Abstract
Schizophrenia is a highly heritable neuropsychiatric disorder affecting ∼1% of the world's population. Linkage and association studies have identified multiple candidate schizophrenia susceptibility genes whose functions converge on the glutamatergic neurotransmitter system. One such susceptibility gene encoding d-amino acid oxidase (DAO), an enzyme that metabolizes the NMDA receptor (NMDAR) co-agonist d-serine, has the potential to modulate NMDAR function in the context of schizophrenia. To further investigate its cellular regulation, we sought to identify DAO-interacting proteins that participate in its functional regulation in rat cerebellum, where DAO expression is especially high. Immunoprecipitation with DAO-specific antibodies and subsequent mass spectrometric analysis of co-precipitated proteins yielded 24 putative DAO-interacting proteins. The most robust interactions occurred with known components of the presynaptic active zone, such as bassoon (BSN) and piccolo (PCLO). The interaction of DAO with BSN was confirmed through co-immunoprecipitation assays using DAO- and BSN-specific antibodies. Moreover, DAO and BSN colocalized with one another in cultured cerebellar granule cells and in synaptic junction membrane protein fractions derived from rat cerebellum. The functional consequences of this interaction were studied through enzyme assay experiments, where DAO enzymatic activity was significantly inhibited as a result of its interaction with BSN. Taking these results together, we hypothesize that synaptic d-serine concentrations may be under tight regulation by a BSN-DAO complex. We therefore predict that this mechanism plays a role in the modulation of glutamatergic signaling through NMDARs. It also furthers our understanding of the biology underlying this potential therapeutic entry point for schizophrenia and other psychiatric disorders.
Keywords: Enzymes; Glutamate Receptors Ionotropic (AMPA, NMDA); Neurobiology; Oxidase; Proteomics; Serine; BSN; DAO; DAO Localization
Introduction
Several lines of evidence, including NMDAR2 antagonist studies, pharmacological intervention at the glycine modulatory site, postmortem patient brain analysis, and genetic studies, implicate NMDAR hypofunction in schizophrenia etiology (1). One potential approach for restoring NMDAR signaling is to increase the concentration of NMDAR co-agonists, such as d-serine and d-alanine (2–5). Levels of d-serine have been shown to be reduced in the CSF and serum of schizophrenic patients compared with control subjects (6, 7), which may reflect an increase in the activity of d-amino acid oxidase (DAO), the enzyme that catalyzes d-serine and d-alanine degradation (8, 9). Supporting this hypothesis are reports of increased DAO activity in schizophrenic patients (10–12).
Genetic association of DAO with schizophrenia has been demonstrated in several (13–17) but not all (18–21) linkage and association studies (22). Nevertheless, association studies have identified several single-nucleotide polymorphisms (SNPs) within the gene encoding G72, a putative DAO-interacting protein, which are associated with schizophrenia (14, 15, 17, 23).
Data from DAO functional knock-out mice provide further support for the role of DAO in schizophrenia (24–27). Mice with a naturally occurring mutation within the gene express a functionally inactive form of DAO due to mutation of glycine 181 with arginine (G181R). Studies of these mutant mice suggest that DAO is involved in regulating d-serine levels in vivo (28–30), with d-serine concentrations increased approximately 10-fold in the cerebellum and medulla oblongata of mutant mice compared with wild type mice. Levels of d-alanine, also a NMDAR co-agonist (4, 5) and substrate for DAO (9), were elevated 4-fold in all brain regions tested in G181R mutant mice compared with wild type mice (30). The presence of elevated d-serine and d-alanine levels was accompanied by an increased occupancy of the NMDAR glycine modulatory site, as demonstrated by attenuated effects of L-701,324, a NMDAR glycine site antagonist (25). Moreover, the DAO G181R mutant mice display behavioral phenotypes consistent with altered NMDA receptor signaling, including diminution of stereotypy and ataxia elicited by MK-801 (24) and enhanced spatial learning and long term potentiation in the hippocampus (26) compared with wild type mice. Consistent with these data, pharmacological inhibition of DAO with AS057278 (5-methylpyrazole-3-caroboxylic acid), CBIO (6-chlorobenzo[d]isoxazol-3-ol), or Merck Compound 8 (4H-thieno[3,2-b]pyrrole-5-carboxylic acid) in rodents increased d-serine levels and enhanced NMDAR function (31–33). Taken together, modulation of DAO activity impinges on NMDAR function in a manner that might be relevant for the treatment of schizophrenia.
DAO expression and distribution has been extensively studied. DAO expression in humans shows predominant neuronal expression in the dorsolateral prefrontal cortex and hippocampus, neuronal and glial expression in substantia nigra pars compacta, and strong glial expression in cerebellum (12). In rodents, DAO message and protein have been reported in the same brain regions as in humans (34). On a subcellular level, DAO was first reported to be localized within liver peroxisomes, consistent with the presence of a peroxisomal targeting sequence located within the DAO polypeptide (35). However, in the brain, expression may not be limited to peroxisomes because in samples from rat dorsolateral prefrontal cortex, cerebellum, and hippocampus, DAO exhibits pericellular immunolocalization (12), which does not appear to overlap with peroxisomal markers (36). Expression outside of the peroxisome may have very important consequences for the regulation of DAO because it exposes the protein to an alternative set of putative interacting proteins. An extraperoxisomal localization also suggests an altered function or regulatory role for DAO, which may be elucidated by the identification of its interacting proteins.
To identify novel DAO-interacting proteins, we generated a DAO-specific antibody and conducted co-immunoprecipitation experiments from rat cerebellar detergent extracts. Mass spectrometric analysis of anti-DAO immunoprecipitates resulted in the identification of 24 putative DAO-interacting proteins. Here, we report the identity of these novel DAO-interacting proteins as well as describing an investigation into the interaction of DAO with bassoon (BSN), a component of the presynaptic active zone and one of the most abundant proteins in these immunoprecipitates. Our data suggest a novel localization for DAO as a result of its interaction with BSN, which is likely to be relevant in influencing synaptic d-serine concentrations and in understanding more fully the role of DAO in normal neuronal function and in disease.
EXPERIMENTAL PROCEDURES
Construction of Expression Plasmids
The open reading frames of human and rat DAO were amplified by PCR and cloned into pcDNA3.1 (Invitrogen). To purify hDAO protein, the cDNA encoding hDAO was cloned into pTYB2 (New England Biolabs) with XbaI (5′) and NotI (3′). The EGFP-BSN 95–3938 construct expressed GFP-bassoon under the control of the CMV promoter (37). The BSN truncation mutants, including 95–609, 95–3263, 1692–3263, and 2715–3263, were amplified by PCR and cloned into fusion to EGFP under CMV promoter control with HindIII (5′) and SacII (3′).
Antibodies
The primary antibodies used in this study for Western blotting include catalase (Sigma), BSN (StressGen), PSD-95 (NeuroMab), MAP2 (Sigma), GFP (Clontech), mouse IgG (Santa Cruz Biotechnology, Inc.), and rabbit IgG (Abcam). DAO antiserum was generated in rabbits immunized with the rat DAO peptide 49GLWQPYLSDPSNPQEAEWNQQ69 conjugated to keyhole limpet hemocyanin (OpenBiosystems). The DAO antibody was affinity-purified from serum using a DAO peptide column and by adsorption to immobilized human DAO (hDAO).
Preparation and Separation of DAO Complexes
Dynal protein A and G magnetic beads (Invitrogen) were used to capture rabbit and mouse antibody, respectively, according to the manufacturer's instructions. Cerebellar lysate was used from approximately 6-week-old male Sprague-Dawley rats. The beads were washed with PBS and/or detergent-containing wash solutions (modified radioimmune precipitation assay buffer containing 0.2% SDS and 650 mm NaCl). Samples were resolved by SDS-PAGE followed by Western blotting using the indicated primary antibodies. Secondary fluorescent antibodies (Invitrogen) were used at a 1:1000 dilution. Blots were imaged with the Odyssey (Li-Cor, Lincoln, NE).
Mass Spectrometry
Immunoprecipitates were loaded and separated on 10–20% Tricine SDS-polyacrylamide gel (Invitrogen). Each gel lane was cut into 20 pieces of about 1 × 1 mm2. Proteins within the gel pieces were digested with trypsin and extracted from the gel and reconstituted in 20 μl of 2% acetonitrile, 0.1% fatty acid.
An Agilent 1100 nanoflow system connected to a linear ion trap mass spectrometer (LTQ, ThermoFinnigan) was used for the mass spectrometric analysis. The peptides were eluted with a gradient from 4 to 60% solvent B (90% acetonitrile and 0.1% fatty acid) over 70 min with a flow rate of 250 nl/min. The fragment ion spectra (MS/MS scan) were acquired in a data-dependent manner in which each fragment ion scan was followed by consecutive MS/MS scan on the first three most intense ions from the MS scan.
Maintenance and Transfection of HEK293 Cells
HEK293 were grown at 37 °C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) containing 10% (v/v) fetal calf serum (FCS; Invitrogen) and 1% (v/v) penicillin and streptomycin (Invitrogen). Lipofectamine 2000 (Invitrogen) was used to transform cells according to the manufacturer's instructions (Invitrogen). The cells were harvested 24–72 h post-transfection.
Stable Cell Line Generation
The HEK293 cells were transfected with 10 μg of total plasmid DNA containing an hDAO or rDAO insert by electroporation (400 V, 250 microfarads ohms). Transfected cells were cultured in the presence of hygromycin or G418. Individual colonies were screened for the presence of recombinant DAO.
Amplex Red DAO Functional Assay
The DAO functional assays were carried out in 384-well Matrix plates (Thermo Scientific). DAO stable line activity was ascertained in wells containing 20,000 cells/well (in a 30-μl volume), which were incubated at room temperature for 5 min. Subsequently, 10-μl samples of d-alanine (5 mm) (VWR International), Amplex Red (100 μm) (Invitrogen), HRP (1 unit/ml) (Sigma) solutions were added. Reactions took place in the dark at room temperature for 30 min. Fluorescent signals were measured with a Victor2 plate reader (PerkinElmer) using 544-nm excitation and 590-nm emission filters.
Subcellular Fractionation Experiments
The subcellular fractionation procedure was synthesized from three published methods (38–40).
Preparation, Culture, and Immunolabeling of Cerebellar Granule Neurons
Animal handling was in accordance with appropriate animal welfare regulations as outlined in the Institutional Animal Care and Use Committee (IACUC) guidelines. Cerebella were dissected from postnatal day 14–21 rat pups. The tissue was dissociated using papain solution according to the manufacturer's instructions (Worthington). The cells were plated at 50,000 cells/24-well poly-d-lysine-coated coverslip and incubated for 2 weeks in neurobasal complete medium (500 ml of neurobasal medium without l-glutamine, 10 ml of B27 supplement (50×), 0.75 g of KCl, 200 mm glutamine, 6.6 mg/ml aphidicolin, and 1% penicillin and streptomycin).
The cerebellar granule neurons (CGNs) were fixed with 4% paraformaldehyde or 100% ice-cold methanol. The cells were washed with 1% sodium borohydride, permeabilized with 0.5% Triton X-100 in PBS followed by a 1-h incubation in normal goat serum (Vector) containing 0.2% Triton X-100. Primary antibodies were added to the blocking solution at a 1:100 dilution for 1–3 h. Secondary rabbit and mouse antibodies (Odyssey) at 1:200 dilution suspended in the blocking reagent were then applied to the coverslips for 1 h. The coverslips were mounted onto slides with ProLong Gold (Invitrogen) and visualized under fluorescence microscopy.
RESULTS
Generation and Characterization of a DAO-specific Antibody
To generate an antibody that recognizes rat DAO, an immunogenic peptide was chosen based on a previously published antibody raised against mouse DAO (25). The anti-rat DAO antibody was affinity-purified from crude serum against the immunogenic peptide and purified hDAO.
Purified anti-DAO antibody detected two immunoreactive bands at the expected molecular masses of 37 and 40 kDa in immunoblots of lysates from HEK293 cells stably expressing human DAO. A single DAO band at 38 kDa was detected from rat cerebellum lysates (Fig. 1A), the brain region where DAO has been shown to be expressed at especially high levels (12, 41). In contrast, no immunoreactive bands were detected by anti-DAO antibody in immunoblots of lysates from untransfected HEK293 or from rat spleen lysates (Fig. 1A), highlighting the specificity of the purified anti-DAO antibody because DAO is not expressed in spleen (42). Furthermore, the DAO-specific signal was completely blocked by preadsorption of 1.4 μg/ml immunogenic peptide (Fig. 1B). In order to assess the utility of this anti-DAO antibody for downstream interaction studies, we also determined that this anti-DAO antibody can specifically immunoprecipitate DAO from rat cerebellum (Fig. 1C).
FIGURE 1.
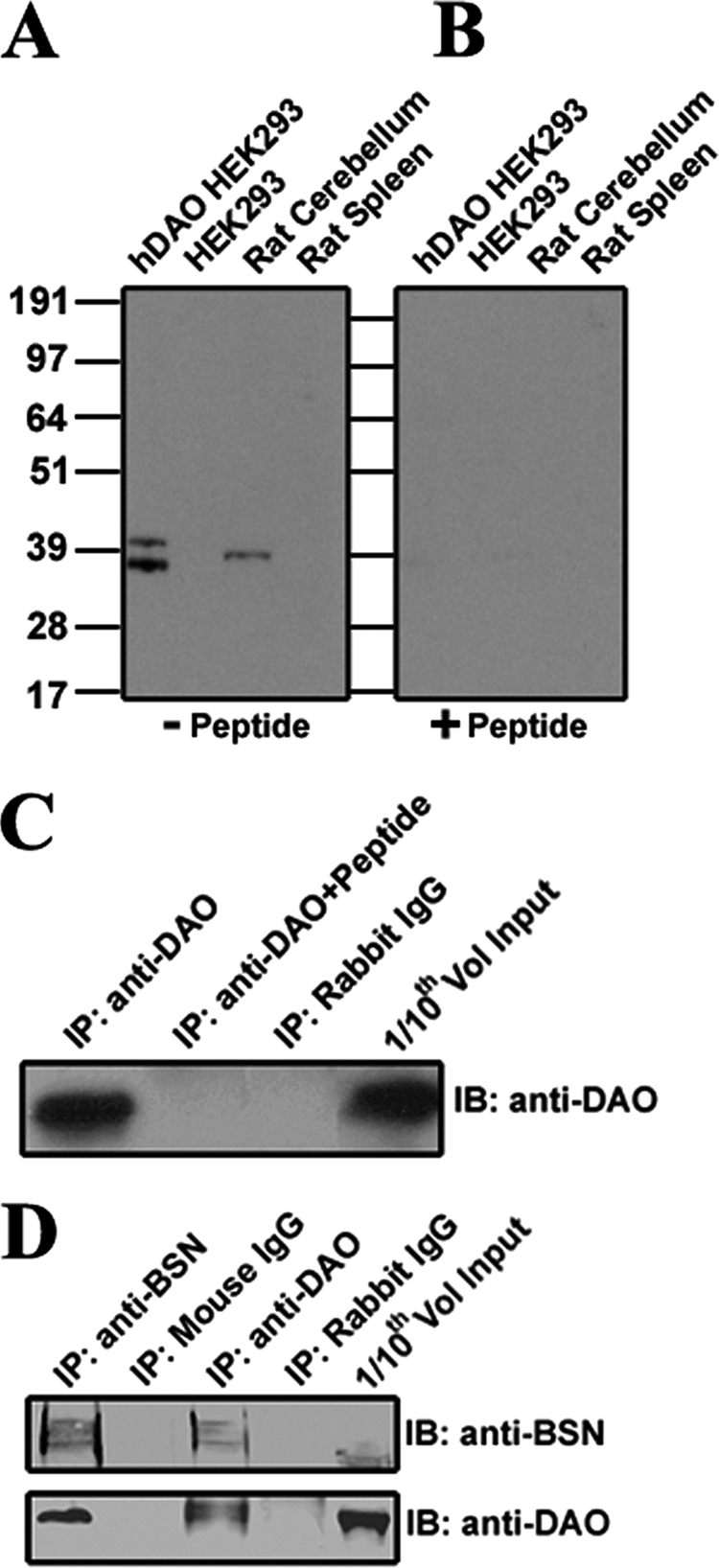
Characterization of a DAO-specific polyclonal antibody. A, an anti-DAO antibody revealed two immunoreactive bands at ∼37 and 40 kDa in immunoblots of lysates from a stable HEK293 cell line, expressing hDAO (lane 1), but not of control HEK293 lysates (lane 2). The anti-DAO antibody detected a single immunoreactive band at ∼39 kDa in immunoblots of rat cerebellar lysates (lane 3) but not of lysates from rat spleen (lane 4), the latter of which is not expected to express DAO. B, when the anti-DAO antibody was preincubated with the immunizing peptide, no immunoreactive bands were detected in lysates of either the hDAO stable line or rat cerebellum (lanes 1 and 3). C, anti-DAO, covalently coupled to Dynal magnetic beads, immunoprecipitated (IP) DAO protein from rat cerebellar lysates, as determined by Western blotting (IB) with anti-DAO antibody (lane 1). In contrast, DAO was not immunoprecipitated when anti-DAO was preincubated with the immunogenic peptide (lane 2) or when beads were covalently coupled to nonspecific rabbit IgG in place of anti-DAO (lane 3). Rat cerebellar lysate (Input; lane 4) was used at one-tenth the volume to verify that DAO was present in starting material. D, DAO and BSN coimmunoprecipitate with one another from rat cerebellar lysates. BSN was immunoprecipitated from rat cerebellar lysates by both anti-BSN (lane 1) and anti-DAO (lane 3) antibodies but not by nonspecific mouse (lane 2) or rabbit (lane 4) IgG controls. DAO was immunoprecipitated from rat cerebellar lysates by both anti-BSN (lane 1) and anti-DAO (lane 3) antibodies but not by nonspecific mouse (lane 2) or rabbit (lane 4) IgG controls.
Identification of Novel DAO-interacting Proteins
To the best of our knowledge, there have been no reported DAO-interacting proteins identified to date other than the primate-specific protein G72 (14). Thus, in order to identify novel DAO-interacting proteins, we used the anti-DAO antibody to immunoprecipitate DAO-containing complexes from rat cerebellar lysates for mass spectrometric analysis. We initially used a “PBS-only” washing condition, which resulted in the identification of 198 distinct proteins that were co-immunoprecipitated by anti-DAO but not by rabbit IgG or by anti-DAO incubated with 1.4 μg/ml immunogenic peptide (supplemental Table S1). The latter two control immunoprecipitation conditions did not precipitate DAO from rat cerebellar lysate (Fig. 1C), suggesting that the 198 putative DAO interactors were specifically co-immunoprecipitated with DAO from rat cerebellar lysates. To limit the number of DAO-interacting proteins, the immunoprecipitation experiment was repeated with a high stringency washing condition utilizing modified radioimmune precipitation assay buffer. Under these more stringent conditions, a total of 24 putative DAO-interacting proteins were revealed by mass spectrometric analysis (Table 1).
TABLE 1.
DAO interactome
Shown are DAO-interacting proteins retained by the DAO antibody column after washing with modified radioimmune precipitation assay buffer containing 0.2% SDS and 650 mm NaCl.
| Abbreviation | Protein name |
|---|---|
| DYNC1H1 | Dynein heavy chain |
| BSN | Bassoon |
| BAT2D1 | HBxAg transactivated protein 2 |
| PCLO | Piccolo |
| PRKCG | Protein kinase C, γ |
| SNIP | SNAP-25-interacting protein |
| PC | Pyruvate carboxylase, mitchondrial |
| RAPGEF4 | Exchange factor directly activated by cAMP 2 |
| CEP97 | Centrosomal protein 97 |
| PHYHIP | Phyhip protein |
| MAP1B | Microtubule-associated protein 1B |
| PABPC1 | Poly(A)-binding protein, cytoplasmic 1 |
| YLPM1 | YLP motif-containing 1 |
| AP1B1 | Adopted-related protein complex 1, β1 |
| PPP1CB | Protein phosphate 1 |
| CRMP1 | Crmp1 protein |
| ERC1 | ELKS/RAB6-interacting/CAST family member |
| CLINT1 | Clathrin interactor 1 |
| DYNLL2 | Dynein, light chain |
| NCOA6 | Peroxisome proliferator activater receptor interacting protein |
| DSP | Desmoplakin 1 |
| CHAINB | Chain B, perchloric acid-soluble protein-a translational inhibitor |
| HRNR | Homeric precursor |
| PFN1 | Profilin 1 |
DAO Interacts with Bassoon, a Protein Enriched in the Presynaptic Active Zone
Based on the two immunoprecipitation experiments, presynaptic active zone proteins, including bassoon (BSN) and piccolo (PCLO) (supplemental Table S1), were found to be highly enriched among the pool of co-precipitating proteins because these proteins were represented by the greatest number of unique and total tryptic peptide fragments as well as by the largest percentage of protein coverage as revealed by mass spectrometric analysis of anti-DAO immunoprecipitates (supplemental Table S2). Of the presynaptic proteins identified, peptides corresponding to BSN were the most prevalent, with 29% of the entire BSN protein represented in the DAO immunoprecipitates, a value very close to that of DAO itself (30%; supplemental Table S2). This observation suggested that BSN was the most abundant protein present in the immunoprecipitates, second only to DAO itself (Table 1 and supplemental Table S2).
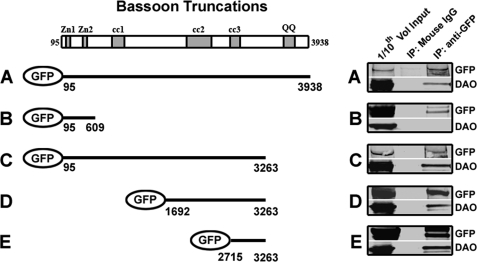
The DAO-BSN interaction was confirmed through mutual co-immunoprecipitation experiments with DAO- and BSN-specific antibodies from rat cerebellar lysates (Fig. 1D). To identify the region of BSN responsible for the interaction with DAO, a rat DAO-expressing stable HEK293 cell line was transfected with cDNAs encoding full-length BSN or selected fragments of BSN fused to GFP at the N terminus (Fig. 2, left). These BSN constructs (encoding amino acids 95–609, 95–3263, 1692–3263, and 2715–3263), were based on previously identified BSN functional domains (37). Immunoprecipitation with a mouse anti-GFP antibody, but not mouse IgG, from the transiently transfected HEK293 lysates resulted in immunoprecipitation of full-length GFP-BSN as well as co-immunoprecipitation of DAO (Fig. 2A). DAO did not co-immunoprecipitate with the BSN fragment composed of amino acids 95–609, containing the BSN zinc finger domain (Fig. 2B). However, DAO did co-immunoprecipitate with the BSN fragment composed of amino acids 95–3263 (Fig. 2C). DAO also co-immmunoprecipitated with the BSN fragment composed of amino acids 1692–3263 (Fig. 2D) and with a BSN fragment composed of amino acids 2715–3263 containing a single coiled-coiled domain (Fig. 2E). Together, these data suggest that the region of BSN required for its interaction with DAO is localized between amino acids 2715 and 3263, containing the third coiled-coil domain.
FIGURE 2.
Determination of the DAO-binding domain within BSN. Shown is a schematic BSN diagram depicting the functional domains: double zinc finger domains (Zn1 and Zn2) and predicted coiled-coil regions (cc1, cc2, and cc3). A stable HEK293 cell line expressing DAO was transfected with either BSN 95–3938 (A), BSN 95–609 (B), BSN 95–3263 (C), BSN 1692–3263 (D), or BSN 2715–3263 (E), each fused to GFP at its N terminus. Lysates were subjected to immunoprecipitation (IP) with a mouse anti-GFP antibody (lane 3) or nonspecific mouse IgG (lane 2) and immunoblotted with an anti-GFP antibody (top) or an anti-DAO antibody (bottom). The combined data suggest that the domain required for the interaction with DAO resides between amino acids 2715 and 3263 of the BSN polypeptide.
DAO Colocalizes with BSN in Cultured Cerebellar Granule Neurons and in Synaptic Fractions of Rat Cerebellum
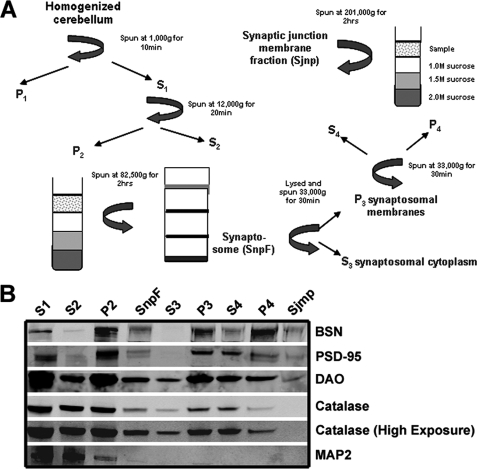
The interaction with BSN, a major component of the presynaptic active zone, suggested that DAO might colocalize with BSN in this subcellular compartment. To test this hypothesis, we generated subcellular fractions from rat cerebellum and immunoblotted these fractions with the anti-DAO antibody (Fig. 3). DAO was detected in the synaptic junction membrane fraction containing both the pre- and postsynaptic markers BSN and PSD-95, respectively (Fig. 3B), suggesting that DAO and BSN may colocalize in vivo. The absence of catalase in the synaptic junction membrane fraction suggested that the DAO found in this fraction was outside of the peroxisome. This finding is important in light of the historical association of DAO as a peroxisomally expressed enzyme (35). BSN, PSD-95, and DAO were all found in the pure synaptic membrane protein fraction, without catalase (Fig. 3, lane 9). This suggests that a pool of DAO may localize outside of the peroxisome at the synaptic junction, where it may be accessible to interact with BSN presynaptically.
FIGURE 3.
DAO and BSN, but not catalase, cofractionate in a cerebellar synaptic junction membrane protein fraction. A, schematic diagram of the fractionation protocol employed to determine the subcellular localization of DAO in rat cerebellum. B, DAO is present in a fraction containing synaptic junction membrane proteins (Sjmp), including PSD-95 and BSN but not catalase. Catalase, a peroxisomal marker, was not detected in the Sjmp fraction even when the immunoblot was overexposed, confirming that the DAO detected in this fraction is not derived from peroxisomal membranes. Due to a low yield, the Sjmp sample was loaded at half the amount found in all of the other lanes. SnpF, synaptosomal fraction.
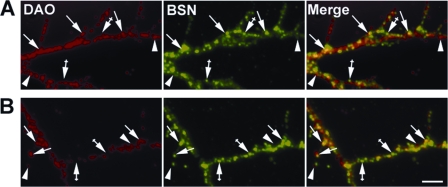
To further examine DAO localization in relation to BSN, we compared their localization patterns by immunofluorescence microscopy in primary cultures of CGNs. DAO appeared to be localized within the neuronal processes (supplemental Fig. S1A), with no signal detected in these processes when neurons were labeled with the secondary antibody alone (supplemental Fig. S1B) or when the immunogenic peptide was used to block the purified anti-DAO antibody (supplemental Fig. S1C). Given that DAO has been reported to be highly localized to the peroxisomal compartments, we anticipated partial DAO colocalization with BSN at the presynaptic active zone. As expected, when paraformaldehyde-fixed CGNs were double-labeled with antibodies to both DAO and BSN (Fig. 4), we observed partial colocalization between DAO and BSN (Fig. 4A, arrows). The colocalization was retained when CGNs were fixed with methanol, to wash out soluble proteins (Fig. 4B, arrows), suggesting that DAO is associated with synaptic membranes. These data suggest that a pool of DAO may be localized to the presynaptic active zone, by virtue of its colocalization with presynaptically localized BSN.
FIGURE 4.
DAO partially colocalizes with BSN in cultured cerebellar granule neurons. CGNs were cultured for 2 weeks, fixed with either paraformaldehyde (A) or methanol (B), and subjected to double-label immunofluorescence microscopy with rabbit anti-DAO (red) and mouse anti-BSN (green) antibodies. Images of neuronal processes were collected with a 100× oil immersion objective. Arrows, anti-DAO immunofluorescent puncta that colocalize with that of anti-BSN; arrowheads, anti-DAO immunofluorescent puncta that do not colocalize with that of anti-BSN. Crossed arrows, anti-BSN immunofluorescent puncta that do not colocalize with that of anti-DAO. Colocalizing puncta appear yellow in the merged images (right panels). Size bar, 2 μm.
BSN Does Not Act as a Scaffold for a DAO·SRR·Asc-1 Complex
Given the interaction between DAO and BSN, we hypothesized that BSN may act as a scaffolding protein for DAO, alanine-serine-cysteine transporter-1 (Asc-1), and serine racemase (SRR) to collectively modulate d-serine release from the presynaptic termini. To support this hypothesis, Asc-1 and SRR have been localized to neuronal synapses (43, 44). To directly test this hypothesis, rat cerebellar lysates were immunoprecipitated with the anti-BSN antibody, and the immunoprecipitates were probed with Asc-1 and SRR antibodies (supplemental Fig. S2); however, neither Asc-1 nor SRR were found to interact with BSN. Thus, our data suggest that BSN does not act as a scaffold for the three d-serine-related proteins.
BSN Inhibits DAO Enzymatic Activity
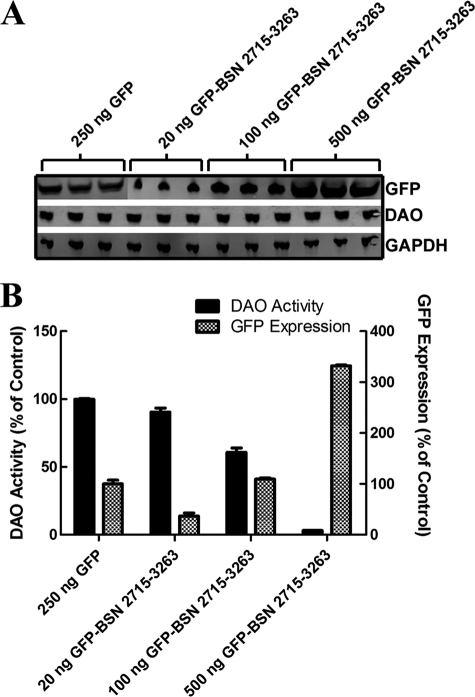
We reasoned that a specific interaction with DAO might reflect a regulatory role for BSN as a modulator of DAO functional activity. To test this hypothesis, we transfected the rDAO stable cell line with cDNA encoding the aforementioned truncated fragments of BSN and measured DAO enzymatic activity using Amplex Red. Indeed, we found that BSN constructs that were previously found to interact with DAO significantly inhibited DAO enzymatic activity. Specifically, the full-length BSN and DAO-interacting BSN fragments 95–3263, 1692–3263, and 2715–3263 (Fig. 2, A and C–E) led to a 30% reduction of DAO enzymatic activity when transiently transfected into the rDAO stable line (supplemental Fig. S3). GFP and BSN 95–609, which did not co-immunoprecipitate with DAO (Fig. 2B), did not alter the enzymatic activity of DAO. We were unable to conduct standard enzymology experiments because we could not produce a sufficient quantity of purified BSN 2715–3263. Therefore, we took an alternative cell-based approach and carefully titrated cDNA encoding GFP-BSN 2715–3263, the shortest BSN fragment retaining the ability to interact with DAO, into the rDAO stable line. We measured the relative expression level of GFP and related it back to inhibition of the enzyme (Fig. 5). This BSN fragment inhibited DAO enzymatic activity in a dose-dependent manner when transiently expressed in the rDAO stable cell line and, when expressed at a high level, led to a 97% reduction in DAO activity (Fig. 5). These data suggest that BSN negatively regulates the enzymatic activity of DAO within the presynaptic active zone.
FIGURE 5.
BSN inhibition of DAO enzymatic activity. A stable HEK293 cell line expressing DAO was transfected with cDNA encoding GFP-BSN 2715–3263 at a concentration of 20, 100, or 500 ng/30,000 cells. A, BSN 2715–3263 protein expression increased with increasing cDNA concentrations, as measured by GFP immunoreactivity on Western blots. Neither DAO nor GAPDH protein expression was altered by the BSN 2713–3263 expression. B, DAO enzymatic activity decreased in concert with increasing BSN 2713–3263 expression relative to GFP-transfected DAO HEK293 cells. *, p < 0.05; ***, p < 0.001 by one-way analysis of variance followed by Dunnett's multiple comparison against GFP-transfected cells.
DISCUSSION
Through a co-immunoprecipitation and mass spectrometry approach, we have identified 24 putative DAO-interacting proteins from rat cerebellum (Table 1). Many of these proteins, including BSN, PCLO, SNIP, ERC1, and RAPGEF4, are enriched in the presynaptic active zone (37, 45–48), suggesting a subcellular localization for DAO alternative to that of the widely accepted astrocytic, peroxisome-bound DAO (49–51). Our findings are supported by recent observations of DAO expression 1) in neurons, as exemplified by reports of DAO expression in Golgi and Purkinje cells of the rat cerebellum (52) and pyramidal neurons of human hippocampus and cerebral cortex (12), and 2) in the extraperoxisomal space, as exemplified by a pericellular distribution of DAO in the human brain combined with the lack of an overlap between DAO and peroxisomal markers in human astrocyte cultures (12, 36). Thus, because published reports hint at DAO localization outside of the peroxisome, we explored an alternative localization for DAO at the presynaptic active zone by virtue of its interaction with BSN.
BSN was confirmed to interact with DAO in rat cerebellar extracts through co-immunoprecipitation combined with Western blotting using DAO- and BSN-specific antibodies (Fig. 1D). Subcellular fractionation studies of rat cerebellum showed that DAO is present in the synaptic junction membrane fraction that contained BSN but not catalase (Fig. 3B). This finding suggests that DAO is localized outside of the peroxisome, where it was historically believed to be enriched (35). In light of our immunoprecipitation findings, the fractionation data suggest that DAO may be found in the presynaptic “bouton,” where it may interact with BSN in vivo. Immunocytochemistry of cultured CGNs confirmed the localization of DAO in neuronal processes partially colocalizing with BSN (Fig. 4).
BSN was co-immunoprecipitated with DAO from HEK293 cells expressing both proteins (Fig. 2), suggesting that the two may directly interact because none of the major members of the presynaptic active zone are expressed in the HEK293 cells (data not shown). Whereas the C terminus of BSN consisting of amino acids 3601–3942 is responsible for an interaction with many of the other presynaptic active zone proteins (53), the BSN region spanning amino acids 2715–3263, representing a predicted single coiled-coil domain, was found to co-immunoprecipitate with DAO (Fig. 2). This finding suggests that an interaction of BSN with other members of the presynaptic active zone would not be disrupted by a DAO-BSN interaction because it involves a different region of BSN.
The presynaptic cytoskeletal protein matrix, called the “cytometrix assembled at the active zones” (CAZ) (54), is a specialized subcellular domain where synaptic vesicles are anchored and primed prior to membrane fusion and neurotransmitter release (55). BSN and PCLO play a major role in the organization of the CAZ, probably as scaffolding elements (38, 56). Some of the CAZ members, including PCLO, have been shown to be up-regulated, in terms of both mRNA and protein, in patients with schizophrenia. Although BSN did not reach statistical significance in this study, the BSN mRNA was found to be elevated in patients with schizophrenia (57, 58). Another member of the presynaptic terminus, Asc-1, a d-serine and d-alanine uptake transporter, plays a major role in synaptic d-serine/d-alanine clearance in the forebrain and cerebellum (43, 59–63). The presence of DAO at the CAZ suggests that some of the reabsorbed d-serine/d-alanine may be metabolized, further fine tuning their concentration within the synapse and influencing NMDAR activation. Our findings are important in light of a recent report of d-serine release from neurons via a nonvesicular mechanism (64), suggesting that this nonvesicular pool of d-serine might be readily metabolized by presynaptically localized DAO.
The interaction of DAO with BSN may be especially relevant in the forebrain, where DAO has been reported to be predominantly expressed in neurons (12, 34, 36, 65). Such expression, in conjunction with our observation of an inhibitory effect of BSN on the activity of DAO, may in part explain why the enzymatic activity of DAO has been consistently undetectable in the forebrain (66, 67). We found that a fraction of the total DAO partitions to the CAZ (Figs. 3 and 4) and that a molar excess of BSN is required for complete inhibition of DAO enzymatic activity (Fig. 5). Because BSN is predominantly found within the active zone (37), it is likely to be in molar excess to DAO, resulting in an inhibitory effect on the DAO enzymatic activity. In the hindbrain, however, where DAO is more likely to be expressed in glia rather than in neurons, BSN would not be expected to influence the activity of DAO because BSN expression has not been detected in glia. In fact, within the brain, DAO activity is most robust in hindbrain when compared with all other brain regions (10, 41, 67–69).
A loss of the inhibitory effect of BSN on neuronal DAO activity may also explain observations made in BSN functional knock-out mice (70). These mice express a fragment of BSN that lacks amino acids 505–2889, a region that partially overlaps with the DAO binding site identified in the present study (Fig. 2). Moreover, this BSN fragment only partially partitions (between 10 and 30%) with the synaptic protein fraction. Reduced long term potentiation in striatal medium spiny neurons of the BSN mutant mice has been observed (71). Because long term potentiation requires an activation of NMDAR (72), it is tempting to speculate that presynaptic DAO, in the absence of the inhibitory influence of BSN, as would be the case in these BSN mutant mice, is functionally overactive, metabolizing synaptic d-serine/d-alanine and resulting in NMDAR hypoactivity. Other alternatives for the altered LTP in the BSN functional knock-out mice exist, including, for example, an altered NMDAR expression.
The functional importance of our finding that DAO enzymatic activity is inhibited by BSN remains unknown. Nonetheless, given the role of d-serine as an NMDAR co-agonist (73), a model whereby DAO, localized to the presynaptic active zone, is inhibited by colocalizing BSN may hold important implications for the maintenance of healthy synapses. According to this model, the levels of synaptic d-serine/d-alanine would probably be influenced by the interplay of DAO and BSN because our findings show that DAO is poised to impact presynaptic concentrations of these obligatory co-agonists. The altered synaptic d-serine/d-alanine levels due to unbalanced interaction of DAO and BSN may manifest as of psychotic symptoms because of modified NMDAR activation.
The precise role DAO plays at the presynaptic active zone is also unknown. For example, it has not yet been determined if presynaptically localized DAO is enzymatically active or whether this activity is modulated by an interaction with BSN in vivo. However, our studies are consistent with a model whereby BSN might play an important homeostatic role, inhibiting excessive DAO activity at the presynaptic active zone.
Supplementary Material
Acknowledgments
We thank Andrew Hill and the Pfizer protein informatics group for creating the Spotfire guide and helping with the analysis of the mass spectrometry data.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S3.
- NMDAR
- NMDA receptor
- DAO
- d-amino acid oxidase
- hDAO and rDAO
- human and rat DAO, respectively
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine
- CGN
- cerebellar granule neuron
- CAZ
- cytometrix assembled at the active zones.
REFERENCES
- 1. Coyle J. T. (2006) Cell. Mol. Neurobiol. 26, 365–384 [DOI] [PubMed] [Google Scholar]
- 2. Panatier A., Theodosis D. T., Mothet J. P., Touquet B., Pollegioni L., Poulain D. A., Oliet S. H. (2006) Cell 125, 775–784 [DOI] [PubMed] [Google Scholar]
- 3. Andersen J. D., Pouzet B. (2004) Neuropsychopharmacology 29, 1080–1090 [DOI] [PubMed] [Google Scholar]
- 4. Sakata K., Fukushima T., Minje L., Ogurusu T., Taira H., Mishina M., Shingai R. (1999) Biochemistry 38, 10099–10106 [DOI] [PubMed] [Google Scholar]
- 5. Tanii Y., Nishikawa T., Hashimoto A., Takahashi K. (1994) J. Pharmacol. Exp. Ther. 269, 1040–1048 [PubMed] [Google Scholar]
- 6. Hashimoto K., Fukushima T., Shimizu E., Komatsu N., Watanabe H., Shinoda N., Nakazato M., Kumakiri C., Okada S., Hasegawa H., Imai K., Iyo M. (2003) Arch. Gen. Psychiatry 60, 572–576 [DOI] [PubMed] [Google Scholar]
- 7. Bendikov I., Nadri C., Amar S., Panizzutti R., De Miranda J., Wolosker H., Agam G. (2007) Schizophr. Res. 90, 41–51 [DOI] [PubMed] [Google Scholar]
- 8. Leighton F., Poole B., Beaufay H., Baudhuin P., Coffey J. W., Fowler S., De Duve C. (1968) J. Cell Biol. 37, 482–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. D'Aniello A., Vetere A., Petrucelli L. (1993) Comp. Biochem. Physiol. B 105, 731–734 [DOI] [PubMed] [Google Scholar]
- 10. Madeira C., Freitas M. E., Vargas-Lopes C., Wolosker H., Panizzutti R. (2008) Schizophr. Res. 101, 76–83 [DOI] [PubMed] [Google Scholar]
- 11. Burnet P. W., Eastwood S. L., Bristow G. C., Godlewska B. R., Sikka P., Walker M., Harrison P. J. (2008) Mol. Psychiatry 13, 658–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Verrall L., Walker M., Rawlings N., Benzel I., Kew J. N., Harrison P. J., Burnet P. W. (2007) Eur. J. Neurosci. 26, 1657–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wirgenes K. V., Djurovic S., Agartz I., Jonsson E. G., Werge T., Melle I., Andreassen O. A. (2009) Neuropsychobiology 60, 31–36 [DOI] [PubMed] [Google Scholar]
- 14. Chumakov I., Blumenfeld M., Guerassimenko O., Cavarec L., Palicio M., Abderrahim H., Bougueleret L., Barry C., Tanaka H., La Rosa P., Puech A., Tahri N., Cohen-Akenine A., Delabrosse S., Lissarrague S., Picard F. P., Maurice K., Essioux L., Millasseau P., Grel P., Debailleul V., Simon A. M., Caterina D., Dufaure I., Malekzadeh K., Belova M., Luan J. J., Bouillot M., Sambucy J. L., Primas G., Saumier M., Boubkiri N., Martin-Saumier S., Nasroune M., Peixoto H., Delaye A., Pinchot V., Bastucci M., Guillou S., Chevillon M., Sainz-Fuertes R., Meguenni S., Aurich-Costa J., Cherif D., Gimalac A., Van Duijn C., Gauvreau D., Ouellette G., Fortier I., Raelson J., Sherbatich T., Riazanskaia N., Rogaev E., Raeymaekers P., Aerssens J., Konings F., Luyten W., Macciardi F., Sham P. C., Straub R. E., Weinberger D. R., Cohen N., Cohen D., Ouelette G., Realson J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 13675–13680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu X., He G., Wang X., Chen Q., Qian X., Lin W., Li D., Gu N., Feng G., He L. (2004) Neurosci. Lett. 369, 228–233 [DOI] [PubMed] [Google Scholar]
- 16. Schumacher J., Jamra R. A., Freudenberg J., Becker T., Ohlraun S., Otte A. C., Tullius M., Kovalenko S., Bogaert A. V., Maier W., Rietschel M., Propping P., Nöthen M. M., Cichon S. (2004) Mol. Psychiatry 9, 203–207 [DOI] [PubMed] [Google Scholar]
- 17. Corvin A., McGhee K. A., Murphy K., Donohoe G., Nangle J. M., Schwaiger S., Kenny N., Clarke S., Meagher D., Quinn J., Scully P., Baldwin P., Browne D., Walsh C., Waddington J. L., Morris D. W., Gill M. (2007) Am. J. Med. Genet. B. Neuropsychiatr. Genet. 144B, 949–953 [DOI] [PubMed] [Google Scholar]
- 18. Yamada K., Ohnishi T., Hashimoto K., Ohba H., Iwayama-Shigeno Y., Toyoshima M., Okuno A., Takao H., Toyota T., Minabe Y., Nakamura K., Shimizu E., Itokawa M., Mori N., Iyo M., Yoshikawa T. (2005) Biol. Psychiatry 57, 1493–1503 [DOI] [PubMed] [Google Scholar]
- 19. Fallin M. D., Lasseter V. K., Avramopoulos D., Nicodemus K. K., Wolyniec P. S., McGrath J. A., Steel G., Nestadt G., Liang K. Y., Huganir R. L., Valle D., Pulver A. E. (2005) Am. J. Hum. Genet. 77, 918–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Liu Y. L., Fann C. S., Liu C. M., Chang C. C., Wu J. Y., Hung S. I., Liu S. K., Hsieh M. H., Hwang T. J., Chan H. Y., Chen J. J., Faraone S. V., Tsuang M. T., Chen W. J., Hwu H. G. (2006) Schizophr. Res. 87, 15–20 [DOI] [PubMed] [Google Scholar]
- 21. Vilella E., Costas J., Sanjuan J., Guitart M., De Diego Y., Carracedo A., Martorell L., Valero J., Labad A., De Frutos R., Nájera C., Moltó M. D., Toirac I., Guillamat R., Brunet A., Vallès V., Pérez L., Leon M., de Fonseca F. R., Phillips C., Torres M. (2008) J. Psychiatr. Res. 42, 278–288 [DOI] [PubMed] [Google Scholar]
- 22. Shi J., Badner J. A., Gershon E. S., Liu C. (2008) Schizophr. Res. 98, 89–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Korostishevsky M., Kremer I., Kaganovich M., Cholostoy A., Murad I., Muhaheed M., Bannoura I., Rietschel M., Dobrusin M., Bening-Abu-Shach U., Belmaker R. H., Maier W., Ebstein R. P., Navon R. (2006) Am. J. Med. Genet. B. Neuropsychiatr. Genet. 141B, 91–95 [DOI] [PubMed] [Google Scholar]
- 24. Hashimoto A., Yoshikawa M., Niwa A., Konno R. (2005) Brain. Res. 1033, 210–215 [DOI] [PubMed] [Google Scholar]
- 25. Almond S. L., Fradley R. L., Armstrong E. J., Heavens R. B., Rutter A. R., Newman R. J., Chiu C. S., Konno R., Hutson P. H., Brandon N. J. (2006) Mol. Cell. Neurosci. 32, 324–334 [DOI] [PubMed] [Google Scholar]
- 26. Maekawa M., Watanabe M., Yamaguchi S., Konno R., Hori Y. (2005) Neurosci. Res. 53, 34–38 [DOI] [PubMed] [Google Scholar]
- 27. Labrie V., Wang W., Barger S. W., Baker G. B., Roder J. C. (2010) Genes Brain Behav. 9, 11–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Konno R., Yasumura Y. (1983) Genetics 103, 277–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sasaki M., Konno R., Nishio M., Niwa A., Yasumura Y., Enami J. (1992) Biochim. Biophys. Acta 1139, 315–318 [DOI] [PubMed] [Google Scholar]
- 30. Hamase K., Konno R., Morikawa A., Zaitsu K. (2005) Biol. Pharm. Bull. 28, 1578–1584 [DOI] [PubMed] [Google Scholar]
- 31. Hashimoto K., Fujita Y., Horio M., Kunitachi S., Iyo M., Ferraris D., Tsukamoto T. (2009) Biol. Psychiatry 65, 1103–1106 [DOI] [PubMed] [Google Scholar]
- 32. Marino M. J., Knutsen L. J., Williams M. (2008) J. Med. Chem. 51, 1077–1107 [DOI] [PubMed] [Google Scholar]
- 33. Smith S. M., Uslaner J. M., Yao L., Mullins C. M., Surles N. O., Huszar S. L., McNaughton C. H., Pascarella D. M., Kandebo M., Hinchliffe R. M., Sparey T., Brandon N. J., Jones B., Venkatraman S., Young M. B., Sachs N., Jacobson M. A., Hutson P. H. (2009) J. Pharmacol. Exp. Ther. 328, 921–930 [DOI] [PubMed] [Google Scholar]
- 34. Ono K., Shishido Y., Park H. K., Kawazoe T., Iwana S., Chung S. P., Abou El-Magd R. M., Yorita K., Okano M., Watanabe T., Sano N., Bando Y., Arima K., Sakai T., Fukui K. (2009) J. Neural Transm. 116, 1335–1347 [DOI] [PubMed] [Google Scholar]
- 35. De Duve C., Baudhuin P. (1966) Physiol. Rev. 46, 323–357 [DOI] [PubMed] [Google Scholar]
- 36. Sacchi S., Bernasconi M., Martineau M., Mothet J. P., Ruzzene M., Pilone M. S., Pollegioni L., Molla G. (2008) J. Biol. Chem. 283, 22244–22256 [DOI] [PubMed] [Google Scholar]
- 37. Dresbach T., Hempelmann A., Spilker C., tom Dieck S., Altrock W. D., Zuschratter W., Garner C. C., Gundelfinger E. D. (2003) Mol. Cell. Neurosci. 23, 279–291 [DOI] [PubMed] [Google Scholar]
- 38. tom Dieck S., Sanmartí-Vila L., Langnaese K., Richter K., Kindler S., Soyke A., Wex H., Smalla K. H., Kämpf U., Fränzer J. T., Stumm M., Garner C. C., Gundelfinger E. D. (1998) J. Cell Biol. 142, 499–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wyneken U., Smalla K. H., Marengo J. J., Soto D., de la Cerda A., Tischmeyer W., Grimm R., Boeckers T. M., Wolf G., Orrego F., Gundelfinger E. D. (2001) Neuroscience 102, 65–74 [DOI] [PubMed] [Google Scholar]
- 40. Carlin R. K., Grab D. J., Cohen R. S., Siekevitz P. (1980) J. Cell Biol. 86, 831–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Horiike K., Tojo H., Arai R., Nozaki M., Maeda T. (1994) Brain Res. 652, 297–303 [DOI] [PubMed] [Google Scholar]
- 42. Wang L. Z., Zhu X. Z. (2003) Acta Pharmacol. Sin. 24, 965–974 [PubMed] [Google Scholar]
- 43. Shao Z., Kamboj A., Anderson C. M. (2009) J. Neurosci. Res. 87, 2520–2530 [DOI] [PubMed] [Google Scholar]
- 44. Kartvelishvily E., Shleper M., Balan L., Dumin E., Wolosker H. (2006) J. Biol. Chem. 281, 14151–14162 [DOI] [PubMed] [Google Scholar]
- 45. Wang Y., Liu X., Biederer T., Südhof T. C. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 14464–14469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li Y., Asuri S., Rebhun J. F., Castro A. F., Paranavitana N. C., Quilliam L. A. (2006) J. Biol. Chem. 281, 2506–2514 [DOI] [PubMed] [Google Scholar]
- 47. Wang X., Kibschull M., Laue M. M., Lichte B., Petrasch-Parwez E., Kilimann M. W. (1999) J. Cell Biol. 147, 151–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chin L. S., Nugent R. D., Raynor M. C., Vavalle J. P., Li L. (2000) J. Biol. Chem. 275, 1191–1200 [DOI] [PubMed] [Google Scholar]
- 49. Cristiano L., Bernardo A., Cerù M. P. (2001) J. Neurocytol. 30, 671–683 [DOI] [PubMed] [Google Scholar]
- 50. Usuda N., Yokota S., Hashimoto T., Nagata T. (1986) J. Histochem. Cytochem. 34, 1709–1718 [DOI] [PubMed] [Google Scholar]
- 51. Wanders R. J., Waterham H. R. (2006) Annu. Rev. Biochem. 75, 295–332 [DOI] [PubMed] [Google Scholar]
- 52. Moreno S., Nardacci R., Cimini A., Cerù M. P. (1999) J. Neurocytol. 28, 169–185 [DOI] [PubMed] [Google Scholar]
- 53. Wang X., Hu B., Zieba A., Neumann N. G., Kasper-Sonnenberg M., Honsbein A., Hultqvist G., Conze T., Witt W., Limbach C., Geitmann M., Danielson H., Kolarow R., Niemann G., Lessmann V., Kilimann M. W. (2009) J. Neurosci. 29, 12584–12596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Garner C. C., Kindler S., Gundelfinger E. D. (2000) Curr. Opin. Neurobiol. 10, 321–327 [DOI] [PubMed] [Google Scholar]
- 55. Owald D., Sigrist S. J. (2009) Curr. Opin. Neurobiol. 19, 311–318 [DOI] [PubMed] [Google Scholar]
- 56. Fenster S. D., Chung W. J., Zhai R., Cases-Langhoff C., Voss B., Garner A. M., Kaempf U., Kindler S., Gundelfinger E. D., Garner C. C. (2000) Neuron 25, 203–214 [DOI] [PubMed] [Google Scholar]
- 57. Weidenhofer J., Scott R. J., Tooney P. A. (2009) J. Psychiatr. Res. 43, 282–290 [DOI] [PubMed] [Google Scholar]
- 58. Weidenhofer J., Bowden N. A., Scott R. J., Tooney P. A. (2006) Mol. Cell. Neurosci. 31, 243–250 [DOI] [PubMed] [Google Scholar]
- 59. Fukasawa Y., Segawa H., Kim J. Y., Chairoungdua A., Kim D. K., Matsuo H., Cha S. H., Endou H., Kanai Y. (2000) J. Biol. Chem. 275, 9690–9698 [DOI] [PubMed] [Google Scholar]
- 60. Helboe L., Egebjerg J., Møller M., Thomsen C. (2003) Eur. J. Neurosci. 18, 2227–2238 [DOI] [PubMed] [Google Scholar]
- 61. Matsuo H., Kanai Y., Tokunaga M., Nakata T., Chairoungdua A., Ishimine H., Tsukada S., Ooigawa H., Nawashiro H., Kobayashi Y., Fukuda J., Endou H. (2004) Neurosci. Lett. 358, 123–126 [DOI] [PubMed] [Google Scholar]
- 62. Nakauchi J., Matsuo H., Kim D. K., Goto A., Chairoungdua A., Cha S. H., Inatomi J., Shiokawa Y., Yamaguchi K., Saito I., Endou H., Kanai Y. (2000) Neurosci. Lett. 287, 231–235 [DOI] [PubMed] [Google Scholar]
- 63. Rutter A. R., Fradley R. L., Garrett E. M., Chapman K. L., Lawrence J. M., Rosahl T. W., Patel S. (2007) Eur. J. Neurosci. 25, 1757–1766 [DOI] [PubMed] [Google Scholar]
- 64. Rosenberg D., Kartvelishvily E., Shleper M., Klinker C. M., Bowser M. T., Wolosker H. (2010) FASEB J. 24, 2951–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kapoor R., Lim K. S., Cheng A., Garrick T., Kapoor V. (2006) Brain Res. 1106, 205–210 [DOI] [PubMed] [Google Scholar]
- 66. Neims A. H., Zieverink W. D., Smilack J. D. (1966) J. Neurochem. 13, 163–168 [DOI] [PubMed] [Google Scholar]
- 67. Weimar W. R., Neims A. H. (1977) J. Neurochem. 29, 649–656 [DOI] [PubMed] [Google Scholar]
- 68. Horiike K., Tojo H., Arai R., Yamano T., Nozaki M., Maeda T. (1987) Brain. Res. Bull. 19, 587–596 [DOI] [PubMed] [Google Scholar]
- 69. Arnold G., Liscum L., Holtzman E. (1979) J. Histochem. Cytochem. 27, 735–745 [DOI] [PubMed] [Google Scholar]
- 70. Altrock W. D., tom Dieck S., Sokolov M., Meyer A. C., Sigler A., Brakebusch C., Fässler R., Richter K., Boeckers T. M., Potschka H., Brandt C., Löscher W., Grimberg D., Dresbach T., Hempelmann A., Hassan H., Balschun D., Frey J. U., Brandstätter J. H., Garner C. C., Rosenmund C., Gundelfinger E. D. (2003) Neuron 37, 787–800 [DOI] [PubMed] [Google Scholar]
- 71. Ghiglieri V., Picconi B., Sgobio C., Bagetta V., Barone I., Paillè V., Di Filippo M., Polli F., Gardoni F., Altrock W., Gundelfinger E. D., De Sarro G., Bernardi G., Ammassari-Teule M., Di Luca M., Calabresi P. (2009) Eur. J. Neurosci. 29, 1979–1993 [DOI] [PubMed] [Google Scholar]
- 72. Bortolotto Z. A., Collingridge G. L. (1998) Neuropharmacology 37, 535–544 [DOI] [PubMed] [Google Scholar]
- 73. Wolosker H. (2006) Sci. STKE 2006, pe41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.