Abstract
γ-Glutamyl transpeptidase (GGT) is a heterodimeric membrane enzyme that catalyzes the cleavage of extracellular glutathione and other γ-glutamyl-containing compounds. GGT is synthesized as a single polypeptide (propeptide) that undergoes autocatalytic cleavage, which results in the formation of the large and small subunits that compose the mature enzyme. GGT is extensively N-glycosylated, yet the functional consequences of this modification are unclear. We investigated the effect of N-glycosylation on the kinetic behavior, stability, and functional maturation of GGT. Using site-directed mutagenesis, we confirmed that all seven N-glycosylation sites on human GGT are modified by N-glycans. Comparative enzyme kinetic analyses revealed that single substitutions are functionally tolerated, although the N95Q mutation resulted in a marked decrease in the cleavage efficiency of the propeptide. However, each of the single site mutants exhibited decreased thermal stability relative to wild-type GGT. Combined mutagenesis of all N-glycosylation sites resulted in the accumulation of the inactive propeptide form of the enzyme. Use of N-glycosylation inhibitors demonstrated that binding of the core N-glycans, not their subsequent processing, is the critical glycosylation event governing the autocleavage of GGT. Although N-glycosylation is necessary for maturation of the propeptide, enzymatic deglycosylation of the mature wild-type GGT does not substantially impact either the kinetic behavior or thermal stability of the fully processed human enzyme. These findings are the first to establish that co-translational N-glycosylation of human GGT is required for the proper folding and subsequent cleavage of the nascent propeptide, although retention of these N-glycans is not necessary for maintaining either the function or structural stability of the mature enzyme.
Keywords: Cell Surface Enzymes, Enzyme Processing, Glycoprotein, Glycoprotein Structure, Glycosylation, Post-translational Modification, Protein Folding, GGT, gamma-Glutamyl Transpeptidase
Introduction
In humans, γ-glutamyl transpeptidase (GGT,2 EC 2.3.2.2) is a single pass type II membrane glycoprotein that localizes to the apical surfaces of cells that align ducts and glands throughout the body, where it functions to initiate the breakdown of extracellular glutathione into its constituent amino acids, glutamate, cysteine, and glycine (1–3). This activity allows the body to reclaim the amino acids that would otherwise be excreted as intact glutathione (2). Knocking out GGT in mice results in gross postnatal mortality within 10–18 weeks, whereas overexpression and mislocalization of GGT in cancer and other diseases have been shown to contribute to drug resistance and disease progression (4–7). Human GGT is a member of the N-terminal nucleophile hydrolase family and is initially translated as an inactive 569-amino acid propeptide that, during the course of its maturation, is cleaved into the mature heterodimeric form of the active enzyme (8–10). The resolution of the propeptide into the large (amino acids 1–380) and small (amino acids 381–569) subunits is predicted to occur through an autocatalytic cleavage event that is dependent on Thr-381 (9, 11, 12). However, when human GGT is expressed in bacteria, the enzyme fails to autocleave and achieve proper functional maturity, suggesting that post-translational modifications are also required for active heterodimer formation (13).
It has long been appreciated that GGT exhibits a heterogeneous pattern of glycosylation that varies in a tissue-specific manner, yet the role that N-glycosylation plays in modulating the functional maturation and kinetic behavior of the enzyme has yet to be ascertained (14, 15). Human GGT possesses seven potential N-glycosylation sites (sequon NX(S/T), X≠P): six in the large subunit and one in the small subunit. Previous analyses conducted on GGT isolated from normal human kidney tissue have demonstrated that all seven of these sites are capable of being glycosylated (16). Although variations in the compositional features of the glycans themselves have been shown to modulate the reaction kinetics of the mature heterodimer, little is known regarding the impact that glycosylation plays in governing the structural integrity and autocatalytic cleavage of the nascent enzyme (17). N-Glycosylation has been shown to modulate the intramolecular structural properties of the proteins to which they are conjugated by contributing to their solubility, stability, and protease resistance (18, 19). Although the proper folding and maturation of nascent polypeptides is primarily dictated by their amino acid sequence, the co-translational conjugation and subsequent processing of N-glycans on glycoproteins in the endoplasmic reticulum (ER) help to govern the proper rate and spatial order of their conformational maturity. This property is often mediated by resident ER lectin chaperones that bind to the immature glycoproteins and shepherd them through the folding process or, when this process fails, direct improperly folded glycoproteins for targeted degradation (20, 21). Thus, perturbation of the N-glycosylation pattern of a protein can have dramatic effects on its maturation and functional fate.
Therefore, in an effort to better understand the contextual role of N-glycosylation on human GGT, we conducted a series of studies to evaluate the manner in which these modifications affect the functional maturation, kinetic activity, and structural integrity of the enzyme. Using a combination of mutagenesis and inhibitors of glycosylation, we confirm that all seven N-glycosylation sites on human GGT are utilized in HEK293 cells. We found that no single glycosylation site is strictly required for formation of the mature active enzyme, yet each of the single site mutations decreased the thermal stability of the mature heterodimeric enzyme. Moreover, in the absence of glycosylation at Asn-95, the enzyme exhibited a marked defect in its autocatalytic cleavage efficiency. In silico modeling of the glycosylated human GGT propeptide predicts that this defect may be attributable to a loss of stabilizing intramolecular hydrogen bonding between the glycan on Asn-95 and residues within a structurally adjacent helix-turn-helix motif that typically promotes disulfide bond formation between cysteines 50 and 74, an event that has been shown previously to be essential for the autocatalytic cleavage of mammalian GGT (10). When N-glycosylation was eliminated throughout the entire protein, GGT exhibited no enzymatic activity and failed to mature into its constituent subunits, accumulating in its inactive propeptide form. Conversely, enzymatic deglycosylation of fully matured human GGT demonstrated that N-glycans are largely dispensable for the catalytic activity and structural stability of the heterodimeric enzyme. These findings indicate that the primary role of N-glycosylation on human GGT is to govern its proper folding and autocatalytic cleavage rather than acting to modulate the activity or inherent stability of the mature enzyme. In addition to their functional implications for the enzyme in vivo, these new insights will assist in the strategic design of refined strategies aimed at achieving the long anticipated crystal structure of human GGT.
EXPERIMENTAL PROCEDURES
Construction of Plasmids Bearing GGT N-Glycosylation Mutants for Expression in HEK293 Cells and Pichia pastoris
Full-length wild-type human GGT (EC 2.3.2.2) cDNA in a pcDNA3.1(+) plasmid served as the template for PCR mutagenesis (2). Asparagine residues within GGT that were predicted (NetNGlyc 1.0 Server) to undergo glycosylation (Asn-95, -120, -230, -266, -297, -344, and -511) were individually targeted for mutagenesis into glutamine codons with the QuikChange Site-directed Mutagenesis kit from Stratagene (La Jolla, CA), according to the manufacturer's protocol with the primer sets listed in supplemental Table 1. The total N-glycosylation knock-out mutant, GGT(N95Q,N120Q,N230Q, N266Q,N297Q,N344Q,N511Q), was generated by the iterative addition of point mutations within the plasmid containing the N297Q mutation, using the QuikChange Lightning Multi Site-directed Mutagenesis kit from Stratagene. The product from each round of mutagenesis was sequenced (DNA Sequencing Facility, Oklahoma Medical Research Foundation, Oklahoma City, OK), which confirmed the point mutations. The construction of the plasmid for the expression of the soluble form of human GGT in a P. pastoris expression vector has been described previously (22). The total N-glycosylation knock-out mutant for expression in P. pastoris was constructed with the primers MBW-P1 and MBW-P2 (see supplemental Table 1) and the total knock-out construct described above for full-length GGT as the template for PCR amplification. The PCR product was cloned into the yeast expression plasmid as described previously (22). The GGT(G61L), GGT(G62L), and GGT(G61LG62L) mutants were constructed, using the primer sets listed in supplemental Table 1 and the QuikChange Site-directed Mutagenesis kit from Stratagene as described above.
Transient Expression of GGT N-Glycosylation Mutants in HEK293 Cells
HEK293 cells (human embryonic kidney, ATCC CRL-1573) were plated at 106 cells/P100 plate and cultured in 10 ml of Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum and penicillin/streptomycin (50 units/ml, 50 μg/ml) at 37 °C in the presence of 5% CO2. The following day, the cells were transfected with 10 μg of the appropriate expression plasmid, according to the calcium phosphate method (23). After 3 h, the transfection medium was removed, and 10 ml of fresh complete growth medium was added to each plate of cells. Transfectant cells were cultured at 37 °C in the presence of 5% CO2 until they were harvested.
Transfectant cells were resuspended in ice-cold phosphate-buffered saline (PBS) (10 mm Na2HPO4, 1.8 mm KH2PO4, 137 mm NaCl, 2.7 mm KCl, pH 7.4) in the presence of protease inhibitors (1 μg/ml aprotinin, 1 μm leupeptin) by scraping with a rubber policeman. Cells were pelleted by low speed centrifugation (1000 × g) at 4 °C for 5 min and then resuspended in ice-cold PBS containing 0.5% CHAPS and protease inhibitors. The suspensions were incubated on a rotary wheel at 4 °C for 30 min, and the insoluble material was pelleted at 15,000 × g for 15 min. The concentration of protein in the soluble fraction was determined with the Pierce BCA protein assay kit (ThermoScientific, Rockford, IL).
SDS-PAGE and Western Analysis
Cell lysates were incubated at 100 °C for 10 min in Laemmli sample buffer (2% SDS, 5% glycerol, 5% 2-mercaptoethanol, 0.002% bromphenol blue, 62.5 mm Tris-HCl, pH 6.8) and resolved on either 8 or 10% SDS-polyacrylamide gels. Resolved proteins were then electroblotted onto nitrocellulose membranes and blocked for 30 min at room temperature with Tris-buffered saline solution containing 0.1% Tween 20 (TBST) and 3% dry milk. Western blotting was then conducted using the appropriate primary antibodies diluted in TBST followed by incubation with HRP-conjugated secondary antibodies. The blots were washed extensively in TBST and visualized by chemiluminescence according to the manufacturer's protocol (ECL Plus, GE Healthcare). For Western analyses against GGT, the large subunit of the heterodimer was detected by the GGT129 antibody (3) at a 1:1,000-dilution in TBST. The GGT small subunit was detected by the GGT1 (M01) antibody (Abnova, Taipei City, Taiwan) at a 1:5,000-dilution in TBST. Additional immunoblots were carried out against glyceraldehydes-3-phosphate dehydrogenase (anti-GAPDH, Imgenex, San Diego) at a 1:5,000 dilution and calnexin (SPA-860, Ann Arbor, MI) at a 1:5,000 dilution, according to the same protocols.
Deglycosylation of GGT by Peptide:N-Glycosidase F
Aliquots of cell lysates containing 35 μg of total protein were diluted to 100 μl with PBS + 0.5% CHAPS and supplemented with SDS to 0.5%. The samples were then heat-denatured at 100 °C for 10 min and cooled to room temperature. Each sample was then supplemented with protease inhibitors (1 μg/ml aprotinin, 1 μm leupeptin) and 50 units of peptide:N-glycosidase F (PNGaseF, EC 3.5.1.52; New England Biolabs, Ipswich, MA) and incubated for 18 h at 37 °C. The reactions were stopped by boiling in Laemmli sample buffer, and the deglycosylated samples were then subjected to SDS-PAGE and Western analyses as described above.
For densitometric analyses, transfectant cell extracts were incubated with PNGaseF as described above to eliminate differences in the density of the banding patterns imposed by N-glycan microheterogeneity. Immunoblots against the small subunit of GGT were conducted as described above, and digital scans of triplicate blots were subjected to quantitative band density measurements, using the ImageJ processing software developed by the National Institutes of Health. The reported expression unit values represent an average of the measured densities from three independent blots relative to those calculated for the wild-type GGT standard.
GGT Activity Assays
The kinetic assays were conducted in a 96-well plate format as described previously (22). The same assay buffer was used for both the hydrolysis reaction and the transpeptidation reaction. The assay buffer contained 100 mm Na2HPO4, 3.2 mm KCl, 1.8 mm KH2PO4, and 27.5 mm NaCl, pH 7.4. The concentration of the substrate for the transpeptidation reaction, l-γ-glutamyl-paranitroanilide (l-GpNA, Sigma), was varied from 0.25 to 3 mm. Reactions were carried out in the presence of 40 mm glycyl-glycine (Gly-Gly, Sigma), which acted as the acceptor substrate. To initiate the transpeptidation reaction, 5 μg of cell extract were added to each well. The cleavage of l-GpNA was carried out at 37 °C and monitored continuously at 405 nm by a Bio-Rad model 680 microplate reader with Microplate Manager 5.2 software (Bio-Rad). For the hydrolysis reactions, the concentration of the d-γ-glutamyl-paranitroanilide (d-GpNA, Bachem, Torrance, CA) substrate was varied from 0.25 to 3 mm d-GpNA in the absence of an acceptor substrate. The reaction was initiated with the addition of 15 μg of the appropriate cell extract. The cleavage of d-GpNA was monitored as described above for l-GpNA. For both the transpeptidation and hydrolysis reactions, 1 unit of GGT activity was defined as the amount of enzyme that released 1 μmol of paranitroaniline/min at 37 °C, pH 7.4. For thermal stability analysis, 5 μg of lysate was incubated for 15 min at 37–77 °C, allowed to re-equilibrate to 37 °C, and then assayed for activity.
Data Analysis
Samples in each experiment were run in triplicate. Each GGT glycosylation mutant was evaluated in two or more independent experiments. Data adhering to Michaelis-Menten kinetics were fitted to Equation 1.
 |
Graphs, averages, and standard deviations were calculated using Prism GraphPad software (San Diego). Data are presented as the mean ± S.D. of at least three independent determinations. The statistical relevance of each kinetic parameter was determined by unpaired, two-tailed Student's t tests (p < 0.05 categorized as statistically significant).
Inhibition of Glycosylation
P35 plates seeded with 2 × 105 HEK293 cells/plate were transiently transfected with 2 μg of the wild-type GGT expression plasmid as described above. After 3 h, the medium on each plate was replaced with complete DMEM containing either tunicamycin (0.025–1.0 μg/ml), castanospermine (100 or 200 μg/ml), or deoxynojirimycin (1 mm), and the cells were cultured in the presence of the glycosylation inhibitors for 42 h prior to harvesting.
Immunoprecipitations
CHAPS-solubilized extracts were rotated for 30 min at 4 °C in a 1.5-ml tube rotator. The lysate was then cleared by centrifugation at 13,000 × g in a microcentrifuge for 15 min at 4 °C. The protein concentrations of the clarified lysates were then measured by the BCA assay. 500 μg of total protein from each sample were diluted to a 500-μl final volume with PBS, 0.5% CHAPS buffer, and 3 μl of anti-calnexin (AF8 (24)) antibody was added to each. The lysates were incubated with the anti-calnexin antibody for 45 min at 4 °C on a 1.5-ml tube rotator prior to adding 20 μl of pre-washed, 0.1% BSA-blocked protein G-Sepharose beads (GE Healthcare) to each sample. The suspensions were incubated at 4 °C on the tube rotator for an additional 1.5 h. The beads were then washed three times with 1 ml of PBS, 0.5% CHAPS buffer, and the immunoprecipitated material in each tube was eluted with 50 μl of Laemmli sample buffer at 100 °C. 20 μl from each immunoprecipitate was subjected to SDS-PAGE and immunoblotting with either anti-calnexin or anti-GGT antibodies as described above.
Enzyme Activity of Deglycosylated GGT
For the deglycosylation of human GGT expressed in HEK293 cells, 20 μg of total protein from cell extracts was incubated in the presence or absence of an endoglycosidase mixture containing 50 units of PNGaseF and 100 units of endoglycosidase Hf (New England Biolabs, Ipswich, MA) in PBS + 0.5% CHAPS at 37 °C for up to 48 h. For the deglycosylation of human GGT lacking the transmembrane domain, the enzyme was expressed in P. pastoris and isolated as described previously (22). The specific activity of the purified enzyme from P. pastoris was 406.5 units/mg. Duplicate 585-milliunit aliquots of GGT were incubated in PBS + 0.5% CHAPS at 37 °C for 18 h in the presence or absence of the endoglycosidase mixture described above. Kinetic assays were then performed on 4-milliunit aliquots from each reaction as described above. For the thermal stability assays, 4-milliunit aliquots of glycosylated or deglycosylated GGT were incubated for 15-min intervals at 37–77 °C, allowed to re-equilibrate to 37 °C, and then assayed for transpeptidation activity with 3 mm l-GpNA in the presence of 40 mm glycyl-glycine as described above.
Human GGT Structural Modeling
A model of human GGT in its precursor form (residues 36–569) was built using the MODELLER software (25). The modeling was completed in four iterations, based on two crystallographic templates (Protein Data Bank codes 2E0W (26) and 3A75 (27)). An initial set of 500 models was built and evaluated based on the DOPE scoring function available as part of the MODELLER program (see supplemental Fig. 1) (28). Subsequent iterations of modeling refined specific loop regions and incorporated the top scoring model from the previous iteration as an additional modeling template.
Glycans were built with the LEaP module of the Amber suite of programs and manually linked to Asn-95, although some glycan torsions were manually altered to avoid clashes with the protein (29). Once the glycosylated model was complete, the system was solvated in a truncated TIP3P water octahedron and charge-neutralized with Na+ ions. A 15-Å solvent buffer surrounded the final system. Molecular dynamics (MD) simulations were carried out with the particle mesh Ewald (PME) method in Amber using the ff99SBildn force field for the protein (30) and the GLYCAM06 force field for the glycans (31). Prior to simulation, the system was energy-minimized with 500 steps of steepest descent followed by 1500 steps of conjugate gradient minimization.
MD production runs were preceded by two stages of equilibration. In the first stage, all protein and glycan atoms were restrained in their energy-minimized positions, whereas the water molecules were free to equilibrate around the surface of the system for 100 ps. During this stage, the temperature of the system was raised from 0 to 300 K with a heat bath coupling constant of 2.0 ps. Following water equilibration, the restraints on the glycan were released to allow the removal of any strain in the manually built chain. After 500 ps of glycan equilibration, all restraints were released, and several nanoseconds of MD were run with a 2.0-ps time step using the SHAKE algorithm at a constant 300 K and 1 atm with a pressure relaxation time of 2.0 ps.
Three mutant GGT models (G61L, G62L, and G61L,G62L substitutions) were created based on the equilibrated glycosylated form of the wild-type GGT propeptide model. Fifty models of each mutation were built with MODELLER, and the best model for each mutation was selected based on the calculated DOPE scores. For each mutant model, a 10-ns MD simulation was run to evaluate any changes in the interactions between the glycan and the GGT protein. In particular, stable hydrogen bonds that were observed between the glycan and protein in the wild-type MD simulation were tracked in the simulations of the mutant models to determine whether any of the mutations destabilized the protein/glycan interaction.
RESULTS
SDS-PAGE Analysis of Single N-Glycosylation Site Mutants of Human GGT
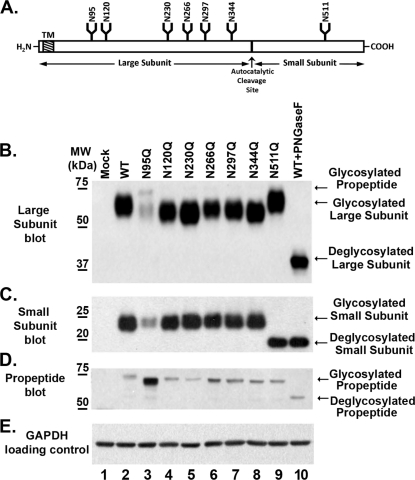
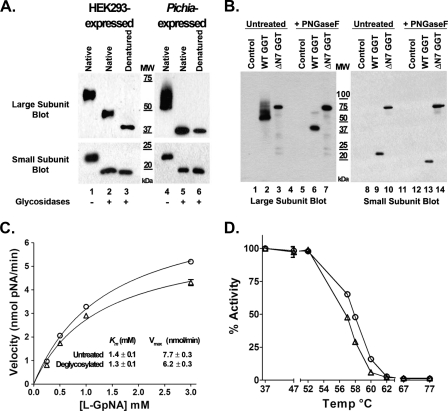
The primary amino acid sequence of human GGT contains seven consensus N-glycosylation sites (Fig. 1A). The large subunit of GGT contains six of these sites (Asn-95, -120, -230, -266, -297, and -344), and the small subunit contains a single site (Asn-511) (Fig. 1A). To determine whether each of these sites is glycosylated, all seven asparagine residues were independently converted into glutamines by site-directed mutagenesis. The resulting mutant GGT alleles were then transiently transfected into HEK293 cells and analyzed by SDS-PAGE (Fig. 1, B–D). HEK293 cells do not endogenously express GGT (Fig. 1B, lane 1). When wild-type GGT was expressed in HEK293 cells, it resolved as a heterodimer with a large and small subunit with molecular masses of 64 and 22 kDa, respectively. Western analysis revealed that each of the GGT alleles bearing single N-glycosylation site mutations in the large subunit migrated faster than the large subunit from the wild-type protein (Fig. 1B, lanes 2–8). These data indicate that each site in the large subunit was glycosylated. None of the single substitutions in the large subunit altered the migration pattern of the small subunit of GGT relative to the wild-type allele (Fig. 1C, lanes 2–8). The N511Q mutation, which deleted the single N-glycosylation site in the small subunit of GGT, resulted in a small subunit that migrated faster than that from wild-type GGT (Fig. 1C, lanes 2 and 9), whereas the large subunit in the N511Q mutant protein migrated identically to that of the wild-type allele (Fig. 1B, lanes 2 and 9).
FIGURE 1.
All seven N-glycosylation sites on human GGT are glycosylated in HEK293 cells. A, diagrammatic representation of the human GGT propeptide and its seven potential N-glycosylation sites. The positions of the transmembrane domain (TM), N-glycosylation consensus sites (N), propeptide cleavage site (↑), and relative sizes of the large and small subunits of the mature enzyme are indicated. B–E, Western analyses against wild-type GGT and single N-glycosylation mutants expressed in HEK293 cells. Total cell lysates from HEK293 cells transfected with either empty vector (lane 1), wild-type GGT (lane 2), or mutant GGT alleles (lanes 3–9) were resolved by SDS-PAGE and electroblotted onto nitrocellulose. Fully deglycosylated (PNGaseF-treated) wild-type GGT expressed in HEK293 cells is included as a visual reference (lane 10). Immunoblotting was conducted using antibodies specific to either the large (B) or small (C and D) subunits of GGT. An immunoblot against GAPDH (E) is included as a loading control. MW, molecular weight markers.
The substitution at Asn-95 in the large subunit of GGT resulted in a marked decrease (∼8-fold) in the expression levels of both the large and small subunits of the enzyme (Fig. 1, B and C, lane 3). Relative to the other mutant alleles, this mutant also exhibited an increase in the amount of the immature (i.e. uncleaved) propeptide form of GGT, which was unambiguously detected by the small subunit antibody at an apparent molecular mass of 73 kDa. These observations suggest that glycosylation at this site facilitates the rate or extent of the autocatalytic cleavage of GGT (Fig. 1D). Like the N95Q mutant, each of the single site mutants resulted in a propeptide band that migrated more rapidly than the wild-type glycosylated propeptide (73 versus 75 kDa, respectively). These data are consistent with the loss of a single oligosaccharide on each propeptide (Fig. 1D).
A comparison of the migration pattern of the large subunit from each of the N-glycosylation site mutants to the fully deglycosylated large subunit from wild-type GGT demonstrated that mutating individual glycosylation sites in the large subunit did not inhibit N-glycosylation at the remaining sites (Fig. 1B, lanes 3–10). In addition, this analysis revealed that the Asn-511 site is the only site on the small subunit modified by N-glycosylation, as the small subunit of the N511Q mutant migrated identically to the fully deglycosylated small subunit from wild-type GGT (Fig. 1C, lanes 9 and 10). These results confirm that each of the seven predicted N-glycosylation sites on human GGT is modified by carbohydrates in HEK293 cells. When equal expression units of GGT (see supplemental Fig. 2A) were loaded in each well and subjected to SDS-PAGE analysis, the large subunits of the mutant enzymes bearing substitutions at the Asn-95 and -266 sites consistently exhibited a lesser migrational shift than those bearing mutations at the remaining four sites in the large subunit, whereas the N297Q mutation exhibited an intermediate migrational shift (see supplemental Fig. 2B). This observation suggests that the mature N-glycans that typically occupy these sites are smaller or compositionally distinct from those at the other sites.
Effects of Single N-Glycosylation Site Mutations on the Enzyme Activity of Human GGT
To investigate whether blocking carbohydrate occupancy at individual N-glycosylation sites alters the enzymatic activity of GGT, extracts from HEK293 cells expressing either wild-type or single site mutant GGT alleles were assayed in both transpeptidation and hydrolysis reactions, using the glutathione substrate analogs l-GpNA and d-GpNA, respectively. Initial assays were conducted with transfected cell extracts standardized for total protein. A refined set of experiments, using normalized GGT protein levels (i.e. expression units, based on relative levels of the small subunit as determined by immunoblot, see supplemental Fig. 2A), were then conducted to gain insight into the comparative reaction kinetics exhibited by the mutant enzymes (Table 1). The enzyme kinetic parameters were fitted to standard Michaelis-Menten equations. No GGT activity was detected in mock-transfected extracts, consistent with our inability to detect GGT expression in the host HEK293 cell line (Fig. 1B).
TABLE 1.
Kinetic parameters for wild-type and N-glycosylation mutants of human GGT
Data represent the mean ± S.D.
| GGT allele | Transpeptidation |
Hydrolysis |
||
|---|---|---|---|---|
| Km | Vmaxa | Km | Vmaxa | |
| mm | nmol/min | μm | nmol/min | |
| Wild type | 1.3 ± 0.1 | 9.5 ± 0.2 | 159.0 ± 18 | 0.37 ± 0.01 |
| N95Q | 1.2 ± 0.1 | 6.2 ± 0.2 | 150.1 ± 20 | 0.25 ± 0.01 |
| N120Q | 1.3 ± 0.1 | 7.5 ± 0.7 | 169.6 ± 13 | 0.29 ± 0.03 |
| N230Q | 1.3 ± 0.1 | 6.3 ± 0.8 | 188.4 ± 20 | 0.29 ± 0.04 |
| N266Q | 1.1 ± 0.1 | 8.2 ± 0.2 | 179.3 ± 14 | 0.31 ± 0.01 |
| N297Q | 1.3 ± 0.1 | 8.6 ± 1.3 | 184.2 ± 27 | 0.31 ± 0.05 |
| N344Q | 1.3 ± 0.1 | 6.8 ± 0.5 | 152.0 ± 27 | 0.23 ± 0.02 |
| N511Q | 1.3 ± 0.1 | 7.0 ± 0.8 | 162.6 ± 19 | 0.26 ± 0.03 |
| ΔN7 | NDb | ND | ND | ND |
a Equivalent amounts of mature enzyme (based on protein level of the small subunit) were used for each sample assay. Units are nanomoles of paranitroaniline per min. 1 nmol of paranitroaniline/min is equivalent to 1 milliunit of GGT activity.
b ND indicates no detectable GGT activity in the specified assay.
The transpeptidation reaction, in which GGT cleaves l-GpNA and transfers the γ-glutamyl group to the dipeptide acceptor glycyl-glycine (Gly-Gly), is the canonical assay for quantifying GGT activity. Using variable concentrations of l-GpNA and a constant concentration of 40 mm Gly-Gly, the Km value for the transpeptidation reaction catalyzed by wild-type GGT expressed in HEK293 cells was 1.3 ± 0.1 mm, whereas the Vmax value for wild-type GGT in the transpeptidation reaction was 9.5 ± 0.2 milliunits of GGT activity per expression unit of protein. The Km value for l-GpNA in the transpeptidation reaction for wild-type GGT expressed in HEK293 cells is consistent with the Km values for l-GpNA reported previously for human GGT isolated from a variety of sources when assayed under similar conditions (22, 32, 33).
The mutant alleles exhibited Km values for the transpeptidation reaction that were not significantly different from that of wild-type GGT (Table 1). Despite the increased prevalence of inactive propeptide and the ∼8-fold decrease in the amount of the mature heterodimer, the subpopulation of the N95Q mutant of GGT that reached functional maturity exhibited a Michaelis constant (Km = 1.2 ± 0.06) that was equivalent to that of the wild-type allele in the transpeptidation reaction (Fig. 1B and Table 1). Despite their conserved Michaelis constants, single N-glycosylation site substitutions generally resulted in a decreased maximal rate of l-GpNA turnover (Vmax), with only one mutant, N297Q, exhibiting a kinetic profile that was comparable with the native enzyme (Table 1).
In vivo, the primary reaction catalyzed by GGT is the hydrolysis reaction, in which water, rather than amino acids, acts as the acceptor molecule during the cleavage of the γ-glutamyl bond of glutathione (34, 35). The l-stereoisomer of GpNA can act as a weak acceptor in the transpeptidation reaction (1, 36, 37). d-GpNA does not act as an acceptor and therefore was used to specifically examine the effects of the site mutations on the kinetic parameters of GGT in the hydrolysis reaction. The Km value of the wild-type allele of GGT for d-GpNA was 159.0 ± 18 μm (Table 1), whereas the Vmax value for the hydrolysis reaction was 0.37 ± 0.01 milliunits of GGT activity per expression unit. As observed for the transpeptidation reaction, the Km values for each of the single site mutants in the hydrolysis reaction did not differ significantly from that measured for wild-type GGT (Table 1), and each of the individual glutamine substitutions, except for the one at asparagine 297, decreased the rate of d-GpNA hydrolysis, as observed by their lower Vmax values relative to wild-type GGT. These data demonstrate that the absence of any single carbohydrate modification affects both the transpeptidation and hydrolysis reactions in a similar fashion. All of the glycosylation sites, with the exception of Asn-297, must be glycosylated to produce a mature enzyme that exhibits maximal activity.
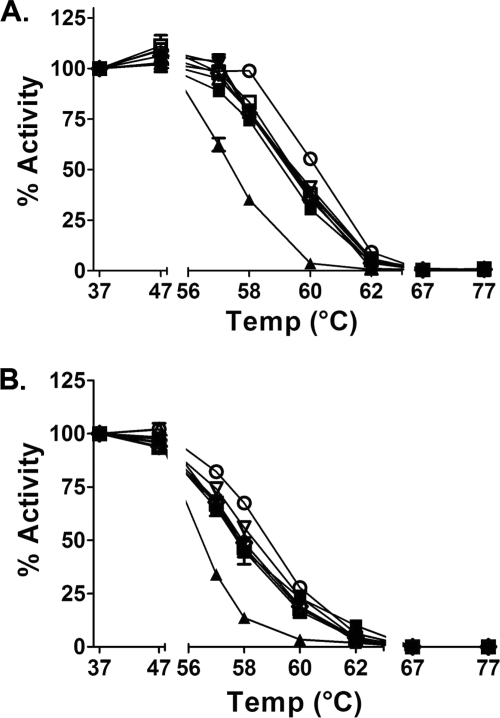
Thermal Stability Analysis of N-Glycosylation Site Mutants
To discern whether site occupancy at individual N-glycosylation sites contributes to the structural integrity of the enzyme, we evaluated the thermal stability of the mutant alleles relative to wild-type GGT. In both the transpeptidation and hydrolysis reactions, wild-type GGT exhibited full activity up to 47 °C (Fig. 2). At temperatures greater than 62 °C, the wild-type enzyme was rendered completely inactive. Performing these analyses on the glycosylation site mutants demonstrated that, like wild-type GGT, the mutant enzymes retained full activity up to 47 °C. However, at temperatures exceeding 47 °C, all of the mutants exhibited decreased thermal stability relative to the wild-type control, with the N297Q mutant exhibiting the least sensitivity and the N511Q mutant exhibiting the greatest sensitivity to increased temperature (Fig. 2). Within the transpeptidation data set (Fig. 2A), the most pronounced distinction in the thermal stability patterns was observed at 58 °C, where the residual activity was 98.8% for wild-type GGT, 77–83% for the large subunit glycosylation mutants (N95Q, N120Q, N230Q, N266Q, N297Q, and N344Q), and only 35% activity for the small subunit glycosylation mutant (N511Q). A similar trend was observed in the hydrolysis reaction (Fig. 2B). At 58 °C, the residual d-GpNA hydrolytic activity was 69% for wild-type GGT, 45–55% for the large subunit glycosylation mutants, and 13% for GGT bearing the small subunit mutation. These results suggest that carbohydrate occupancy at each N-glycosylation site contributes to the structural stability of the enzyme, with occupancy at the single N-glycosylation site in the small subunit (Asn-511) eliciting the greatest effect.
FIGURE 2.
Thermal stability of wild-type and mutant GGT alleles. A and B, cell extracts from HEK293 cells transfected with either wild-type GGT (○) or the N-glycosylation mutants, N95Q (■), N120Q (△), N230Q (▾), N266Q (♢), N297Q (▿), N344Q (□), and N511Q (▴), were preincubated at 37, 47, 56, 58, 60, 62, 67, and 77 °C for 15 min. A, residual transpeptidation activity was assayed in the presence of 3 mm l-GpNA and 40 mm glycyl-glycine at 37 °C for 30 min. B, residual hydrolysis activity was assayed in the presence of 3 mm d-GpNA in the absence of glycyl-glycine. Data presented here represent the mean ± S.D. of triplicate assays.
SDS-PAGE Analysis of N-Glycan-deficient Human GGT
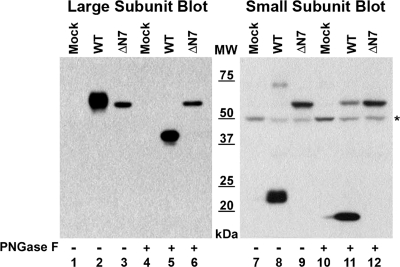
We next sought to determine whether GGT matured into a heterodimer with enzymatic activity in the absence of all N-glycosylation. To this end, we generated an N-glycan-deficient mutant (ΔN7), bearing glutamine substitutions at all seven of its N-glycosylation sites. When this mutant allele was expressed in HEK293 cells, a single band with an apparent molecular mass of 61 kDa was detected by Western analysis in the whole cell extract, using antibodies against either the large or small subunits of GGT (Fig. 3, lanes 3 and 9). This contrasted markedly with the wild-type allele, in which the large and small subunits of GGT resolved separately with apparent molecular masses of 64 kDa (Fig. 3, lane 2) or 22 kDa (Fig. 3, lane 8), respectively. These data indicate that the novel 61-kDa protein band is composed of both the large and small subunits of GGT and that it is the unglycosylated, unprocessed propeptide that has not been resolved into the mature heterodimer.
FIGURE 3.
SDS-PAGE analysis of the N-glycosylation-deficient GGT mutant. Total cell lysates from HEK293 cells transiently transfected with either empty vector, wild-type GGT, or the total N-glycosylation site knock-out mutant (ΔN7) were incubated in the absence or presence of PNGaseF and resolved on 10% SDS-polyacrylamide gels. Resolved proteins were electroblotted onto nitrocellulose and immunoblotted using antibodies specific to the large (left panel) or small (right panel) subunits of GGT. MW, molecular weight markers. * denotes position of nonspecific band that cross-reacts with the small subunit antibody in HEK293 cell extracts.
To confirm this conclusion, whole cell extracts from HEK293 cells expressing wild-type GGT and the ΔN7 mutant were subjected to total N-glycan deglycosylation with the N-glycosidase PNGaseF. As shown in Fig. 3, incubating the ΔN7 mutant of GGT with PNGaseF did not alter the migration of the mutant protein by SDS-PAGE and subsequent Western analysis against either the large or small subunits of GGT (lanes 3, 6, 9, and 12), indicating that the ΔN7 mutant is not N-glycosylated. Moreover, because of the sensitivity of the GGT small subunit antibody, trace amounts of the glycosylated immature propeptide (apparent molecular mass of 75 kDa) could also be detected in extracts prepared from HEK293 extracts expressing the wild-type GGT allele (Fig. 3, lane 8). When wild-type extracts were incubated with PNGaseF, this population of unprocessed propeptide quantitatively shifted to resolve at an apparent mass that was indistinguishable from that of the ΔN7 mutant by SDS-PAGE (Fig. 3, lanes 11 and 12). These results are consistent with the accumulation of an unglycosylated inactive propeptide form of GGT in the absence of N-glycosylation. No GGT activity was observed in extracts transfected with the ΔN7 mutant (Table 1), indicating that N-glycosylation is required for the formation of the mature, catalytically active enzyme.
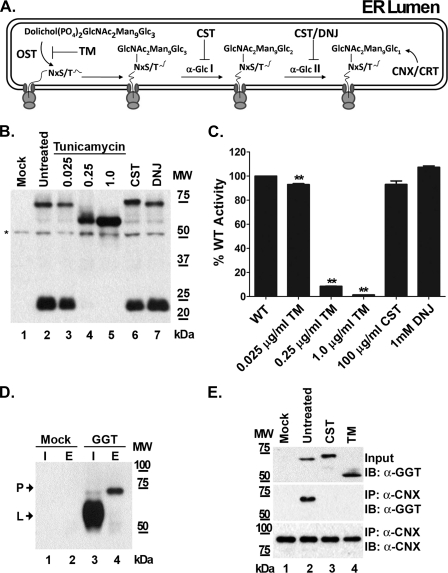
Characterization of the Effects of N-Glycosylation Inhibitors on the Processing and Enzymatic Maturation of Wild-type Human GGT
To independently verify that N-glycosylation is required for the autocatalytic cleavage of human GGT and to circumvent the possibility of unforeseen structural perturbations introduced by our glutamine substitutions, wild-type GGT was expressed in HEK293 cells in the presence of the N-glycosylation inhibitor tunicamycin. Tunicamycin inhibits the initial step of protein N-glycosylation in the ER by blocking the activity of oligosaccharyltransferase, the enzyme complex responsible for conjugating the primary core oligosaccharide to asparagine residues on nascent polypeptides (Fig. 4A). Therefore, tunicamycin treatment is capable of acting as a surrogate for simultaneously mutating all of the N-glycosylation sites on GGT, while preserving the fidelity of the primary sequence of the wild-type allele. When wild-type GGT was expressed in the presence of increasing concentrations of tunicamycin, the enzyme failed to mature into a heterodimer (Fig. 4B, lanes 3–5). At a tunicamycin concentration of 1 μg/ml, the small subunit of GGT was detected exclusively as part of the unglycosylated propeptide (Fig. 4B, lane 5). The propeptide that accumulated under these conditions migrated with an apparent molecular mass of 60–61 kDa, which is consistent with the size of the unglycosylated propeptide that was observed for the ΔN7 mutant (Fig. 3). The GGT expressed in the presence of tunicamycin exhibited a concomitant decrease in enzyme activity in a dose-dependent manner, resulting in a complete loss of activity at a concentration of 1 μg/ml (Fig. 4C). These results indicate that N-glycosylation of the nascent GGT polypeptide is required for its cleavage into the active, mature heterodimer.
FIGURE 4.
Effects of N-glycosylation inhibitors on the autocatalytic cleavage of GGT. A, diagrammatic representation depicting relevant steps of N-glycan processing in the endoplasmic reticulum. The drug tunicamycin (TM) blocks the first step in the pathway, which is the covalent attachment of the common precursor oligosaccharide to the peptide backbone by oligosaccharyltransferase (OST). The drugs castanospermine (CST) and deoxynojirimycin (DNJ) inhibit processing of the common precursor oligosaccharide structure by blocking the activities of α-glucosidases I (α-Glc I) and II (α-Glc II), disrupting N-glycan-dependent interaction with the chaperones, calnexin (CNX), and calreticulin (CRT). B, SDS-PAGE analysis of GGT expressed in the presence of N-glycosylation inhibitors. Total cell lysates from HEK293 cells transiently transfected with wild-type GGT and cultured in the presence of either tunicamycin (0.025–1 μg/ml), castanospermine (100 μg/ml), or deoxynojirimycin (1 mm) were subjected to Western analysis, using an antibody against the small subunit of GGT. C, transpeptidation activities were assayed at 37 °C in the presence of 3 mm l-GpNA and 40 mm glycyl-glycine using 5 μg of total protein from the cell lysates from B. Data are the mean ± S.D. from triplicate assays. ** denotes p < 0.01. D, co-immunoprecipitation of the GGT propeptide with anti-calnexin. Cell extracts from HEK293 cells transiently transfected with either empty vector or wild-type GGT were subjected to immunoprecipitation (IP) with a monoclonal anti-calnexin antibody. Total cell extracts (I) and immunoprecipitation eluates (E) were subjected to Western analysis against the large subunit of GGT. The mature large subunit (L) and immature propeptide (P) of GGT are indicated by arrows. E, co-immunoprecipitation analysis of GGT expressed in the presence of N-glycosylation inhibitors. Cell extracts from HEK293 cells expressing GGT in the absence of inhibitors or in the presence of tunicamycin (1 μg/ml) or castanospermine (100 μg/ml) were immunoblotted (IB) against the small subunit of GGT to detect uncleaved propeptide (upper panel). Samples were immunoprecipitated as in D, and the immunoprecipitation eluates were immunoblotted with either the GGT small subunit antibody (middle panel) or a polyclonal calnexin antibody (lower panel).
To determine whether core oligosaccharide-protein conjugation or subsequent N-glycan processing is required for cleavage of GGT into the active heterodimer, wild-type GGT was expressed in the presence of the glycosidase inhibitors castanospermine and deoxynojirimycin. These inhibitors block consecutive steps in the early processing of N-glycans on glycoproteins in the ER by interfering with the activities of α-1,2-glucosidase I and α-1,3-glucosidase II (Fig. 4A). As a result, inhibition by either castanospermine or deoxynojirimycin leads to the persistence of immature N-glycans on nascent glycopeptides (38, 39). Western analysis of wild-type GGT expressed in the presence of castanospermine (100 μg/ml) revealed that the inhibitor did not block its subsequent processing into the mature heterodimer (Fig. 4B, lanes 2 and 6). Similar results were observed in extracts prepared from cells in which GGT was expressed in the presence of 1 mm deoxynojirimycin (Fig. 4B, lane 7). GGT expressed in the presence of either castanospermine or deoxynojirimycin exhibited transpeptidation activities that were comparable with that of untreated controls (Fig. 4C, 89 and 100% of control, respectively). Similar data were obtained for the hydrolysis reaction (data not shown). These results indicate that N-glycan processing by α-glucosidases I or II is not necessary for the enzymatic maturation of GGT.
Inhibition of the terminal processing of immature N-glycans by castanospermine and deoxynojirimycin prevents the association of nascent glycoproteins with the resident ER chaperones calnexin and calreticulin, which assist in the folding of proteins bearing monoglucosylated oligosaccharides (39, 40). Co-immunoprecipitation analysis demonstrated that the propeptide of GGT interacts with calnexin during its transient maturation through the ER (Fig. 4D). Tunicamycin treatment disrupted the association of the GGT propeptide with this chaperone, consistent with an N-glycan-dependent mode of substrate recognition by calnexin (Fig. 4E, lane 4). This interaction was also significantly attenuated when GGT was expressed under conditions in which N-glycan terminal processing was suppressed (e.g. in the presence of castanospermine, Fig. 4E, middle panel, lane 3). Castanospermine treatment resulted in a GGT propeptide that migrated more slowly on SDS-PAGE than the wild-type propeptide, consistent with the persistence of larger precursor oligosaccharides (Fig. 4E, upper panel, lanes 2 and 3). However, the data also show that the GGT propeptide autocleaved into the functional heterodimer even when the N-glycan processing-dependent interaction of calnexin and calreticulin with the nascent glycopeptide was disrupted (Fig. 4, B, lanes 6 and 7, and C). Therefore, the conjugation of core oligosaccharide units to nascent GGT polypeptides, rather than the subsequent processing of the N-glycans, appears to be the requisite factor for facilitating the series of events that lead to the proper folding and autocatalytic processing of GGT into the mature heterodimer.
Characterization of Relative Activity and Thermal Stability of Deglycosylated Human GGT following Enzyme Maturation
Our data indicate that N-glycosylation plays a critical role in facilitating the proper folding and autocatalytic cleavage of human GGT. To investigate whether N-glycosylation is required for the enzymatic activity of the mature protein, wild-type human GGT was deglycosylated, and its enzymatic activity was characterized. HEK293 extracts from cells transfected with wild-type GGT were incubated with the endoglycosidases PNGaseF and endoglycosidase H to enzymatically remove the N-glycans from the mature enzyme (Fig. 5A, left panel). Although this strategy was sufficient for the quantitative removal of N-glycans on the small subunit of human GGT (Fig. 5A, lower panel), it did not result in the complete deglycosylation of the large subunit, even after 48 h of incubation (Fig. 5A, upper panel). The apparent molecular mass of the HEK293-expressed large subunit following treatment of the native enzyme with glycosidases was 48.5 kDa, consistent with the removal of ∼66% of the N-glycans. Denaturation of the enzyme prior to deglycosylation was necessary to remove all the N-glycans from the large subunit (Fig. 5A, upper panel, lanes 2 and 3); however, enzymatic activity could not be assessed following denaturation.
FIGURE 5.
N-Glycans are not required for the enzymatic activity and thermal stability of mature human GGT. A, either homogenates from HEK293 cells transfected with wild-type human GGT or purified human GGT expressed in P. pastoris X33 cells were incubated in the absence or presence of glycosidases at 37 °C. SDS-PAGE and Western analysis against the large (upper panel) and small (lower panel) subunits of GGT were then carried out among the untreated and deglycosylated samples. Fully deglycosylated subunits from denatured human GGT expressed in either HEK293 cells or P. pastoris were resolved in parallel as a visual reference (lanes 3 and 6). B, cell extracts from P. pastoris X33 cells induced to express either empty vector, wild-type human GGT, or the total N-glycosylation site knock-out mutant (ΔN7) were incubated in the absence or presence of PNGaseF and then subjected to SDS-PAGE and Western analysis against the large (lanes 1–7) and small (lanes 8–14) subunits of human GGT. C, transpeptidation activity was assayed using 4 milliunits of untreated (○) or deglycosylated (△) human GGT expressed in P. pastoris in the presence of 0.25–3 mm l-GpNA and 40 mm glycyl-glycine. D, thermal stability analysis was conducted as described for Fig. 2, using 4 milliunits of untreated (○) or deglycosylated (△) human GGT expressed in P. pastoris. The residual transpeptidation activity was assayed in the presence of 3 mm l-GpNA and 40 mm glycyl-glycine at 37 °C for 30 min.
To produce fully deglycosylated mature human GGT, we used a truncated form of GGT that lacked the transmembrane domain. Structural models of the human enzyme predict that two of the N-glycosylation sites on the large subunit (Asn-95 and -266) are located within small clefts on the surface of the enzyme adjacent to the transmembrane domain (16). A well characterized soluble form of human GGT (amino acids 29–569) that retains full enzymatic activity yet lacks this transmembrane domain can be expressed and purified from the yeast P. pastoris (12, 22). To circumvent a potential steric barrier to deglycosylation imposed by the transmembrane domain, we utilized this soluble form of human GGT to address whether full deglycosylation could be achieved on the native enzyme. The soluble form of the enzyme retains all seven of its N-glycosylation sites, which are modified with N-glycans (Fig. 5B, lanes 2, 6, 9, and 13) (12).3 As was observed for human GGT expressed in HEK293 cells, comprehensive substitution of these asparagines for glutamine residues resulted in the accumulation of the inactive, unglycosylated propeptide when the mutant form of the human enzyme was expressed in yeast (Fig. 5B, lanes 3, 7, 10, and 14). This indicates that the requirement of N-glycosylation for cleavage of human GGT into the active heterodimer is a conserved feature across heterologous eukaryotic expression systems.
In contrast to the membrane-bound form of the enzyme, wild-type human GGT lacking its transmembrane domain was capable of being fully deglycosylated in its native conformation (Fig. 5A, right panel). A comparative analysis between the glycosylated and deglycosylated forms of the enzyme revealed that the reaction kinetics of the two populations were strikingly similar (Fig. 5C). The Km value in the transpeptidation reaction for the deglycosylated GGT showed no significant difference from that measured for the untreated sample, whereas the Vmax value of the enzyme following deglycosylation was ∼80% of its glycosylated counterpart (Fig. 5C), which contrasted dramatically from the complete loss of enzymatic activity observed when N-glycosylation was blocked on the nascent polypeptide (Table 1 and Fig. 4C). Moreover, the glycan-deficient form of the enzyme exhibited a thermal stability profile that closely mimicked that of the untreated enzyme across the entire temperature range that was examined (Fig. 5D). Taken together, these results indicate that although N-glycosylation plays a critical role in the proper folding and autocatalytic cleavage of the nascent enzyme, it is not a significant factor governing the enzymatic activity or structural stability of GGT after it has reached full maturation.
DISCUSSION
In this study, we provide several lines of evidence demonstrating that N-glycosylation plays a fundamental role in the proper folding and autocatalytic cleavage of human GGT. Through mutational analysis, we demonstrate that all seven N-glycosylation sites (asparagines 95, 120, 230, 266, 297, 344, and 511) on human GGT are glycosylated when the enzyme is expressed in human embryonic kidney (HEK293) cells. We found that single site mutants decrease the structural stability of the enzyme yet are functionally tolerated, despite the fact that the N95Q mutant exhibited a marked decrease in the expression levels of the mature heterodimer. However, blocking N-glycosylation at all of these sites simultaneously, either by mutation or inhibition of glycosylation by tunicamycin, resulted in the expression of an unglycosylated GGT propeptide that failed to mature into an enzymatically active heterodimer. Downstream inhibition of N-glycan processing on GGT by the drugs castanospermine and deoxynojirimycin revealed that initial glycan attachment, not subsequent processing of the carbohydrates, is the required feature for conferring cleavage competence. Conversely, we observed that enzymatic removal of all N-glycans from mature heterodimeric GGT had little effect on the kinetic behavior or thermal stability of the enzyme, indicating that the aberrant behavior of the total knock-out mutant was attributable to defects in the maturation of the propeptide during its translocation through the secretory pathway.
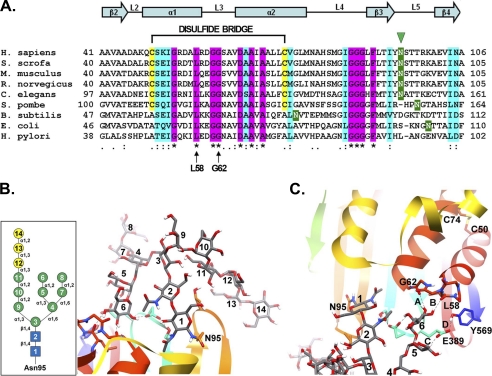
The observation that all seven potential N-glycosylation sites are modified by glycans in the HEK293 cell line correlates well with our previous mass spectrometric characterization of GGT glycopeptides from normal human kidney tissue, in which we identified carbohydrates occupying each of these sites (16). The size of the N-glycans or extent of glycosylation at the Asn-95 and -266 sites on GGT expressed in HEK293 cells appeared to differ from the other sites in the large subunit of the enzyme, as mutation of either of these sequons had a lesser effect on the apparent molecular mass than mutation of the other N-glycosylation sites (see supplemental Fig. 2B). The indication of smaller N-glycans at the Asn-95 and -266 sites is consistent with our previous findings that, on GGT from human kidney, these sites are primarily decorated with uncharged N-glycans that are smaller than the glycans on the other four sites on the large subunit of GGT (16). Homology modeling in our previous publication suggested that the smaller N-glycans at these sites might be attributable to their localization to small clefts on the surface of the enzyme adjacent to the plasma membrane (16). In this study, we confirmed that the single predicted N-glycosylation site on the small subunit is quantitatively occupied with N-glycans and is indeed the only N-glycosylation site on this subunit (Fig. 1C). Sequence alignment with other GGT orthologs demonstrates that four of the N-glycosylation sites on human GGT (asparagines 95, 120, 344, and 511) are highly conserved among eukaryotes, and two additional sites (asparagines 230 and 297) are conserved among mammalian GGTs (Fig. 6A and supplemental Fig. 3). One site, asparagine 266, is unique to human GGT. Despite being well conserved among eukaryotic GGTs, only one of these N-glycosylation motifs, asparagine 104 in Escherichia coli GGT, is observed among the well characterized bacterial GGTs (Fig. 6A). None of these species of bacteria employs this mode of glycosylation, suggesting that the proliferation of these sequons has evolved in parallel with the advent of eukaryotic N-glycosylation.
FIGURE 6.
Comparative sequence and substructural analysis of the Asn-95 glycosylation site on human GGT. A, amino acid sequence alignment of residues 41–106 from human (Homo sapiens) GGT with the corresponding sequences from pig (Sus scrofa), mouse (Mus musculus), rat (Rattus norvegicus), nematode (Caenorhabditis elegans), fission yeast (Schizosaccharomyces pombe), and the bacteria Bacillus subtilis, E. coli, and Helicobacter pylori with the ClustalW program (48). Shading is used to indicate the occurrence of similar amino acids (identical residues, magenta; conserved substitutions, aquamarine) or putative N-glycosylation sites (green). Also indicated are the positions of the Asn-95 glycosylation site in human GGT (green arrowhead), the conserved glycine and leucine residues (Leu-58 and Gly-62 in human GGT) predicted to form hydrogen bonds with the N-glycan at asparagine 95 (arrows), and the C2-C3 (Cys-50 and Cys-74 in human GGT) disulfide bridge in mammalian GGT. Conserved α-helices and β-sheets are also indicated. B, homology model of human GGT propeptide showing the structural orientation of the primary oligomannosyl N-glycan (Glc3Man9GlcNAc2) conjugated to asparagine 95. Inset, structural schematic of the primary Glc3Man9GlcNAc2 carbohydrate moiety. Symbols used are as follows: blue box, N-acetylglucosamine; green circle, mannose; yellow circle, glucose. C, model of the stable hydrogen bonds (bonds A and B, dashed lines) predicted to form between a terminal mannose residue (mannose 6) in the primary N-glycan and the carbonyl oxygens of both Leu-58 and Gly-62 within the peptide loop (L3) connecting α-helices 2 and 3. Additionally, mannose 5 is predicted to hydrogen bond with Glu-389 (bond C), which is further stabilized by Tyr-569 (bond D). The position of the disulfide bond (between cysteines 50 and 74) interconnecting α-helices 2 and 3 is shown for reference.
Despite their relatively high degree of conservation, individual N-glycosylation site mutations did not disrupt the functionality of the mature enzyme, with each exhibiting Km values for both transpeptidation and hydrolysis that were not significantly different from the wild-type enzyme (Table 1). However, the N95Q mutant did exhibit an ∼8-fold reduction in the levels of mature heterodimer with a corresponding increase in the level of the propeptide, suggesting that there is a partial kinetic barrier to heterodimer formation for this mutant (Fig. 1). This observation indicates that glycosylation at this site is critical for optimal cleavage of the propeptide. Three-dimensional homology modeling of human GGT predicts that Asn-95 is located in a small cleft on the surface of one of two parallel β-sheets that form a hairpin structure and create a channel through which substrates access the active site of the enzyme (16). Our molecular modeling studies indicate that the N-glycans that are conjugated to this asparagine residue are unique among glycans at the other N-glycosylation sites in that they are predicted to project outward in such a way as to create intramolecular interactions with the protein backbone (Fig. 6, B and C). A terminal mannose residue (mannose 6) in the common precursor oligosaccharide (Glc3Man9GlcNAc2) conjugated to the Asn-95 site of the propeptide by oligosaccharyltransferase in the ER is predicted to form stable hydrogen bonds with the carbonyl groups of conserved leucine and glycine residues (Leu-58 and Gly-62 in human GGT) that are present within a structurally adjacent peptide loop (loop3, L3) that bridges two parallel α-helices in the large subunit of GGT (Fig. 6, B and C). It has been previously shown that the spatial orientation of this α-helical pair is maintained internally by a disulfide bond that forms between cysteines C2 and C3 in mammalian GGT (Cys-50 and -74 in human GGT, Fig. 6C) (10). These two cysteine residues are conserved among eukaryotic GGTs and are absent in the bacterial orthologs (Fig. 6A). Systematic mutagenesis of the cysteine residues present in rat GGT has shown that this highly conserved disulfide bond is essential for the proper folding of the propeptide in order for it to undergo autocatalytic cleavage (Fig. 6A) (10). Thus, a likely explanation for the autoprocessing defect in the N95Q mutant of human GGT is that the formation of the critical disulfide bond between Cys-50 and Cys-74 is partially impeded when the glycan is missing. N-Glycosylation of Asn-95 may therefore stabilize the nascent polypeptide in a conformation that is advantageous for this disulfide bond to form. Our model predicts that single leucine substitutions at Gly-61 and Gly-62 would induce mild conformational changes in loop3 that interrupt the putative interaction between mannose 6 and Gly-62 and Leu-58, respectively, whereas the GGT(G61L,G62L) double mutant would simultaneously destabilize both of these hydrogen bonds (supplemental Fig. 4). In fact, analysis of the G61L mutant showed that the protein exhibited a processing defect that closely resembled that observed for the N95Q mutant (supplemental Fig. 5). Remarkably, both the G62L allele and the G61L,G62L double mutant arrest in the propeptide state, with no discernible enzymatic activity or heterodimer formation, indicating that these mutations might act by conferring a greater impact on the loop3 structure than that elicited by an absence of glycosylation at Asn-95 alone (supplemental Fig. 5). These observations indicate that loop3 is highly sensitive to structural perturbations, which is consistent with our hypothesis that this portion of the protein must assume the proper conformation to promote propeptide cleavage, presumably by maintaining the precise relative orientation of Cys-50 and -74 for disulfide bond formation. Stabilization of loop3 by hydrogen bonding between the common oligosaccharide precursor moiety on Asn-95 and the carbonyl groups of Leu-58 and Gly-62 may ensure that this conformational state is optimally achieved. There is precedent for glycans facilitating disulfide bond formation in viral proteins and prions, but this is the first report of a glycan functioning in this manner in the folding and processing of any member of the GGT family of enzymes (41, 42).
Although glycosylation on Asn-95 appears to play an important role in the optimal cleavage of the GGT propeptide, deletion of all N-glycosylation sites except the Asn-95 site (ΔN6, GGTN120Q,N230Q,N266Q,N297Q,N344Q,N511Q) resulted in the accumulation of an inactive propeptide form of GGT with no detectable levels of mature heterodimer, analogous to that which was observed for the total knock-out mutant (Fig. 3 and data not shown). Therefore, although important, glycosylation at Asn-95 is not sufficient for the activation of human GGT. This indicates that a complex level of coordination among the N-glycosylation sites on GGT is required to ensure the proper folding pattern necessary for inducing the autocleavage event. This conclusion is further supported by an apparent loss of structural fidelity associated with each individual site substitution, as measured during our thermal stability profiling of the single N-glycosylation mutants (Fig. 2).
In contrast to the single site mutants, mutagenesis of all seven N-glycosylation sites resulted in a quantitative block in heterodimer formation and the accumulation of an inactive, unglycosylated propeptide form of the enzyme (Fig. 3). Formation of the mature, enzymatically active heterodimer is induced by a nucleophilic attack mediated by the active site threonine (Thr-381 in human GGT) after the propeptide acquires the appropriate folding conformation for the autocatalytic cleavage event in the ER (10, 43). Therefore, the inability of the glycan-deficient mutant to reach functional maturity indicates that the propeptide fails to fold properly in the absence of glycosylation. These results were recapitulated when wild-type GGT was expressed in the presence of the potent N-glycosylation inhibitor tunicamycin, demonstrating that the maturational block was specific for the unglycosylated propeptide and not the result of a change in the primary structure of the mutant allele (Fig. 4). Taken together, these results indicate that N-glycosylation is a rate-limiting processing event that dictates the maturational fate of the human propeptide. This observation may account for the inability of human GGT to autocleave and reach functional maturity when expressed in E. coli in the absence of N-glycosylation (13).3
We observed a quantitative block in human GGT autocatalytic cleavage when cells were incubated in 1 μg/ml tunicamycin (Fig. 4B). Barouki et al. (43) reported that rat GGT autocleaved even when N-glycosylation was blocked by 4 μg/ml tunicamycin. We are unable to account for the apparent discrepancy between our data and those of Barouki et al. (43), which is unexpected given the conservation of N-glycosylation sites and high sequence identity between rat and human GGT. Barouki et al. (43) did find that in vitro translation of rat GGT mRNA produced a propeptide that failed to autocleave. This observation is consistent with our conclusion that the unmodified peptide backbone of human GGT is not competent for auto-maturation. We provide here independent mutational confirmation for the central role of N-glycosylation in the maturation of human GGT that is independent of the eukaryotic expression system (Figs. 3 and 5B).
Although tunicamycin treatment resulted in a complete block in the autocleavage of GGT, inhibition of subsequent N-glycan processing by the inhibitors castanospermine and deoxynojirimycin did not significantly alter either the processing of the propeptide or the kinetic behavior of the mature enzyme (Fig. 4). These inhibitors are known to block the glycan-dependent interaction of the ER-resident glycoprotein chaperones, calnexin and calreticulin, with nascent glycoproteins (39). These results suggest that the addition of the core N-glycan by oligosaccharyltransferase is the critical glycosylation event governing the functional maturation of GGT. The subsequent trimming of the immature glycans is apparently not required for N-glycan-dependent chaperone interaction with the newly synthesized polypeptide. This distinction has been observed for other glycoproteins whose proper folding and functional maturation appear to be dependent upon primary glycosylation and independent of glycan processing (44, 45). One of these glycoproteins, human polycystin-1 (PC1), undergoes an autoproteolytic cleavage event in the early secretory pathway that is analogous to GGT and other N-terminal nucleophile hydrolases, perhaps indicative of a shared mode of N-glycan-dependent structural maturation among these cell surface proteins (44).
Despite the central role that N-glycans play in the functional activation of human GGT, they proved to be dispensable once the enzyme reaches maturity. We show here that the deglycosylated form of mature wild-type human GGT exhibits comparable enzyme kinetics to untreated controls (Fig. 5C). These results are consistent with those reported by Smith and Meister (46) for enzymatically deglycosylated GGT isolated from pig and rat kidneys. These observations suggest that, although N-glycosylation is a critical event in the early maturation and corresponding autocatalytic activation of the propeptide, it is not a requisite feature for the function of the mature enzyme. However, in vivo, the persistence of N-glycans on GGT may provide steric protection from proteases and nonspecific interactions (47). Although mutation of the Asn-511 glycosylation site resulted in significantly decreased thermal stability relative to the wild-type allele (Fig. 2), mature deglycosylated GGT exhibited a profile similar to glycosylated controls (Fig. 5D). This discrepancy likely indicates that the enhanced heat sensitivity exhibited by this point mutation results from structural insults in the folding of the nascent mutant propeptide in the absence of glycosylation at this site that persist within the heterodimer upon reaching maturation.
In conclusion, we found that impeding co-translational glycosylation on human GGT resulted in a complete loss of propeptide cleavage and enzyme function. This study clearly demonstrated that N-glycosylation plays a fundamental role in the folding and subsequent processing of human GGT during its early maturation in the secretory pathway. It is noteworthy that the bacterial orthologs of GGT, which also undergo autocleavage maturational events, lack both disulfide bonds and N-glycosylation. Therefore, it is possible that these two structural features have co-evolved in eukaryotic systems to facilitate the proper folding of the nascent GGT propeptide in a manner that promotes its autocatalytic processing.
Supplementary Material
Acknowledgments
We acknowledge the assistance of Dr. Leonidas Tsiokas at OUHSC and Dr. Paul F. Cook at the University of Oklahoma for their thoughtful insights and Dr. Michael Brenner for the generous gift of the anti-calnexin antibody (AF8) (Harvard Medical School).
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 CA57530 and R56 CA57530 (to M. H. H.) and F32 CA128338 (to M. B. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table and Figs. 1–5.
M. B. West and M. H. Hanigan, unpublished results.
- GGT
- γ-glutamyl transpeptidase
- PNGaseF
- peptide:N-glycosidase F
- GpNA
- γ-glutamyl-paranitroanilide
- ER
- endoplasmic reticulum
- MD
- molecular dynamics.
REFERENCES
- 1. Allison R. D. (1985) Methods Enzymol. 113, 419–437 [DOI] [PubMed] [Google Scholar]
- 2. Hanigan M. H., Ricketts W. A. (1993) Biochemistry 32, 6302–6306 [DOI] [PubMed] [Google Scholar]
- 3. Hanigan M. H., Frierson H. F., Jr. (1996) J. Histochem. Cytochem. 44, 1101–1108 [DOI] [PubMed] [Google Scholar]
- 4. Hiramatsu K., Asaba Y., Takeshita S., Nimura Y., Tatsumi S., Katagiri N., Niida S., Nakajima T., Tanaka S., Ito M., Karsenty G., Ikeda K. (2007) Endocrinology 148, 2708–2715 [DOI] [PubMed] [Google Scholar]
- 5. Hanigan M. H., Frierson H. F., Jr., Swanson P. E., De Young B. R. (1999) Hum. Pathol. 30, 300–305 [DOI] [PubMed] [Google Scholar]
- 6. Lieberman M. W., Wiseman A. L., Shi Z. Z., Carter B. Z., Barrios R., Ou C. N., Chévez-Barrios P., Wang Y., Habib G. M., Goodman J. C., Huang S. L., Lebovitz R. M., Matzuk M. M. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 7923–7926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hanigan M. H., Gallagher B. C., Townsend D. M., Gabarra V. (1999) Carcinogenesis 20, 553–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brannigan J. A., Dodson G., Duggleby H. J., Moody P. C., Smith J. L., Tomchick D. R., Murzin A. G. (1995) Nature 378, 416–419 [DOI] [PubMed] [Google Scholar]
- 9. Suzuki H., Kumagai H. (2002) J. Biol. Chem. 277, 43536–43543 [DOI] [PubMed] [Google Scholar]
- 10. Kinlough C. L., Poland P. A., Bruns J. B., Hughey R. P. (2005) Methods Enzymol. 401, 426–449 [DOI] [PubMed] [Google Scholar]
- 11. Inoue M., Hiratake J., Suzuki H., Kumagai H., Sakata K. (2000) Biochemistry 39, 7764–7771 [DOI] [PubMed] [Google Scholar]
- 12. Castonguay R., Halim D., Morin M., Furtos A., Lherbet C., Bonneil E., Thibault P., Keillor J. W. (2007) Biochemistry 46, 12253–12262 [DOI] [PubMed] [Google Scholar]
- 13. Angele C., Oster T., Visvikis A., Michels J. M., Wellman M., Siest G. (1991) Clin. Chem. 37, 662–666 [PubMed] [Google Scholar]
- 14. Nemesánszky E., Lott J. A. (1985) Clin. Chem. 31, 797–803 [PubMed] [Google Scholar]
- 15. Rademacher T. W., Parekh R. B., Dwek R. A. (1988) Annu. Rev. Biochem. 57, 785–838 [DOI] [PubMed] [Google Scholar]
- 16. West M. B., Segu Z. M., Feasley C. L., Kang P., Klouckova I., Li C., Novotny M. V., West C. M., Mechref Y., Hanigan M. H. (2010) J. Biol. Chem. 285, 29511–29524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Meredith M. J. (1991) Biochem. Int. 25, 321–330 [PubMed] [Google Scholar]
- 18. Helenius A., Aebi M. (2004) Annu. Rev. Biochem. 73, 1019–1049 [DOI] [PubMed] [Google Scholar]
- 19. Skropeta D. (2009) Bioorg. Med. Chem. 17, 2645–2653 [DOI] [PubMed] [Google Scholar]
- 20. McCracken A. A., Brodsky J. L. (2003) BioEssays 25, 868–877 [DOI] [PubMed] [Google Scholar]
- 21. Williams D. B. (2006) J. Cell Sci. 119, 615–623 [DOI] [PubMed] [Google Scholar]
- 22. King J. B., West M. B., Cook P. F., Hanigan M. H. (2009) J. Biol. Chem. 284, 9059–9065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Graham F. L., van der Eb A. J. (1973) Virology 52, 456–467 [DOI] [PubMed] [Google Scholar]
- 24. Hochstenbach F., David V., Watkins S., Brenner M. B. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 4734–4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sali A., Blundell T. L. (1993) J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 26. Okada T., Suzuki H., Wada K., Kumagai H., Fukuyama K. (2007) J. Biol. Chem. 282, 2433–2439 [DOI] [PubMed] [Google Scholar]
- 27. Wada K., Irie M., Suzuki H., Fukuyama K. (2010) FEBS J. 277, 1000–1009 [DOI] [PubMed] [Google Scholar]
- 28. Shen M. Y., Sali A. (2006) Protein Sci. 15, 2507–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Case D. A., Cheatham T. E., 3rd, Darden T., Gohlke H., Luo R., Merz K. M., Jr., Onufriev A., Simmerling C., Wang B., Woods R. J. (2005) J. Comput. Chem. 26, 1668–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J. L., Dror R. O., Shaw D. E. (2010) Proteins Struct. Funct. Bioinformat. 78, 1950–1958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kirschner K. N., Yongye A. B., Tschampel S. M., González-Outeiriño J., Daniels C. R., Foley B. L., Woods R. J. (2008) J. Comput. Chem. 29, 622–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shaw L. M., London J. W., Petersen L. E. (1978) Clin. Chem. 24, 905–915 [PubMed] [Google Scholar]
- 33. Sener A., Yardimci T. (2005) J. Biochem. Mol. Biol. 38, 343–349 [DOI] [PubMed] [Google Scholar]
- 34. Elce J. S., Broxmeyer B. (1976) Biochem. J. 153, 223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Curthoys N. P., Hughey R. P. (1979) Enzyme 24, 383–403 [DOI] [PubMed] [Google Scholar]
- 36. Thompson G. A., Meister A. (1976) Biochem. Biophys. Res. Commun. 71, 32–36 [DOI] [PubMed] [Google Scholar]
- 37. Keillor J. W., Castonguay R., Lherbet C. (2005) Methods Enzymol. 401, 449–467 [DOI] [PubMed] [Google Scholar]
- 38. Gross V., Tran-Thi T. A., Schwarz R. T., Elbein A. D., Decker K., Heinrich P. C. (1986) Biochem. J. 236, 853–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Herscovics A. (1999) Biochim. Biophys. Acta 1473, 96–107 [DOI] [PubMed] [Google Scholar]
- 40. Elbein A. D. (1987) Annu. Rev. Biochem. 56, 497–534 [DOI] [PubMed] [Google Scholar]
- 41. Hebert D. N., Zhang J. X., Chen W., Foellmer B., Helenius A. (1997) J. Cell Biol. 139, 613–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bosques C. J., Imperiali B. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 7593–7598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Barouki R., Finidori J., Chobert M. N., Aggerbeck M., Laperche Y., Hanoune J. (1984) J. Biol. Chem. 259, 7970–7974 [PubMed] [Google Scholar]
- 44. Wei W., Hackmann K., Xu H., Germino G., Qian F. (2007) J. Biol. Chem. 282, 21729–21737 [DOI] [PubMed] [Google Scholar]
- 45. Nakajima M., Koga T., Sakai H., Yamanaka H., Fujiwara R., Yokoi T. (2010) Biochem. Pharmacol. 79, 1165–1172 [DOI] [PubMed] [Google Scholar]
- 46. Smith T. K., Meister A. (1994) FASEB J. 8, 661–664 [DOI] [PubMed] [Google Scholar]
- 47. Rudd P. M., Elliott T., Cresswell P., Wilson I. A., Dwek R. A. (2001) Science 291, 2370–2376 [DOI] [PubMed] [Google Scholar]
- 48. Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.