent reaction (Fig. 4); however, different substrate side chains of 3-malate derivatives (Fig. 1B) must fit in the plant IPMDH active site for production of aliphatic glucosinolates with six different chain lengths (C3-C8). Thus, we hypothesize that this single amino acid exchange from the leucine found in AtIPMDH2 and AtIPMDH3 to the phenylalanine in the active site of AtIPMDH1 may contribute to the functional divergence of this isoform for glucosinolate biosynthesis.
FIGURE 4.
Proposed mechanism for AtIPMDH. The conversion of 3-isopropylmalate to 4-methyl-2-oxovalerate in leucine synthesis and the conversion of 3-(2′-methylthio)ethylmalate to 5-methylthio-2-oxopentoate in glucosinolate synthesis utilize the same chemistry but with different R-groups.
Biochemical Analysis of Wild-type and Mutant AtIPMDHs
Previous studies on the AtIPMDHs demonstrate that each isoform accepts 3-isopropylmalate as a substrate (11, 13), but a kinetic comparison with a glucosinolate pathway substrate has not been reported. Using both 3-isopropylmalate and 3-(2′-methylthio)ethylmalate, the steady-state kinetic parameters for each AtIPMDH were determined (Table 2). Comparison of the catalytic efficiencies shows that AtIPMDH2 and AtIPMDH3 favor 3-isopropylmalate over 3-(2′-methylthio)ethylmalate by 14,900- and 29,600-fold, respectively. Moreover, these isoforms were ∼20-fold more active with the leucine biosynthesis substrate than AtIPMDH1. In comparison, AtIPMDH1 accepts both substrates with comparable kcat/Km values, but was ∼500-fold more efficient with the glucosinolate substrate than the other two isoforms. These catalytic efficiencies agree with the observed in vivo roles of the AtIPMDH isoforms in glucosinolate and leucine synthesis pathways (11–13).
TABLE 2.
Kinetic parameters of wild-type and mutant AtIPMDHs
All reactions were performed as described under “Experimental Procedures.” All kcat and Km values are expressed as a mean ± S.E. for an n = 3.
| 3-Isopropylmalate |
3-(2′-Methylthio)ethylmalate |
|||||
|---|---|---|---|---|---|---|
| kcat | Km | kcat/Km | kcat | Km | kcat/Km | |
| min−1 | μm | m−1s−1 | min−1 | μm | m−1s−1 | |
| AtIPMDH1 | 37 ± 4 | 25.2 ± 2.3 | 24,471 | 51 ± 5 | 45.3 ± 3.6 | 18,763 |
| AtIPMDH1-F137L | 230 ± 14 | 11.4 ± 1.7 | 336,257 | 2.0 ± 0.2 | 323 ± 21 | 103 |
| AtIPMDH2 | 373 ± 33 | 10.9 ± 1.3 | 570,336 | 1.0 ± 0.2 | 435 ± 32 | 38.3 |
| AtIPMDH2-L133F | 37 ± 5 | 30.3 ± 2.5 | 20,352 | 22 ± 1 | 77.0 ± 8.5 | 4,761 |
| AtIPMDH3 | 543 ± 36 | 9.2 ± 1.4 | 983,696 | 1.0 ± 0.1 | 502 ± 35 | 33.2 |
| AtIPMDH3-L134F | 44 ± 5 | 28.5 ± 1.5 | 25,731 | 16 ± 1 | 103 ± 10 | 2,589 |
To investigate the significance of the active site difference in the AtIPMDH, a series of point mutants (AtIPMDH1-F137L, AtIPMDH2-L133F, and AtIPMDH3-L134F) were generated. Kinetic analysis of these mutants demonstrates the critical role of this active site change in determining substrate specificity (Table 2). In AtIPMDH1, substitution of Phe-137 with a leucine reduced the kcat/Km of the mutant for 3-(2′-methylthio)ethylmalate to values comparable to those observed for AtIPMDH2 and AtIPMDH3. This was also accompanied by improved catalytic efficiency with 3-isopropylmalate, as the AtIPMDH1-F137L mutant was only 2- to 3-fold less efficient with this substrate than AtIPMDH2 and AtIPMDH3. The complementary mutation in either AtIPMDH2 (L133F) or AtIPMDH3 (L134F) yields mutant enzymes that were ∼30-fold less active with 3-isopropylmalate than the corresponding wild-type proteins, but still comparable to wild-type AtIPMDH1. Moreover, AtIPMDH2-L133F and AtIPMDH3-L134F displayed nearly a 100-fold improvement in activity with 3-(2′-methylthio)ethylmalate as a substrate to kcat/Km values that were 4- and 7-fold less than those observed with AtIPMDH1. In addition, based on the currently available plant sequences in GenBankTM, AtIPMDH1 is the only IPMDH with the unique phenylalanine instead of leucine, which is present in all other species (Fig. 5). These results demonstrate the critical role of the residue at position 133 (AtIPMDH2 numbering) in the evolution of AtIPMDH1 for the methionine chain-elongation reactions of glucosinolate biosynthesis.
FIGURE 5.
Alignment of IPMDH sequences from different plant species. Amino acid residues with high consensus value (90%) are colored in red. The residues interacting with the malate backbone of the substrates are shaded in yellow and residues interacting with the γ moiety of the substrates are highlighted in light blue. The LF residues in AtIPMDH1 and conserved LL residues in other IPMDH homologs were marked with asterisks. Species abbreviation: Bn, Brassica napus (rape); Ca, Capsicum annuum (pepper); Gm, Glycine max (soybean); Os, Oryza sativa (rice); Pp, Physcomitrella patens (moss); Pt, Populus trichocarpa (poplar); Rc, Ricinus communis (castor); Sb, Sorghum bicolor (sorghum); Vv, Vitis vinifera (grape); and Zm; Zea mays (corn).
In Vivo Analysis of AtIPMDH Mutant Function
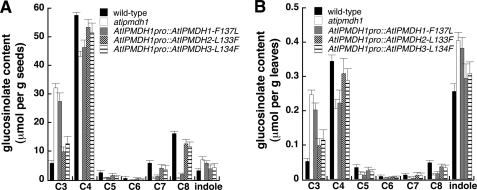
To test whether the amino acid substitution that occurred in AtIPMDH1 contributes to its specific function in vivo, atipmdh1 mutant plants were transformed with each of the mutant AtIPMDH genes driven by the AtIPMDH1 promoter. After isolation of homozygous lines, the glucosinolate profile in each mutant was examined. In comparison to the results shown in Fig. 2, the pronounced glucosinolate phenotype in the atipmdh1 mutant could not be rescued by AtIPMDH1-F133L (Fig. 6), indicating that the active site substitution impaired AtIPMDH1 function for glucosinolate synthesis in vivo. In contrast, the glucosinolate phenotype could be restored to the wild-type profile by expression of either AtIPMDH2-L133F or AtIPMDH3-L134F under the control of AtIPMDH1 native promoter (Fig. 6). The in planta findings corroborate the conclusion drawn from the biochemical analysis of recombinant proteins and provide evidence for the evolution of AtIPMDH1 by gene duplication and a single critical amino acid substitution.
FIGURE 6.
Glucosinolate profiles of point mutants. Glucosinolates in seeds (A) and leaves (B) of wild-type, atipmdh1 mutant, and transgenic plants expressing AtIPMDH1-F137L, AtIPMDH2-L133F and AtIPMDH3-L134F driven by AtIPMDH1 native promoter were analyzed. Levels of aliphatic glucosinolates with varied methylene chain length (C3-C8) are shown. All indole glucosinolates are combined into a single group. Data are mean ± S.D. (n = 3).
DISCUSSION
The evolution of specialized metabolism from primary metabolism is a common theme across biochemical pathways in plants (and microbes). Here we explored the molecular basis underlying the divergence of biological function in the IPMDHs in the leucine and glucosinolate biosynthesis pathways of Arabidopsis. Although all three AtIPMDHs accept 3-isopropylmalate, AtIPMDH1 is less efficient than the other isoforms (11, 13). Previous work also showed that knock-out mutants of AtIPMDH1 result in reduced levels of leucine and the C4 to C8 aliphatic glucosinolates (11). In contrast, knock-out mutations of the other isoforms did not alter glucosinolate levels but reduced leucine content (11, 13). Interestingly, a double mutation of AtIPMDH2 and AtIPMDH3 in Arabidopsis plants led to defects in pollen and embryo sac development, suggesting that leucine synthesis is essential for gametophyte formation. Using a combination of structural and functional analysis, this work demonstrates that a single amino acid change in the AtIPMDH active site leads to functional divergence of these enzymes in leucine synthesis (primary metabolism) and aliphatic glucosinolate synthesis (specialized metabolism).
Functional specification of AtIPMDHs in leucine and glucosinolate biosynthesis has been observed (11, 13). To evaluate if altered expression of AtIPMDH isoforms underlies functional specialization, each isoform gene was expressed under control of the AtIPMDH1 promoter in an atimpdh1 mutant background. Because the glucosinolate profile in the mutant was rescued only by expression of AtIPMDH1 (Fig. 2), it appears that gene duplication and subsequent mutation to a new function is the underlying evolutionary mechanism.
The three-dimensional structure of AtIPMDH2 (Fig. 3) and functional analysis (Table 2) of the AtIPMDHs provides insight on the specific changes required to alter the metabolic roles of these enzymes. A common chemical transformation is required to convert 3-isopropylmalate to 4-methyl-2-oxovalerate in leucine synthesis and 3-malate derivatives to 2-oxo acids in glucosinolate synthesis (Fig. 1). The AtIPMDH active site includes invariant residues for binding of either Mg2+ or Mn2+ (Asp-288, Asp-292, Asp-264*) and for charge-charge interactions with the substrate carboxylate groups (Arg-136, Arg-146, and Arg-174). Likewise, Tyr181 and Lys232*, which are proposed to perform general acid-base chemistry in the reaction mechanism (26), are conserved. For both 3-isopropylmalate (leucine synthesis) and 3-malate derivatives (glucosinolate synthesis), the overall reaction (Fig. 4) involves oxidation of the alcohol by deprotonation and hydride transfer to NAD+. This is followed by spontaneous decarboxylation, stabilization of the resulting enolate by the metal ion, and protonation to yield the final product.
Leucine and glucosinolate synthesis requires the same chemistry, but the AtIPMDH active site must accommodate reactants with different side-chains (i.e. isopropyl versus elongated methionine side-chain groups). The AtIPMDH2 structure and sequence analysis reveals a single amino acid difference of a leucine (AtIPMDH2 and AtIPMDH3) versus a phenylalanine (AtIMPDH1) in the active site. This difference occurs in the set of residues proposed to form the substrate interaction surface in the bacterial and plant IPMDH (20, 24, 25). Both in vitro and in vivo functional analysis of AtIPMDH1-F137L, AtIPMDH2-L133F, and AtIPMDH3-L134F demonstrates that switching this amino acid in each isoform is sufficient to interconvert catalytic efficiency (Table 2) and to change the aliphatic glucosinolate profiles in transgenic plants (Fig. 6). These results suggest that gene duplication of AtIPMDH followed by mutation of one active site residue in AtIPMDH1 leads to its specialized role for glucosinolate synthesis in Arabidopsis.
Sequence alignment of AtIPMDH homologs in other plant species revealed that the phenylalanine is not present in other homologs (Fig. 5), suggesting they may be primarily involved in leucine biosynthesis. As more genomic sequences become available, a broad implication of this amino acid substitution can be appreciated. It should be noted that the three AtIPMDHs have overlapping substrate specificity, but with distinct preferences. AtIPMDH1 is primarily involved in methionine chain-elongation of aliphatic glucosinolate biosynthesis. When AtIPMDH1 is mutated, the functions of AtIPMDH2 and AtIPMDH3 in glucosinolate biosynthesis become evident (13). On the other hand, all three AtIPMDHs are involved in leucine biosynthesis, with AtIPMDH2 and AtIPMDH3 exhibiting dominant roles (11, 13). This study has uncovered the molecular basis, i.e. a single amino acid substitution underlying substrate preference and functional divergence of the AtIPMDHs.
The structure-function analysis of the AtIPMDH provides insight on the molecular basis for altered function, but it is unclear how the leucine to phenylalanine mutation allows AtIPMDH1 to accommodate the growing methionine chain in subsequent iterations of the glucosinolate synthesis reactions (Fig. 1A). Multiple structures of IPMDH from bacteria indicate that the structural features around the active site are flexible and that active site dynamics likely plays a potential role in substrate recognition and catalysis (27). Moreover, the effect of the longer side-chain on the kinetics of the various glucosinolate biosynthesis pathway enzymes (i.e. BCAT, MAM, IPMI, and IPMDH) has not been explored. In Arabidopsis, multiple lines of evidence strongly support the evolution of methionine chain-elongation process of glucosinolate biosynthesis from leucine biosynthesis (5–8, 11); however, the molecular underpinnings for this evolution are only beginning to be understood. For example, the substrate specialization of the heterodimeric IPMI is determined by which small subunit associates with the large subunit (2, 8, 10, 12). Recently, the changes needed to convert IPMS from leucine synthesis into a MAM was demonstrated to involve the loss of a C-terminal regulatory domain responsible for feedback inhibition by leucine and a series of amino acid mutations (14). In contrast to large remodeling of protein structure in IPMS and MAM, the substrate specificity of IPMDH requires one amino acid difference.
Interactions between Arabidopsis and its environment may have driven the co-evolution of the pathways needed to synthesize the core glucosinolate structure and the elongation of the methionine side-chain. The biosynthesis of the glucosinolates has been suggested to have evolved from the prevalent system of cyanogenic glucoside biosynthesis (28–30). Evidence for this includes the wide distribution of cyanogenic glucosides in plants and arthropods, and the conservation of cytochrome P450s in the biosynthesis of glucosinolates and cyanogenic glucosides. In addition, metabolic engineering using cytochromes P450 involved in cyanogenic glycoside biosynthesis allows for the generation of acyanogenic plants that also display altered glucosinolate profiles (28–31). It is evident that when environmental challenges such as insect herbivores present themselves, specialization of enzymes from different pathways contributes to the evolution of methionine-derived glucosinolates for plant survival (32–34).
In summary, we have determined a key molecular change responsible for altering substrate specificity of the IPMDHs in Arabidopsis and the recruitment of an IPMDH from leucine biosynthesis for the specialized synthesis of glucosinolates. Future studies need to explore protein level changes in other glucosinolate enzymes to understand how the entire glucosinolate pathway evolved.
Acknowledgment
We thank Bing Chen for technical support.
This work was supported by the National Science Foundation (Grants MCB-0845162 (to S. C.) and MCB-0904215, to (J. M. J.)). Portions of this research were carried out at the Argonne National Laboratory Structural Biology Center at the Advanced Photon Source, a national user facility operated by the University of Chicago for the Dept. of Energy Office of Biological and Environmental Research (DE-AC02-06CH11357).
The atomic coordinates and structure factors (code 3R8W) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- IPMS
- isopropylmalate synthase (EC 2.3.3.13)
- BCAT
- branched-chain aminotransferases
- IPMDH
- isopropylmalate dehydrogenase (EC 1.1.1.85)
- IPMI
- isopropylmalate isomerase (EC 4.2.1.33)
- MAM
- methylthioalkylmalate synthase (EC 2.3.3.-).
REFERENCES
- 1. Kliebenstein D. J., Lambrix V. M., Reichelt M., Gershenzon J., Mitchell-Olds T. (2001) Plant Cell 13, 681–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sawada Y., Kuwahara A., Nagano M., Narisawa T., Sakata A., Saito K., Hirai M. Y. (2009) Plant Cell Physiol. 50, 1181–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Halkier B. A., Gershenzon J. (2006) Annu. Rev. Plant Biol. 57, 303–333 [DOI] [PubMed] [Google Scholar]
- 4. Yan X., Chen S. (2007) Planta 226, 1343–1352 [DOI] [PubMed] [Google Scholar]
- 5. Field B., Cardon G., Traka M., Botterman J., Vancanneyt G., Mithen R. (2004) Plant Physiol. 135, 828–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Textor S., Bartram S., Kroymann J., Falk K. L., Hick A., Pickett J. A., Gershenzon J. (2004) Planta 218, 1026–1035 [DOI] [PubMed] [Google Scholar]
- 7. Schuster J., Knill T., Reichelt M., Gershenzon J., Binder S. (2006) Plant Cell 18, 2664–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Binder S., Knill T., Schuster J. (2007) Physiol. Plant 129, 68–78 [Google Scholar]
- 9. Knill T., Schuster J., Reichelt M., Gershenzon J., Binder S. (2008) Plant Physiol. 146, 1028–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Knill T., Reichelt M., Paetz C., Gershenzon J., Binder S. (2009) Plant Mol. Biol. 71, 227–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. He Y., Mawhinney T. P., Preuss M. L., Schroeder A. C., Chen B., Abraham L., Jez J. M., Chen S. (2009) Plant J. 60, 679–690 [DOI] [PubMed] [Google Scholar]
- 12. He Y., Chen B., Pang Q., Strul J. M., Chen S. (2010) Plant Cell Physiol. 51, 1480–1487 [DOI] [PubMed] [Google Scholar]
- 13. He Y., Chen L., Zhou Y., Mawhinney T. P., Chen B., Kang B. H., Hauser B. A., Chen S. (2011) New Phytol. 189, 160–175 [DOI] [PubMed] [Google Scholar]
- 14. deKraker J. W., Gershenzon J. (2011) Plant Cell 23, 38–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sankoff D. (2001) Curr. Opin. Genet. Dev. 11, 681–684 [DOI] [PubMed] [Google Scholar]
- 16. Textor S., de Kraker J. W., Hause B., Gershenzon J., Tokuhisa J. G. (2007) Plant Physiol. 144, 60–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Galant A., Arkus K. A. J., Zubieta C., Cahoon R. E., Jez J. M. (2009) Plant Cell 21, 3450–3458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 19. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wallon G., Kryger G., Lovett S. T., Oshima T., Ringe D., Petsko G. A. (1997) J. Mol. Biol. 266, 1016–1031 [DOI] [PubMed] [Google Scholar]
- 21. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Acta Crystallogr. D 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) Acta Crystallogr. D 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nozawa A., Takano J., Miwa K., Nakagawa Y., Fujiwara T. (2005) Biosci. Biotech. Biochem. 69, 806–810 [DOI] [PubMed] [Google Scholar]
- 24. Kadono S., Sakurai M., Moriyama H., Sato M., Hayashi Y., Oshima T., Tanaka N. (1995) J. Biochem. 118, 745–752 [DOI] [PubMed] [Google Scholar]
- 25. Imada K., Inagaki K., Matsunami H., Kawaguchi H., Tanaka H., Tanaka N., Namba K. (1998) Structure 6, 971–982 [DOI] [PubMed] [Google Scholar]
- 26. Aktas D. F., Cook P. F. (2009) Biochemistry 48, 3565–3577 [DOI] [PubMed] [Google Scholar]
- 27. Singh R. K., Kefala G., Janowski R., Mueller-Dieckmann C., von Kries J. P., Weiss M. S. (2005) J. Mol. Biol. 346, 1–11 [DOI] [PubMed] [Google Scholar]
- 28. Bak S., Nielsen H. L., Halkier B. A. (1998) Plant Mol. Biol. 38, 725–734 [DOI] [PubMed] [Google Scholar]
- 29. Chen S., Andereason E. (2001) Plant Physiol. Biochem. 39, 743–758 [Google Scholar]
- 30. Bak S., Paquette S. M., Morant M., Morant A. V., Saito S., Bjarnholt N., Zagrobelny M., Jørgensen K., Osmani S., Simonsen H. T., Perez R. S., van Heeswijck T. B., Jorgensen B., Moller B. L. (2006) Phytochem. Rev. 5, 309–329 [Google Scholar]
- 31. Chen S., Glawischnig E., Jørgensen K., Naur P., Jørgensen B., Olsen C. E., Hansen C. H., Rasmussen H., Pickett J. A., Halkier B. A. (2003) Plant J. 33, 923–937 [DOI] [PubMed] [Google Scholar]
- 32. Newton E., Bullock J. M., Hodgson D. (2010) Oecologia 164, 689–699 [DOI] [PubMed] [Google Scholar]
- 33. Kliebenstein D., Pedersen D., Barker B., Mitchell-Olds T. (2002) Genetics 161, 325–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hopkins R. J., van Dam N. M., van Loon J. J. (2009) Annu. Rev. Entomol. 54, 57–83 [DOI] [PubMed] [Google Scholar]