Abstract
Candida albicans is the most common opportunistic fungal pathogen and causes local and systemic disease in immunocompromised patients. Alveolar macrophages (AMs) are pivotal for the clearance of C. albicans from the lung. Activated AMs secrete 5-lipoxygenase-derived leukotrienes (LTs), which in turn enhance phagocytosis and microbicidal activity against a diverse array of pathogens. Our aim was to investigate the role of LTB4 and LTD4 in AM antimicrobial functions against C. albicans and the signaling pathways involved. Pharmacologic and genetic inhibition of LT biosynthesis as well as receptor antagonism reduced phagocytosis of C. albicans when compared with untreated or WT controls. Conversely, exogenous LTs of both classes augmented base-line C. albicans phagocytosis by AMs. Although LTB4 enhanced mainly mannose receptor-dependent fungal ingestion, LTD4 enhanced mainly dectin-1 receptor-mediated phagocytosis. LT enhancement of yeast ingestion was dependent on protein kinase C-δ (PKCδ) and PI3K but not PKCα and MAPK activation. Both LTs reduced activation of cofilin-1, whereas they enhanced total cellular F-actin; however, LTB4 accomplished this through the activation of LIM kinases (LIMKs) 1 and 2, whereas LTD4 did so exclusively via LIMK-2. Finally, both exogenous LTB4 and LTD4 enhanced AM fungicidal activity in an NADPH oxidase-dependent manner. Our data identify LTB4 and LTD4 as key mediators of innate immunity against C. albicans, which act by both distinct and conserved signaling mechanisms to enhance multiple antimicrobial functions of AMs.
Keywords: Eicosanoid Receptor, Eicosanoids Biosynthesis, Eicosanoids Function, Enzyme Mechanisms, G Protein-coupled Receptors (GPCR), Gene Transcription, Immunology, Inflammation, Phagocytosis, Reactive Oxygen Species (ROS)
Introduction
The importance of Candida albicans infection has grown as a result of the increased use of antimicrobial and immunosuppressive agents and of predisposing conditions such as cancer, diabetes, transplantation, HIV infection, and malnutrition (1–5). This pathogenic yeast can cause local infections at portals of entry, such as lung and genitourinary tract as well as disseminated infections. In the lung, alveolar macrophages (AMs)3 are important defenders against opportunistic fungal infections, preventing the hematogenous dissemination of C. albicans in immunocompromised hosts (6). AMs are able to recognize, ingest, and kill C. albicans through a range of pathogen recognition receptors (PRRs) including the C-type lectin-like receptor dectin-1 and the mannose receptor (CD206), representing the major macrophage receptors for β-glucan and mannan, respectively, involved in fungal recognition and ingestion (7). Binding of C. albicans to AMs causes the release of a myriad of proinflammatory mediators, including cytokines and bioactive lipids such as leukotrienes (LTs) (8, 9).
LTs are products of phospholipase A2-derived arachidonic acid metabolism by the enzyme 5-lipoxygenase (5-LO) and the 5-LO activating protein (FLAP) and are synthesized by phagocytes in response to inflammatory or infectious stimuli (10). There are two main classes of LTs, namely LTB4 and the cysteinyl-LTs (cysLTs), which include LTC4, LTD4, and LTE4; these act by ligating the high affinity G protein-coupled receptors BLT1 and cysLT1, respectively (11, 12). LT receptor ligation enhances many aspects of AM activation, including leukocyte accumulation (11), microbial ingestion (13) and killing (14), and generation of proinflammatory mediators (10). We have previously characterized some of the signaling pathways by which LTs enhance AM antimicrobial functions against IgG-opsonized pathogens recognized by the Fcγ receptor (FcR) (15–17). Because of the diversity of signals derived from different phagocytic receptors, the importance of LTs in amplifying phagocytosis could be unique to IgG-coated target ingestion. In addition, during active acute infection, the importance of FcR signaling in the early events of host defense is controversial. Thus, it is of interest to investigate the importance of LTs in mediating AM phagocytosis by non-opsonic receptors.
There is increasing evidence that defects in LT synthesis contribute to impaired innate immunity in a variety of immunosuppressive states, such as malnutrition (18), bone marrow transplantation (19), and HIV infection (20, 21). In view of the importance of LTs in host defense along with the underproduction of LTs observed in immunosuppressive states (22), the present study was undertaken to investigate the role of LTs and the signaling pathways involved in the anti-fungal activity of AMs against the opportunistic pathogen C. albicans. Our data show that both endogenous as well as exogenous LTB4 and LTD4 enhance phagocytosis of C. albicans by promoting F-actin polymerization and assembly and killing via NADPH oxidase activation and reactive oxygen intermediate (ROI) generation.
EXPERIMENTAL PROCEDURES
Reagents
RPMI 1640 was purchased from Invitrogen. LTB4 and LTD4 were purchased from Biomol. The inhibitors of protein kinase Cα (PKCα) (Ro-32-0432) and PKCδ (rottlerin) were supplied by Calbiochem. PI3K inhibitors (LY290042 and wortmannin), 5-LO inhibitors (AA861 and Zileuton), the cysLT1 receptor antagonist MK571, and the NADPH oxidase inhibitor DPI were supplied by Enzo. CP105696 (BLT1 antagonist) was a generous gift of Pfizer. MK0591 (FLAP inhibitor) was from Merck. Alexa488-phalloidin and Alexa594-deoxyribonuclease I (DNase I) were from Molecular Probes. Laminarin (a soluble glucan prepared from Laminaria digitata) and mannan prepared from Saccharomyces cerevisiae were both from Sigma. Compounds requiring reconstitution were dissolved in either ethanol or dimethyl sulfoxide (DMSO). Required dilutions of all compounds were prepared immediately before use, and equivalent quantities of vehicle were added to the appropriate controls.
Animals
Female pathogen-free 5-LO−/− (129-Alox5tm1Fun) mice (23), strain-matched wild-type (WT) sv129 mice, and Wistar rats were obtained from Central Laboratory Animal Medicine of University of São Paulo as well as Charles River Laboratories (Portage, MI). All experiments were in accord with ethical principles in animal research adopted by the Brazilian College of Animal Experimentation and the National Institutes of Health guidelines for the use of experimental animals, with the approval of the Animal Subject Committee of the Biomedical Sciences Institute, University of São Paulo, and the University of Michigan Committee for the Use and Care of Animals.
Cell Isolation and Culture
Rat or murine AMs were obtained by bronchoalveolar lavage as described (24). In general, rat cells were used for pharmacologic and signaling studies, and murine cells were used for comparisons between 5-LO−/− and WT genotypes. Murine resident peritoneal macrophages were obtained as described (25). Cells were cultured overnight in RPMI containing 10% fetal bovine serum and antibiotics and washed twice the next day with warm medium to remove nonadherent cells.
C. albicans Culture
C. albicans strain CHN1 (a human pulmonary clinical isolate) was grown on Sabouraud dextrose agar plates and maintained at 4 °C. 72 h before the experiment, yeast were grown to stationary phase at 37 °C in Sabouraud dextrose broth (1% neopeptone, 2% dextrose (Difco)) with shaking. The cultures were washed in sterile nonpyrogenic PBS, counted with a hemocytometer, and diluted to 2 × 109 colony forming units (CFU)/ml in sterile nonpyrogenic PBS. C. albicans was used either live or killed (through heating for 30 min at 56 °C) as indicated throughout the text and figure legends.
Measurement of LTB4 and CysLTs in the Supernatant of AM Cultures
Levels of LTB4 and cysLTs in the supernatants of AMs (5 × 105) stimulated with 10:1 live or heat-killed C. albicans for different time points were determined using EIA kits (Cayman Chemical Co.) as described (26). In another experimental setting, AMs were pretreated with laminarin or mannan (100 μg/ml each) for 20 min and stimulated with live or heat-killed C. albicans (10:1) for 15 min followed by LTB4 determination.
Phagocytosis Assays
Light Microscopic Assay
2 × 105 WT and 5-LO−/− AMs were plated in 4-well chamber slides (Nunc). In another set of experiments, rat AMs were plated on 8-well glass coverslips in 24-well cell culture-treated dishes (BD Biosciences) and were pretreated or not with inhibitors of PKCδ (6 μm rottlerin), PKCα (9 nm Ro-32-0432), PI3K (10 μm LY 290042 and 10 nm wortmannin), or ERK1/2 (by 2 μm PD98059) and p38 (by 10 μm SB20190) for 20 min before the addition of LTB4 or LTD4 (both at 10 or 100 nm as indicated in the legends). In another set of experiments, AMs were pretreated for 20 min with a 5-LO inhibitor (AA-861, 10 μm), a BLT1 antagonist (CP105,696, 10 μm), or a cysLT1 antagonist (MK571, 10 μm) (24) followed by the addition of 30:1 live C. albicans:AM for 90 min. The inhibitor doses used were based on previously published observations (24, 27–30). After 10 min of incubation with or without LTs, AMs were challenged with a multiplicity of infection of 30:1 live C. albicans for 90 min at 37 °C. Wells were then washed three times with warmed PBS to remove noningested yeast. C. albicans phagocytosis was assessed as described previously (31). Results were expressed as the phagocytic index, which was derived by multiplying the percent of macrophages containing at least one ingested target by the mean number of phagocytosed targets per positive macrophage.
Fluorometric Assay
The ability of AMs to phagocytose C. albicans was also assessed using a previously published protocol for determining the ingestion of fluorescent, heat-killed FITC-labeled C. albicans (FITCC. albicans) (32). Briefly, heat-killed C. albicans were labeled with FITC (33). 4 × 105 murine or rat AMs were seeded in replicates of 5- in 96-well tissue culture plates with opaque sides and optically clear bottoms (Costar, Corning Life Sciences). On the following day AMs were challenged with heat-killed C. albicans using a multiplicity of infection of 10:1 for 90 min to allow phagocytosis to occur. In another set of experiments, AMs were incubated with laminarin and/or mannan (100 μg/ml each) or anti-dectin receptor (clone 2A11; 1 μg/ml) or anti-mannose receptor (clone 15–2; 10 μg/ml) for 30 min before the addition of LTs or C. albicans. Trypan blue (250 μg/ml; Molecular Probes) was added for 10 min to quench the fluorescence of extracellular yeast, and fluorescence was determined using a Spectramax Gemini EM fluorometer at settings of 485 excitation/535 emission (Molecular Devices). The phagocytic index was calculated as previously described in relative fluorescence units (32).
RNA Isolation and Semiquantitative Real Time RT-PCR
RNA from cultured cells was isolated using the RNeasy Mini kit (Qiagen) according to the manufacturer's instructions, and real time RT-PCR was performed as described (34). Cofilin-1 mRNA was normalized to β-actin, and the respective untreated control was set to 100%.
RNA Interference
RNA interference experiments were performed according to a protocol provided by Dharmacon. AMs were transfected using DharmaFECT 1 reagent with 30 nm concentrations of nonspecific control or specific ON-TARGET SMARTpool cofilin-1. After 48 h of transfection, AMs were harvested for mRNA or fluorometric phagocytosis assay.
Confocal Microscopy
A total of 2 × 105 AMs were plated in 4-well chamber slides (Nunc), incubated with or without 10:1 heat-killed C. albicans for 15 min, and then washed with PBS. Slides were fixed in 4% paraformaldehyde in PBS for 30 min at room temperature and permeabilized with 0.5% Triton X-100 in PBS for 3 min. Cells were then blocked with 2% BSA in PBS and incubated with phospho-cofilin-1 (Ser-3, 1:200) in blocking buffer for 1 h at room temperature. Next, slides were washed in PBS and incubated with secondary antibody (1:200) in blocking buffer for 1 h. Cells were also stained with FITC-phalloidin (1 μm) or DNase I-Alexa594 (1 μm) to stain F- or G-actin, respectively, according to the manufacturer's protocol (Molecular Probes). Cells stained with isotype-matched control antibody were used to control for the background from each fluorophore (not shown). Slides were mounted in Prolong Gold mounting media with 4,6 diamidino-2-phenylindole) (Molecular Probes), and cells were imaged on a Zeiss LSM 510 confocal microscope with an inverted Axiovert 100 m microscope stand using a C-apochromat40/1.2 W corr. The 488-nm line from an argon laser and the 543- and 633-nm lines from 2 He/Ne lasers were used for excitation. The images were analyzed using LS 2.5 image analysis software (Zeiss). Photographs were obtained only of AMs containing bound or 1–2 internalized heat-killed C. albicans. Confocal images were taken with identical settings to allow comparison of staining. Single confocal sections of the cells were captured in multitrack. Each set of frames from a given treatment condition depicts a representative AM selected from 20 analyzed cells.
Fluorometric Assay for Quantification of Actin Polymerization
4 × 105 rat AMs were seeded in replicates of 5 in 96-well tissue culture plates with opaque sides and optically clear bottoms (Costar, Corning Life Sciences). On the following day AMs were infected with heat-killed C. albicans using a multiplicity of infection of 10:1 for 30 min. Cells were fixed with 3% paraformaldehyde for 20 min followed by cell blocking with 2% BSA in PBS for 1 h at room temperature. AMs were stained with FITC-phalloidin (1 μm) to stain F-actin, according to the manufacturer's protocol (Molecular Probes). Cells were washed three times in PBS, and fluorescence was determined using a Spectramax Gemini EM fluorometer at settings of 485 excitation/535 emission (Molecular Devices).
Western Blotting
2 × 105 AMs were plated in 6-well tissue culture dishes and were either stimulated or not with 100 nm LTB4 or 100 nm LTD4 for 5 min followed by the addition of 10:1 heat-killed C. albicans for 15 min. AMs were lysed in buffer (50 mm Tris-HCl (pH 7.4), 25 μm KCl, 5 mm MgCl2, and 0.2% Nonidet P-40) supplemented with protease inhibitors (Roche Diagnostics). For immunoblot analysis, protein samples (30 μg) were mixed with loading buffer (50 mm Tris HCl (pH 6.8), 2% SDS, 100 mm DTT, 10% glycerol, and 0.1% bromphenol blue), boiled, applied to 10% SDS-polyacrylamide gels, and subjected to electrophoresis. Immunoblot analysis was performed as previously described (35) using primary antibodies against total and phospho-cofilin-1 (Ser-3) (1:1000, Cell Signaling), phospho-LIMK1 (Thr-508), total and phospho-LIMK-2 (Thr-505) (1:1000, from Abcam), and β-actin (1:10,000, Sigma). Densitometric analysis was as described (36); the intensity of phosphorylation was quantitated as the density of the phosphorylated protein band divided by that of the actin or the respective total protein band, and this ratio was then expressed relative to that of the untreated control, which was set at 100%. In all instances, density values of bands were corrected by subtraction of the background values.
Fungicidal Activity Assay
The ability of AMs to kill C. albicans was evaluated using an assay of CFU as described (37, 38). Briefly, WT and 5-LO−/− mouse AMs or rat AMs (1 × 106) were plated in 12-well tissue culture dishes. The next day live C. albicans yeast cells were added to the macrophages at a multiplicity of infection of 5:1 (C. albicans:AMs) for 1 h to allow phagocytosis to occur. Then cells were treated with compounds of interest for 20 min or as indicated in the figure legends and were incubated at 37 °C for 3 h. Subsequently, all wells were extensively washed to remove extracellular fungi, and the cells were collected and centrifuged at 4500 × g for 5 min. The pellet was resuspended with 1 ml of sterile water to lyse the cells. The contents were vortexed vigorously and then were serially diluted and plated on Sabouraud dextrose agar plates. Inspection of the initial lysate revealed only single colonies, 98% of which were still in the yeast phase. After incubation at 37 °C for 24 h, CFU were counted, and the percentage of C. albicans cells that were killed was calculated by comparison to the CFU obtained from the original inoculum, which also was quantified by serial dilution and plating. Results were expressed as percent survival of ingested yeast, where the survival of ingested yeast = 100% × before inoculum CFU/CFU 3 h after AM incubation.
Measurement of ROI
AMs were added to 96-well plates at a concentration of 4 × 105 cells/well and cultured overnight in RPMI 1640 containing 10% FCS. The next day, medium was replaced with PBS containing 10 μm H2DCF (a cell-permeable oxidant sensitive fluorophore), and the cells were cultured for 1 h. The medium was then replaced with warmed HBSS, and the cells were stimulated with heat-killed C. albicans using a multiplicity of infection of 10:1. ROI production was assessed after 90 min by measuring fluorescence using a Spectramax Gemini XS fluorometer (Molecular Devices) with excitation/emission setting at 493/522 nm. To assess the effect of LTs on ROI production by AMs, LTB4 (100 nm) and LTD4 (100 nm) and heat-killed C. albicans (10:1) were added to this solution before the addition to the AMs.
Statistical Analysis
Graphs represent the mean ± S.E. from three-six independent experiments. The means from different treatments were compared by analysis of variance. When significant differences were identified, individual comparisons were subsequently analyzed with the Bonferroni t test for unpaired values. When two groups were compared, we performed paired Student's t tests. Statistical significance was set at a p value ≤0.05.
RESULTS
Phagocytosis of C. albicans by AMs Requires the Generation of Both Classes of LTs
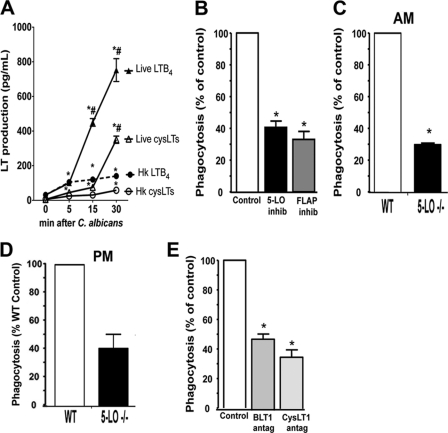
First, we compared the ability of live and heat-killed C. albicans to induce LTB4 and cysLT synthesis in rat AMs at time points relevant to the process of yeast ingestion. Both live and heat-killed C. albicans induced similar levels of LTB4 (∼100 pg/ml, which corresponds to ∼0.5 nm) after 5 min of fungal challenge. However, only live C. albicans further enhanced LTB4 synthesis at the later time point tested (Fig. 1A). The synthesis of cysLTs was also induced by both live and heat-killed C. albicans but at levels lower than LTB4 at all time points studied; this was expected, based on the known capacity for rat AMs to synthesize greater quantities of LTB4 than cysLTs in response to a variety of stimuli (22, 39). Together, our data indicate that LT biosynthesis is a component of the normal macrophage response to this microbe. Next, we asked if LTs produced during C. albicans ingestion have a functional role in AM antimicrobial function. The phagocytic capacity over 90 min of AMs pretreated or not with the 5-LO inhibitor AA861 (10 μm) or the FLAP inhibitor MK0591 (1 μm) was determined microscopically. LT inhibition by both the 5-LO and FLAP inhibitors reduced phagocytosis of live C. albicans by ∼60% when compared with untreated cells (Fig. 1B). To confirm the effects of pharmacologic inhibition of LT synthesis on yeast ingestion determined microscopically, we utilized 5-LO−/− AMs (which cannot synthesize LTs) and control WT AMs in a fluorometric assay of heat-killed FITCC. albicans phagocytosis. We observed a 60% decrease in yeast ingestion in 5-LO−/− cells when compared with WT AMs (Fig. 1C). We also saw a similar reduction in phagocytosis by LT-deficient peritoneal macrophages, suggesting that LTs are necessary for optimal phagocytosis in diverse macrophage populations (Fig. 1D). Thus, LT generation is required for optimal ingestion of both live and heat-killed C. albicans. The importance of individual LT classes in C. albicans ingestion was evaluated using BLT1 and cysLT1 antagonists. Pretreatment with BLT1 antagonist (CP105,696, 10 μm) or cysLT1 antagonist (MK571, 10 μm) resulted in a 45 and 60% reduction, respectively, in live C. albicans ingestion (Fig. 1E). These data suggest that LTs readily produced during C. albicans ingestion are required for optimal yeast phagocytosis.
FIGURE 1.
Endogenously produced LTs are required for optimal phagocytosis of C. albicans. A, LTB4 and cysLT levels were measured in the supernatant from rat AMs infected with 10:1 live or heat-killed C. albicans after 5, 15, and 30 min as described under “Experimental Procedures.” Data are the mean ± S.E. from three separate experiments. *, p < 0.05 compared with unstimulated AMs; #, p < 0.05 compared with live C. albicans. B, AMs were pretreated with or without the 5-LO inhibitor AA 861 (10 μm) or the FLAP inhibitor MK886 (10 μm) for 10 min and challenged with live C. albicans for 90 min, and the phagocytosis was determined microscopically as described. C, AMs from 5-LO−/− and WT mice were challenged with heat-killed FITCC. albicans for 1 h, and phagocytosis was measured by fluorometry. D, peritoneal macrophages (PM) from 5-LO−/− and WT mice were challenged with heat-killed FITCC. albicans for 1 h, and phagocytosis was measured by fluorometry. E, AMs were pretreated or not for 10 min with the BLT1 antagonist CP 105,696 (10 μm) or the cysLT1 antagonist MK 571 (10 μm) before the addition of live C. albicans for 90 min, and phagocytosis was measured microscopically. in C and D, fluorometric assay was performed, and phagocytic indices were calculated and expressed as a percentage of the control. Data are the mean ± S.E. from 3–5 separate experiments. *, p < 0.05 comparing treated to untreated groups or 5-LO−/− to WT AMs.
Having established the importance of endogenously produced LTB4 and cysLTs for the optimal phagocytic capacity of AMs, we next investigated if the addition of LTs to AMs also amplified base-line yeast ingestion. Phagocytosis of live fungus was enhanced by both LTB4 (Fig. 2A) and LTD4 (Fig. 2B) in a dose-dependent fashion. A dose of 100 nm concentrations of each LT caused maximal ingestion of C. albicans, and thus, we chose this dose for subsequent experiments. Exogenous LTB4 and LTD4 each fully restored the deficient phagocytic capability of 5-LO−/− AMs, confirming the importance of LTs for macrophage phagocytosis (Fig. 2C). Interestingly, a combination of LTB4 plus LTD4, each used at a submaximal dose of 10 nm, amplified heat-killed FITCC. albicans ingestion to a greater extent than did either LTB4 or LTD4 alone (Fig. 2C). The same pattern was also observed when testing the effects of exogenous LTs on fungal ingestion employing live C. albicans (Fig. 2D). Consistent with the greater level of LT synthesis in response to live versus heat-killed fungus, the dependence of phagocytosis on LTs as reflected by the magnitude of the phagocytic defect in 5-LO−/− AMs was likewise greater with live than with heat-killed C. albicans (Fig. 2, C and D). Together, these findings show that both classes of LTs are required for optimal C. albicans ingestion in AMs.
FIGURE 2.
Exogenous LTB4 and LTD4 both enhance C. albicans-mediated phagocytosis. Rat AMs were pretreated for 10 min without or with LTB4 (A) or LTD4 (B) in the indicated doses before the addition of live C. albicans for 90 min, and phagocytosis was measured microscopically. C, AMs from WT and 5-LO−/− mice were pretreated or not with 10 nm of LTB4 and/or LTD4 for 10 min before the addition of heat-killed FITCC. albicans for 90 min, and phagocytosis was determined fluorometrically. D, AMs from WT and 5-LO−/− mice were pretreated or not with 10 nm of LTB4 and/or LTD4 for 10 min before the addition of live C. albicans, and phagocytosis was measured microscopically. Data are the mean ± S.E. from three separate experiments. *, p < 0.05 comparing treated to untreated WT groups or 5-LO−/− to WT AMs; #, p < 0.05 comparing untreated 5-LO−/− AMs to LT-activated cells; &, p < 0.05 comparing LTB4 or LTD4 alone.
Identification of the Signaling Pathways Involved in LT Enhancement of Yeast Ingestion
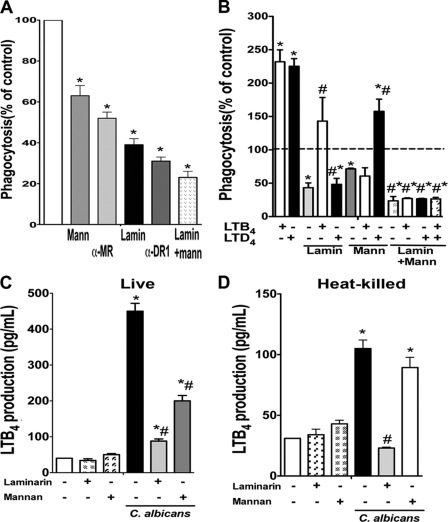
Next, we sought to determine the role of dectin-1 and mannose receptor in yeast phagocytosis and the extent to which ingestion through each was influenced by both classes of LTs. Mannose receptor antagonism by both mannan and anti-mannose receptor antibody inhibited FITCC. albicans phagocytosis by ∼40%, and dectin-1 receptor blockade by both laminarin and anti-dectin-1 antibody inhibited fungal ingestion by ∼60%. The combination of both mannan and laminarin treatment impaired yeast ingestion by ∼80% (Fig. 3A). Although LTB4 enhancement of yeast phagocytosis was abolished when AMs were pretreated with mannan and partially inhibited by laminarin, LTD4 effects were shown to depend primarily on dectin-1 recognition. Conversely, inhibition of both mannose and dectin-1 receptors completely inhibited the stimulatory effect of both classes of LTs (Fig. 3B). We also investigated which PRR is responsible for LTB4 production during C. albicans infection. We pretreated AMs with mannan and laminarin to block mannose receptor and dectin-1, respectively, followed by the addition of either live or heat-killed C. albicans for 15 min. Although LTB4 production induced by live yeast was mainly dependent on dectin-1 and partially dependent on mannose receptor, that induced by heat-killed C. albicans was completely dependent on dectin-1 activation (Fig. 3, C and D).
FIGURE 3.
Role of dectin-1 and mannose receptor in LT-enhanced C. albicans ingestion. A, rat AMs were pretreated with mannan or laminarin (both at 100 μg/ml) or anti-dectin-1 (clone 2A11, Abd Serotec) and anti-mannose receptor (clone 15–1, Biolegend) (both at 10 μg/ml) for 30 min before the addition of 10:1 heat-killed FITC-C. albicans for 90 min, and phagocytosis was determined fluorometrically. Data are the mean ± S.E. from three separate experiments. *, p < 0.05 compared with untreated control. B, rat AMs were pretreated with mannan or laminarin as in A followed by LTB4 or LTD4 for 10 min before the addition of 10:1 heat-killed FITCC. albicans. Phagocytosis was measured by fluorometry after 90 min. Data are the mean ± S.E. from three separate experiments. *, p < 0.05 compared with untreated AMs; #, p < 0.05 versus LTB4 or LTD4 alone. Rat AMs were pretreated with laminarin or mannan as in A followed by 10:1 live (C) or heat-killed (D) C. albicans for 15 min, and LTB4 was determined by ELISA. Data are the mean ± S.E. from three separate experiments. *, p < 0.05 compared with untreated AMs; #, p < 0.05 versus C. albicans alone.
Previously, we showed that BLT1 but not cysLT1 uses Gαi protein to enhance AM antimicrobial functions (17). However, the specific G proteins by which LTs enhance PRR-mediated phagocytosis of unopsonized pathogens is unknown. Here, we sought to investigate the role of Gαi in mediating LTB4/LTD4 effects on C. albicans ingestion. Gαi inhibition by pertussis toxin pretreatment, as described previously (17), abolished the effect of LTB4, but not LTD4, on C. albicans phagocytosis (Fig. 4A). These data show that Gαi mediates BLT1 effects on AM phagocytosis via PRRs.
FIGURE 4.
Inhibition of PI3K and PKCδ but not of PKCα or MAPKs abolishes LT enhancement of C. albicans phagocytosis. A, rat AMs were pretreated with or without pertussis toxin (PTX) for 18 h followed by 10 min of treatment with vehicle or LTB4 and/or LTD4 at a final concentration of 100 nm. Heat-killed FITCC. albicans was added, phagocytosis was measured by fluorometry after 90 min, and phagocytic indices were calculated and expressed as a percentage of the control. Rat AMs were pretreated with selective inhibitors of PKCα (9 nm Ro-32-0432) or PKCδ (6 μm rottlerin) (B), PI3K (10 nm wortmannin and 10 μm LY290042) (D), or ERK1/2 (2 μm PD98059) or p38 (SB20190 10 μm) (E) and then incubated with LTB4 and LTD4 (100 nm for 10 min) before the addition of live C. albicans at a ratio of 30:1. In C, cells were treated overnight with Lipofectin reagent (1% v/v) with or without isotype controls or specific Abs against PKCα and PKCδ (1/500 dilution) as previously described (16) followed by the addition of live C. albicans. In panels B–E, phagocytosis was determined microscopically, and phagocytic indices were calculated and expressed as a percentage of the control. Data are the mean ± S.E. from three-five separate experiments. *, p < 0.05 comparing LT-treated with untreated; #, p < 0.05 versus LTB4 or LTD4 alone.
Next, we investigated the downstream signaling pathways involved in the enhancement of yeast phagocytosis. We focused on the requirement of specific kinases previously shown to be important for LT enhancement of FcR-mediated phagocytosis, including PKCα and -δ, PI3K, and the MAPKs p38 and ERK1/2 (16). Inhibition of PKCδ, but not PKCα, abolished both LTB4 and LTD4 enhancement of C. albicans phagocytosis (Fig. 4B). As a confirmatory approach, we utilized intracellular delivery of a specific mouse monoclonal anti-PKCα or -δ IgG into rat AMs using a liposomal reagent (16). We have previously confirmed the specificity and efficacy of this technique (16, 40, 41). Anti-PKC-δ, but neither anti-PKCα nor nonspecific mouse IgG (not shown), blocked the ability of both LTs to augment phagocytosis (Fig. 4C). We did not observe any effect of the anti-PKCα or -δ Abs on basal phagocytosis (data not shown).
PI3K is pivotal for the engulfment of a variety of phagocytic targets (42). To investigate if PI3K activation is important for LT-enhanced phagocytosis, AMs were treated with two specific PI3K inhibitors, wortmannin (10 nm) and LY290042 (10 μm). We found that the potentiating effects of both LTB4 and LTD4 on yeast phagocytosis were abolished by both inhibitors (Fig. 4D).
Another class of kinases known to participate in C. albicans phagocytosis is the MAPKs, namely, p38 and ERK1/2 (43, 44). We investigated their role in LT amplification of C. albicans phagocytosis using well established pharmacologic inhibitors. The pharmacologic inhibition of ERK1/2 (by 2 μm PD98059) and p38 (by 10 μm SB20190) had no effect on LTB4 or LTD4 potentiation of phagocytosis (Fig. 4E). Moreover, neither ERK1/2 nor p38 influenced basal C. albicans phagocytosis (not shown). Collectively, our data demonstrate that both classes of LTs utilize similar kinase programs to enhance yeast phagocytosis.
LTs Enhance F-actin Assembly during C. albicans Phagocytosis
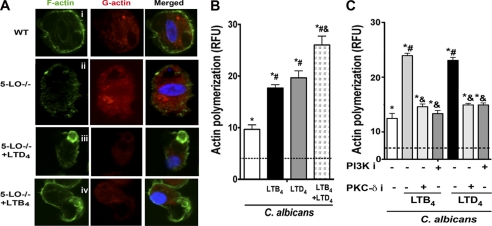
Actin polymerization is required for particle ingestion by phagocytes (45, 46). Both LTB4 and LTD4 are capable of enhancing actin polymerization in leukocytes and epithelial cells (47). However, the role of LTs in assembly of polymerized F-actin during phagocytosis is unknown, and we interrogated this using WT and 5-LO−/− AMs. By using confocal microscopy we observed that 5-LO−/− AMs exhibited higher levels of G-actin and lower levels of F-actin when compared with WT cells (Fig. 5A, i and ii). Interestingly, both exogenously provided LTB4 and LTD4 increased F-actin and reduced G-actin levels during yeast phagocytosis in 5-LO−/− AMs (Fig. 5A, iii and iv). We also determined fluorometrically the importance of LTs on F-actin levels present during live C. albicans ingestion in rat AMs treated with or without LTs; C. albicans enhanced F-actin levels in rat AMs, and both LTB4 and LTD4 potentiated yeast-induced F-actin polymerization. Moreover, the combination of both LTs elicited an additive effect (Fig. 5B). We next determined if kinases involved in potentiation of fungal ingestion by LTB4 and LTD4 also contribute to potentiation of F-actin levels. Indeed, both PI3K and PKCδ inhibition decreased F-actin levels in AMs challenged with both LTB4 and LTD4 during live C. albicans ingestion (Fig. 5C). These results indicate that potentiation of F-actin assembly in AMs is key to the enhancement of C. albicans phagocytosis by both LTB4 and LTD4 and is dependent on both PI3K and PKCδ.
FIGURE 5.
LTB4 and LTD4 enhance actin polymerization through PI3K and PKCδ phosphorylation. A, AMs from WT and 5-LO−/− mice were pretreated or not for 5 min with 100 nm LTB4 or 100 nm LTD4 before the addition of heat-killed C. albicans 10:1 for 15 min. Cells were then incubated with Alexa435-Phalloidin (green) to detect F-actin and Alexa555-DNase (red) to detect G-actin followed by confocal microscopy. Images are from one experiment representative of three independent experiments. B, rat AMs were stimulated and challenged with heat-killed C. albicans as described in A, and cells were incubated with Alexa435-phalloidin (green), after which F-actin was determined by fluorometry. RFU, relative fluorescence units. C, rat AMs were pretreated with the PI3K inhibitor wortmannin (10 nm) or the PKCδ inhibitor rottlerin (6 μm) for 20 min followed by 100 nm LTB4 or LTD4 for 5 min. Heat-killed C. albicans 10:1 was added, and actin polymerization was determined as in B. Each bar represents the mean S.E. from 3–5 individual experiments, each performed in quintuplicate. *, p < 0.05 compared with unstimulated AMs; #, p < 0.05 versus C. albicans; &, p < 0.05 versus LTB4 and LTD4-treated AMs.
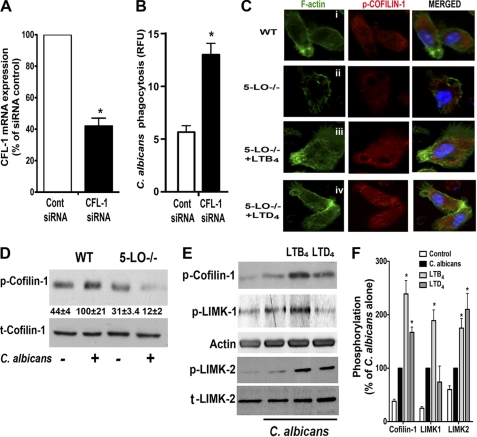
Actin-binding proteins, including cofilin-1, regulate assembly and disassembly of actin filaments (48). Cofilin-1 causes depolymerization at the minus end of filaments, thereby preventing their reassembly (48), and inhibits FcR-mediated phagocytosis (49). However, the importance of this protein in C. albicans phagocytosis is unknown. Thus, we tested the possibility that LTs potentiate F-actin levels by inhibiting the activation of cofilin-1. Cofilin-1 mRNA was silenced with siRNA (Fig. 6A), and yeast ingestion was increased in AMs 2-fold when compared with control siRNA (Fig. 6B), identifying an effect for this actin-binding protein in limiting AM phagocytosis of C. albicans. Cofilin-1 is inactivated by phosphorylation on Ser-3, which results in inhibition of F-actin disassembly, thus allowing actin polymerization. To examine the effect of LTs on cofilin-1 phosphorylation in phagocytosing AMs, we utilized both confocal microscopy in WT and 5-LO−/− AMs (Fig. 6C) and immunoblotting (Fig. 6D). In control WT AMs, C. albicans challenge increased cofilin-1 phosphorylation and, therefore, actin polymerization. As shown in Fig. 6C, i and ii, 5-LO−/− AMs displayed decreased cofilin-1 phosphorylation and concomitantly less actin polymerization upon exposure to C. albicans than did WT cells. That 5-LO−/− AMs were unable to phosphorylate cofilin-1 upon C. albicans challenge was confirmed by immunoblot (Fig. 6D). Next, we sought to investigate the role of specific LTs on cofilin-1 phosphorylation. Treatment of 5-LO−/− AMs with LTD4 and LTB4 restored both cofilin-1 phosphorylation and actin polymerization to levels observed in WT cells infected with C. albicans alone (Fig. 6C, iii and iv). Both classes of LTs also enhanced cofilin-1 phosphorylation in rat AMs, and again LTB4 was more effective than LTD4 in doing so (Fig. 6, E and F). LIM kinases (LIMKs) catalyze the phosphorylation of cofilin-1 on serine 3, and their overexpression reverses cofilin-induced actin depolymerization, leading to accumulation of actin filaments and aggregates (50). Thus, we asked if LTs enhanced activation of LIMK-1 and/or -2. C. albicans challenge itself increased phosphorylation of LIMK-1 but not LIMK-2 (Fig. 6, E and F). Interestingly, although LTB4 increased activation of both LIMK-1 and -2, LTD4 enhanced only the phosphorylation of LIMK-2. These results demonstrate that both LTB4 and LTD4 decreased the activation of cofilin-1, the cellular brake on actin polymerization, thereby enhancing F-actin assembly and optimizing C. albicans ingestion. However, LTB4 exerted stronger effects that correlated with an ability to act via both LIMK-1 and -2, as opposed to the effects of LTD4, which were limited to activation of LIMK-2.
FIGURE 6.
LTs enhance actin polymerization through regulation of cofilin-1 and LIMK-1 and -2 phosphorylation. A, cofilin-1 (CFL-1) mRNA expression by real time RT-PCR was measured in rat AMs incubated with cofilin-1 siRNA or control siRNA for 48 h. B, rat AMs were incubated with cofilin-1 siRNA or control siRNA for 48 h, and heat-killed FITCC. albicans phagocytosis assay was performed fluorometrically. Data represent the mean S.E. from three individual experiments, each performed in quintuplicate. *, p < 0.05 versus control siRNA. RFU, relative fluorescence units. C, AMs from WT and 5-LO−/− mice pretreated or not for 5 min with 100 nm LTB4 or 100 nm LTD4 before the addition of heat-killed C. albicans (10:1 for 10 min) were analyzed by confocal microscopy using phospho-cofilin-1 specific antibody (red) and phalloidin-FITC. Images are from one representative experiment of three independent experiments. D, AMs from WT and 5-LO−/− mice were infected or not with heat-killed C. albicans 10:1 for 10 min, and the lysates were subject to immunoblotting and probed for total and phosphorylated cofilin-1. Numbers under the lanes indicate relative density of phosphorylated cofilin-1 upon C. albicans challenge, expressed as the mean ± S.E. from three independent experiments. E, rat AMs were pretreated or not for 5 min with 100 nm LTB4 or 100 nm LTD4 followed by heat-killed C. albicans (10:1) for 10 min, and the lysates were subject to immunoblotting. Blots are from one experiment representative of three independent experiments. F, relative phosphorylation of cofilin-1, LIMK-1, and LIMK-2 was determined by densitometric analysis of immunoblots from three independent experiments as depicted in panel E and is expressed as the mean ± S.E. In D and E, the intensity of phosphorylation was quantitated as the density of the phosphorylated protein band divided by that of the actin or the respective total protein band, and this ratio was then expressed relative to that of the untreated control, which was set at 100%. *, p < 0.05 versus C. albicans-infected AMs.
LTs Enhance AM Candidacidal Activity
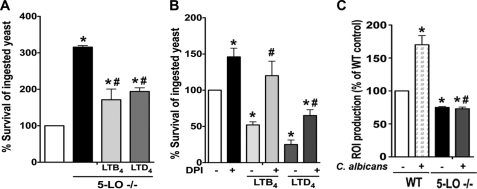
We previously reported that exogenous LTB4 and LTD4 enhanced intracellular killing of IgG-coated Klebsiella pneumoniae as well as Leishmania amazonensis (14, 25). We, therefore, hypothesized that LTs, in addition to enhancing C. albicans phagocytosis, also enhanced AM fungicidal activity. Because ingestion is a prerequisite for intracellular killing and because LTs enhance yeast ingestion, an assay that distinguishes effects on killing from those on phagocytosis was necessary. We accomplished this by adding LTB4 or LTD4 themselves after the ingestion of C. albicans was completed. With such a protocol, exogenous addition of both LTB4 and LTD4 enhanced basal fungicidal activity of LT-deficient AMs (Fig. 7A). Moreover, 5-LO−/− AMs exhibited reduced fungicidal activity (i.e. increased the survival of ingested yeast) than did WT AMs, suggesting that endogenously produced LTs are necessary for the optimal killing of C. albicans by AMs. C. albicans growth was not directly affected by the addition of LTs to macrophage-free cultures (data not shown). These data show that LTs are key mediators that promote two distinct and important steps in the control of yeast infection; that is, ingestion as well as killing.
FIGURE 7.
LTs enhance AM fungicidal activity against C. albicans. A, AMs from 5-LO−/− and WT mice were incubated with live C. albicans at a ratio of 1:5 for 60 min to allow ingestion of the fungi. Cells were then treated with LTB4 (100 nm) or LTD4 (100 nm). The antifungal activity of AMs was measured after 3 h by comparing the CFU of each experimental group compared with control. B, rat AMs were pretreated with the NADPH oxidase inhibitor DPI (10 μm) for 20 min before the addition of live C. albicans (1:5). Thirty minutes later DPI was added back with or without LTB4 (100 nm) or LTD4 (100 nm). Fungicidal activity was determined as described under “Experimental Procedures.” C, AMs were treated with or without LTB4 (100 nm) or LTD4 (100 nm) for 5 min, after which PBS containing 10 μm H2DCF was added, and the cells were cultured for 1 h. The medium was then replaced with warmed HBSS, and the cells were stimulated with heat-killed C. albicans (10:1). ROI production was assessed after 90 min by measuring fluorescence. Data are the mean ± S.E. from 4–8 separate experiments. *, p < 0.05 compared with WT control; #, p < 0.05 when compared with LTB4/LTD4 alone or C.albicans stimulated WT AMs.
LT Enhancement of Candidacidal Activity Involves NADPH Oxidase Activation
ROIs generated by NADPH oxidase activation are important in killing ingested yeast (51, 52), and LTs are known to activate this oxidase in AMs and other cell types (14, 26). Thus, we evaluated the role of NADPH oxidase-derived ROIs in LT enhancement of C. albicans killing. Cells were pretreated with the NADPH oxidase inhibitor DPI (53) followed by the addition of both classes of LTs and then C. albicans. DPI alone inhibited killing (i.e. increased survival of ingested C. albicans) by ∼50% compared with untreated cells (Fig. 7B), indicating an important role for NADPH oxidase in the basal control of C. albicans, as previously reported (51). Exogenous LTB4 failed to overcome the killing defect in DPI-treated cells, whereas exogenous LTD4 did so significantly but incompletely. These results suggest that LTB4 augments candidacidal activity largely via activation of NADPH oxidase, whereas LTD4 accomplishes this partially via oxidase activation and partially via alternative microbicidal mechanisms. The impact of endogenously produced LTs on cellular ROI production in response to live C. albicans infection was examined in WT and LT-deficient AMs. As shown in Fig. 7C, C. albicans infection induced ROI production by WT AMs; this was inhibited in 5-LO−/− AMs, suggesting that endogenous LTs are involved in NADPH oxidase activation and release of ROIs during C. albicans infection. Accordingly, exogenous LTB4 and LTD4 further enhanced C. albicans-induced ROI secretion in 5-LO−/− cells (data not shown). As expected, DPI abolished ROI production in infected AMs (data not shown). Our results show that both classes of LTs enhance yeast killing in a manner-dependent to varying degrees on the activation of NADPH oxidase complex and ROI release.
DISCUSSION
C. albicans is an opportunistic fungal pathogen that causes both local and disseminated infection (3, 54, 55). This pathogen can cause respiratory disease, especially in the setting of immunosuppressive therapy, bone marrow, organ transplants, and HIV infection. It is increasingly apparent that a variety of clinical circumstances are associated with an acquired defect in LT synthesis that itself confers increased susceptibility to infection; examples include HIV infection, bone marrow transplant, vitamin D3 deficiency, and cigarette smoking (10). Here we have explored the role of LTs in phagocytosis and killing of C. albicans by AMs, the resident innate immune defender of the alveolar surface. Overall, our results show that 1) AMs quickly synthesize and release both LTB4 and cysLTs in response to C. albicans, 2) both classes of LTs are required for optimal phagocytosis and killing of the fungi, 3) LTB4 enhances mannose and dectin-1 receptor phagocytosis, whereas LTD4 is required for optimal dectin-1-mediated phagocytosis, 4) LT enhancement of phagocytosis involves the activation of PKCδ and PI3K, with subsequent activation of LIMKs, decreased cofilin-1 activation, and ultimately, F-actin assembly, and 5) LT enhancement of C. albicans killing involves NADPH oxidase activation and ROI generation.
Inhibition of LT synthesis or activity has likewise been reported to impair host defense against a myriad of pathogens, including bacteria (56, 57), viruses (58–60), fungi (20, 61), and parasites (25, 62). That endogenous generation of both classes of LTs is important in the macrophage phagocytic and microbicidal response during C. albicans infection was revealed through both pharmacologic and genetic means. C. albicans can be recognized by a variety of PRRs including dectin-1 and the mannose receptor, each of which can activate specific signaling pathways required for yeast ingestion (7). Here we show that LTB4 production is required for optimal mannose receptor phagocytosis and partially necessary for dectin-1-mediated ingestion. In contrast, LTD4 is essential for optimal dectin-1 phagocytosis. Such findings correlate with the differential role of dectin-1 and mannose receptor in generating LTB4 during C. albicans ingestion. Although live and heat-killed yeast utilize dectin-1 to enhance LTB4 production, mannose receptor participates only in live C. albicans-induced LT synthesis. The greater importance of dectin-1 recognition in mediating heat-killed C. albicans-induced LTB4 production could reflect the exposure of β-glucan in the yeast exposed to high temperatures (63). In addition, our data are in agreement with previous reports showing that dectin-1 is the main receptor involved in both arachidonic acid release and LT secretion in macrophages (9, 64, 65). Little is known about the importance of arachidonic acid metabolites including LTs in enhancing fungal ingestion. Balestrieri et al. (67) identified no defect in phagocytosis of the yeast cell wall product zymosan in LTC4 synthase-deficient peritoneal macrophages. In addition, Okamoto et al. (68) have shown that BLT1−/− bone marrow-derived macrophages exhibit lower phagocytic capability of IgG-opsonized zymosan but not of non-opsonized zymosan. Although our work focused on AMs, we did observe a similar dependence on 5-LO products for phagocytosis by peritoneal macrophages. The discrepancy between these other reports and our own may, therefore, reflect differences in the PRRs recognizing C. albicans versus zymosan or strain of C. albicans and the possibility that LTC4 synthase-deficient cells might overproduce LTB4. Although Balestrieri et al. (38) showed that secretory PLA2 activation was required for optimal C. albicans ingestion and killing, they did not address the role of specific arachidonic acid metabolites. In contrast, our work provides mechanistic insight into the antifungal actions of specific LTs.
Although Gαi signaling was utilized exclusively by LTB4/BLT1, enhanced phagocytosis induced by both LTD4 and LTB4 depended on PKCδ and PI3K, as revealed by both kinase inhibitors and intracellular antibody blockade. We speculate that PI3K is upstream of PKCδ, based on previous work showing that PI3K-induced NADPH oxidase activation is dependent on PKCδ activation (69). Because both PI3K and PKCδ have been previously implicated in phagocytosis as well as in F-actin polymerization (70–72), we explored the effects of LTs on F-actin assembly and the possible signaling programs involved. Both classes of LTs enhanced F-actin assembly and decreased monomeric G-actin levels. Interestingly, LTD4 has been reported to enhance F-actin polymerization in intestinal epithelial cells in a PKCδ- and PI3K-dependent manner (73). It also has been shown that LTB4 enhances actin polymerization in neutrophils (74) and T cells (75). We show that both classes of kinases were required for enhanced F-actin polymerization during C. albicans challenge by LTB4 and LTD4. We also observed that LTB4 enhanced the amount of F-actin surrounding the yeast, which co-localized with phosphorylated (deactivated) cofilin-1. Actin polymerization is regulated by phosphorylation/dephosphorylation of actin depolymerizing factors such as cofilin-1 (48). Cofilin-1 is a major actin-binding protein in leukocytes and plays an important role in restraining the respiratory burst and phagocytosis (49, 76), and its activity is decreased by phosphorylation at serine-3 by LIMKs. LIMKs can be phosphorylated by Rho-family GTPases via the activation of Rho kinase (ROCK) or via Cdc42/Rac-mediated activation of PAK1, -2, or -4 (50, 77). Here, by employing two different approaches (Western blotting and confocal microscopy) we observed that 5-LO-deficient AMs exhibited increased cofilin-1 activation and that LTB4 seems to be more effective than LTD4 in decreasing cofilin-1-mediated F-actin depolymerization. Conversely, LTB4 amplifies LIMK-1 and LIMK-2 phosphorylation, whereas LTD4 only enhances LIMK-2 activity. The greater effect of LTB4 than LTD4 on cofilin-1 phosphorylation may reflect its action via both LIMK isoforms. The underlying reason for differences in actions between the two LTs remains unknown. However, LTB4/BLT1 activates and employs a greater variety of signaling pathways than does LTD4/cysLT1 to enhance AM FcR-mediated phagocytosis (78). Cross-talk between LT signaling and C. albicans signaling could conceivably involve specific cross-activation of dectin-1 and/or mannose receptors by LTD4 and LTB4, respectively, or translocation of those receptors to specific membrane microdomains such as lipid rafts, and association among BLT1 and/or cysLT1 and the fungal PRR(s). In this regard we have shown that BLT1 is more potent than cysLT1 in enhancing FcR-mediated phagocytosis (79). Furthermore, BLT1 associates with FcRI, forming a signaling platform composed of G proteins and kinases, and this platform is required for optimal phagocytosis (78). The observed divergence between BLT1 and cysLT1 in activating LIMKs could be due to activation of different upstream effectors. LIMK-1 is phosphorylated by p21 activated kinases (PAKs) 1–4, whereas LIMK-2 is phosphorylated by PKCδ. However, whether PKCδ is the kinase involved in LTB4 or LTD4-induced LIMK-2 phosphorylation remains to be determined.
Not only did LTs amplify phagocytosis, but they also increased AM candidacidal activity in a manner dependent on NADPH oxidase activation and ROI generation. We have previously confirmed the role of NADPH oxidase in LTB4-mediated killing of K. pneumoniae as suggested by pharmacologic studies with AMs derived from gp91phox−/− mice (14). In addition, LTB4 can promote NADPH oxidase activation by various mechanisms, including enhancement of phosphorylation and membrane translocation of the NADPH oxidase subunit p47phox (14) and also by enhancing the expression of the gp91phox subunit (80). Interestingly, LTD4-induced C. albicans killing was only partially dependent on NADPH oxidase activation. Thus, LTD4 might induce the secretion of candidacidal molecules other than ROIs, such as nitric oxide (81) and defensins (82). Recently, Flamand et al. (66) administered LTB4 intravenously to monkeys and found increased α-defensin plasma levels and enhanced ex vivo antimicrobial activities of plasma.
Previous studies have detailed how signals emanating from G protein-coupled LT receptors enhance AM ingestion and killing of targets recognized by the opsonic FcR (16, 78). The present work extends this concept of LT-mediated phagocytic signal amplification to ingestion via distinct PRRs recognizing C. albicans. We have recently reported that LTB4/BLT1/Gαi signaling also controls NFκB activation downstream of PRRs (36). Together, these studies highlight the emerging importance of cross-talk between LT receptors and PRR signaling.
Acknowledgment
We are grateful to Silvana Aparecida da Silva for technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grants HL-058897 (to M. P.-G.) and HL-103777–01 (C. H. S.). This work was also supported by the Fundação de Amparo a Pesquisa do Estado de São Paulo and the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior.
- AM
- alveolar macrophage
- 5-LO
- 5-lipoxygenase
- PRR
- pathogen recognition receptor
- FLAP
- five-lipoxygenase activating protein
- BLT1
- leukotriene B4 receptor 1
- LT
- leukotriene
- cysLT1
- cysteinyl LT receptor 1
- FcR
- Fcγ receptor
- ROI
- reactive oxygen intermediate
- CFU
- colony forming unit
- LIMK
- LIM kinase.
REFERENCES
- 1. Dictar M. O., Maiolo E., Alexander B., Jacob N., Verón M. T. (2000) Med. Mycol. 38, 251–258 [PubMed] [Google Scholar]
- 2. Kyriazis A. P., Kyriazis A. A. (1993) Diagn. Cytopathol. 9, 487–491 [DOI] [PubMed] [Google Scholar]
- 3. Yao Z., Liao W. (2006) Curr. Opin Pulm. Med. 12, 222–227 [DOI] [PubMed] [Google Scholar]
- 4. Evans S. E. (2010) Proc. Am. Thorac. Soc. 7, 197–203 [DOI] [PubMed] [Google Scholar]
- 5. Jakab G. J., Warr G. A., Astry C. L. (1981) Infect. Immun 34, 610–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sawyer R. T. (1990) Mycopathologia 109, 99–109 [DOI] [PubMed] [Google Scholar]
- 7. Netea M. G., Maródi L. (2010) Trends Immunol 31, 346–353 [DOI] [PubMed] [Google Scholar]
- 8. Castro M., Ralston N. V., Morgenthaler T. I., Rohrbach M. S., Limper A. H. (1994) Infect. Immun. 62, 3138–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Parti R. P., Loper R., Brown G. D., Gordon S., Taylor P. R., Bonventre J. V., Murphy R. C., Williams D. L., Leslie C. C. (2010) Am. J. Respir. Cell Mol. Biol. 42, 415–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Peters-Golden M., Henderson W. R., Jr. (2007) N. Engl. J. Med. 357, 1841–1854 [DOI] [PubMed] [Google Scholar]
- 11. Yokomizo T., Izumi T., Chang K., Takuwa Y., Shimizu T. (1997) Nature 387, 620–624 [DOI] [PubMed] [Google Scholar]
- 12. Lynch K. R., O'Neill G. P., Liu Q., Im D. S., Sawyer N., Metters K. M., Coulombe N., Abramovitz M., Figueroa D. J., Zeng Z., Connolly B. M., Bai C., Austin C. P., Chateauneuf A., Stocco R., Greig G. M., Kargman S., Hooks S. B., Hosfield E., Williams D. L., Jr., Ford-Hutchinson A. W., Caskey C. T., Evans J. F. (1999) Nature 399, 789–793 [DOI] [PubMed] [Google Scholar]
- 13. Mancuso P., Standiford T. J., Marshall T., Peters-Golden M. (1998) Infect. Immun. 66, 5140–5146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Serezani C. H., Aronoff D. M., Jancar S., Mancuso P., Peters-Golden M. (2005) Blood 106, 1067–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Canetti C., Aronoff D. M., Choe M., Flamand N., Wettlaufer S., Toews G. B., Chen G. H., Peters-Golden M. (2006) J. Leukoc. Biol. 79, 1234–1241 [DOI] [PubMed] [Google Scholar]
- 16. Campos M. R., Serezani C. H., Peters-Golden M., Jancar S. (2009) Mol. Immunol. 46, 1204–1211 [DOI] [PubMed] [Google Scholar]
- 17. Peres C. M., Aronoff D. M., Serezani C. H., Flamand N., Faccioli L. H., Peters-Golden M. (2007) J. Immunol. 179, 5454–5461 [DOI] [PubMed] [Google Scholar]
- 18. Skerrett S. J., Henderson W. R., Martin T. R. (1990) J. Immunol. 144, 1052–1061 [PubMed] [Google Scholar]
- 19. Stenke L., Laurén L., Reizenstein P., Lindgren J. A. (1987) Exp. Hematol. 15, 203–207 [PubMed] [Google Scholar]
- 20. Coffey M. J., Phare S. M., George S., Peters-Golden M., Kazanjian P. H. (1998) J. Clin. Invest. 102, 663–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Thorsen S., Busch-Sørensen M., Søndergaard J. (1989) AIDS 3, 651–6532557054 [Google Scholar]
- 22. Peters-Golden M., Canetti C., Mancuso P., Coffey M. J. (2005) J. Immunol. 174, 589–594 [DOI] [PubMed] [Google Scholar]
- 23. Chen X. S., Sheller J. R., Johnson E. N., Funk C. D. (1994) Nature 372, 179–182 [DOI] [PubMed] [Google Scholar]
- 24. Monget P., Fabre S., Mulsant P., Lecerf F., Elsen J. M., Mazerbourg S., Pisselet C., Monniaux D. (2002) Domest. Anim. Endocrinol. 23, 139–154 [DOI] [PubMed] [Google Scholar]
- 25. Serezani C. H., Perrela J. H., Russo M., Peters-Golden M., Jancar S. (2006) J. Immunol. 177, 3201–3208 [DOI] [PubMed] [Google Scholar]
- 26. Serezani C. H., Aronoff D. M., Jancar S., Peters-Golden M. (2005) J. Leukoc. Biol. 78, 976–984 [DOI] [PubMed] [Google Scholar]
- 27. Lee C., Tomkowicz B., Freedman B. D., Collman R. G. (2005) J. Leukoc. Biol. 78, 1016–1023 [DOI] [PubMed] [Google Scholar]
- 28. Dudley D. T., Pang L., Decker S. J., Bridges A. J., Saltiel A. R. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 7686–7689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hu B., Punturieri A., Todt J., Sonstein J., Polak T., Curtis J. L. (2002) J. Leukoc. Biol. 71, 881–889 [PMC free article] [PubMed] [Google Scholar]
- 30. Mugnai S., Ciuffi M., Maurizi M., Bindi D., Franchi-Micheli S., Zilletti L. (1997) Br. J. Pharmacol. 122, 1345–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Smacchia C., Rebulla P., Drago F., Morelati F., Pappalettera M., Sirchia G. (1997) Haematologica 82, 526–531 [PubMed] [Google Scholar]
- 32. Aronoff D. M., Canetti C., Peters-Golden M. (2004) J. Immunol. 173, 559–565 [DOI] [PubMed] [Google Scholar]
- 33. Káposzta R., Maródi L., Hollinshead M., Gordon S., da Silva R. P. (1999) J. Cell Sci. 112, 3237–3248 [DOI] [PubMed] [Google Scholar]
- 34. Serezani C. H., Chung J., Ballinger M. N., Moore B. B., Aronoff D. M., Peters-Golden M. (2007) Am. J. Respir. Cell Mol. Biol. 37, 562–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Canetti C., Hu B., Curtis J. L., Peters-Golden M. (2003) Blood 102, 1877–1883 [DOI] [PubMed] [Google Scholar]
- 36. LeibundGut-Landmann S., Gross O., Robinson M. J., Osorio F., Slack E. C., Tsoni S. V., Schweighoffer E., Tybulewicz V., Brown G. D., Ruland J., Reis e Sousa C. (2007) Nat. Immunol. 8, 630–638 [DOI] [PubMed] [Google Scholar]
- 37. Galès A., Conduché A., Bernad J., Lefevre L., Olagnier D., Béraud M., Martin-Blondel G., Linas M. D., Auwerx J., Coste A., Pipy B. (2010) PLoS Pathog 6, e1000714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Balestrieri B., Maekawa A., Xing W., Gelb M. H., Katz H. R., Arm J. P. (2009) J. Immunol. 182, 4891–4898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Balter M. S., Toews G. B., Peters-Golden M. (1989) J. Immunol. 142, 602–608 [PubMed] [Google Scholar]
- 40. Canetti C., Serezani C. H., Atrasz R. G., White E. S., Aronoff D. M., Peters-Golden M. (2007) J. Immunol. 179, 8350–8356 [DOI] [PubMed] [Google Scholar]
- 41. Chung J., Serezani C. H., Huang S. K., Stern J. N., Keskin D. B., Jagirdar R., Brock T. G., Aronoff D. M., Peters-Golden M. (2008) J. Immunol. 181, 5501–5509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gillooly D. J., Simonsen A., Stenmark H. (2001) J. Cell Biol. 155, 15–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Giraldo E., Martin-Cordero L., Hinchado M. D., Garcia J. J., Ortega E. (2010) Mol. Cell. Biochem. 333, 115–120 [DOI] [PubMed] [Google Scholar]
- 44. Ibata-Ombetta S., Jouault T., Trinel P. A., Poulain D. (2001) J. Leukoc. Biol. 70, 149–154 [PubMed] [Google Scholar]
- 45. Allen L. A., Aderem A. (1996) Curr. Opin Immunol. 8, 36–40 [DOI] [PubMed] [Google Scholar]
- 46. Schindler B., Segal E. (2008) Med. Mycol. 46, 251–258 [DOI] [PubMed] [Google Scholar]
- 47. Deleted in proof.
- 48. Bamburg J. R. (1999) Annu. Rev. Cell Dev. Biol. 15, 185–230 [DOI] [PubMed] [Google Scholar]
- 49. Adachi R., Takeuchi K., Suzuki K. (2002) J. Biol. Chem. 277, 45566–45571 [DOI] [PubMed] [Google Scholar]
- 50. Bernard O. (2007) Int. J. Biochem. Cell Biol. 39, 1071–1076 [DOI] [PubMed] [Google Scholar]
- 51. Takao S., Smith E. H., Wang D., Chan C. K., Bulkley G. B., Klein A. S. (1996) Am. J. Physiol. 271, C1278–C1284 [DOI] [PubMed] [Google Scholar]
- 52. Donini M., Zenaro E., Tamassia N., Dusi S. (2007) Eur. J. Immunol. 37, 1194–1203 [DOI] [PubMed] [Google Scholar]
- 53. Morré D. J. (2002) Antioxid. Redox Signal. 4, 207–212 [DOI] [PubMed] [Google Scholar]
- 54. Ho N. K. (1984) Aust. Paediatr. J. 20, 127–130 [DOI] [PubMed] [Google Scholar]
- 55. Virgili A., Zampino M. R., Mantovani L. (2002) Am. J. Clin. Dermatol. 3, 19–35 [DOI] [PubMed] [Google Scholar]
- 56. Coffey M. J., Phare S. M., Peters-Golden M. (2004) Prostaglandins Leukot. Essent. Fatty Acids 71, 185–190 [DOI] [PubMed] [Google Scholar]
- 57. Bailie M. B., Standiford T. J., Laichalk L. L., Coffey M. J., Strieter R., Peters-Golden M. (1996) J. Immunol. 157, 5221–5224 [PubMed] [Google Scholar]
- 58. Coffey M. J., Phare S. M., Cinti S., Peters-Golden M., Kazanjian P. H. (1999) Blood 94, 3897–3905 [PubMed] [Google Scholar]
- 59. Coffey M. J., Phare S. M., Kazanjian P. H., Peters-Golden M. (1996) J. Immunol. 157, 393–399 [PubMed] [Google Scholar]
- 60. Gaudreault E., Gosselin J. (2007) Viral Immunol. 20, 407–420 [DOI] [PubMed] [Google Scholar]
- 61. Medeiros A. I., Sá-Nunes A., Soares E. G., Peres C. M., Silva C. L., Faccioli L. H. (2004) Infect. Immun. 72, 1637–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Duarte-Escalante E., Zenteno E., Taylor M. L. (2003) Arch. Med. Res. 34, 176–183 [DOI] [PubMed] [Google Scholar]
- 63. Linden J. R., Maccani M. A., Laforce-Nesbitt S. S., Bliss J. M. (2010) Med. Mycol. 48, 355–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Suram S., Gangelhoff T. A., Taylor P. R., Rosas M., Brown G. D., Bonventre J. V., Akira S., Uematsu S., Williams D. L., Murphy R. C., Leslie C. C. (2010) J. Biol. Chem. 285, 30676–30685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hise A. G., Tomalka J., Ganesan S., Patel K., Hall B. A., Brown G. D., Fitzgerald K. A. (2009) Cell Host Microbe 5, 487–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Flamand L., Tremblay M. J., Borgeat P. (2007) J. Immunol. 178, 8036–8045 [DOI] [PubMed] [Google Scholar]
- 67. Balestrieri B., Hsu V. W., Gilbert H., Leslie C. C., Han W. K., Bonventre J. V., Arm J. P. (2006) J. Biol. Chem. 281, 6691–6698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Okamoto F., Saeki K., Sumimoto H., Yamasaki S., Yokomizo T. (2010) J. Biol. Chem. 285, 41113–41121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kilpatrick L. E., Sun S., Li H., Vary T. C., Korchak H. M. (2010) J. Leukoc. Biol. 87, 153–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Waki K., Inanami O., Yamamori T., Nagahata H., Kuwabara M. (2006) Free Radic Res. 40, 359–367 [DOI] [PubMed] [Google Scholar]
- 71. Smallwood N. D., Hausman B. S., Wang X., Liedtke C. M. (2005) Am. J. Physiol. Cell Physiol. 288, C906–C912 [DOI] [PubMed] [Google Scholar]
- 72. Allen L. A., Allgood J. A., Han X., Wittine L. M. (2005) J. Leukoc. Biol. 78, 220–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Massoumi R., Sjölander A. (1998) Eur. J. Cell Biol. 76, 185–191 [DOI] [PubMed] [Google Scholar]
- 74. Omann G. M., Traynor A. E., Harris A. L., Sklar L. A. (1987) J. Immunol. 138, 2626–2632 [PubMed] [Google Scholar]
- 75. Costa M. F., de Souza-Martins R., de Souza M. C., Benjamim C. F., Piva B., Diaz B. L., Peters-Golden M., Henriques M. G., Canetti C., Penido C. (2010) J. Leukoc. Biol. 87, 323–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Bierne H., Gouin E., Roux P., Caroni P., Yin H. L., Cossart P. (2001) J. Cell Biol. 155, 101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lee S., Helfman D. M. (2004) J. Biol. Chem. 279, 1885–1891 [DOI] [PubMed] [Google Scholar]
- 78. Serezani C. H., Aronoff D. M., Sitrin R. G., Peters-Golden M. (2009) Blood 114, 3316–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lee S. P., Serezani C. H., Medeiros A. I., Ballinger M. N., Peters-Golden M. (2009) J. Immunol. 182, 530–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Mancuso P., Lewis C., Serezani C. H., Goel D., Peters-Golden M. (2010) Infect. Immun. 78, 2264–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Vázquez-Torres A., Balish E. (1997) Microbiol. Mol. Biol. Rev. 61, 170–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Hoover D. M., Boulegue C., Yang D., Oppenheim J. J., Tucker K., Lu W., Lubkowski J. (2002) J. Biol. Chem. 277, 37647–37654 [DOI] [PubMed] [Google Scholar]