Abstract
Autophagy pathways in eukaryotic cells mediate the turnover of a diverse set of cytoplasmic components, including damaged organelles and abnormal protein aggregates. Autophagy-mediated degradation is highly regulated, and defects in these pathways have been linked to a number of human disorders. The Atg1 protein kinase appears to be a key site of this control and is targeted by multiple signaling pathways to ensure the appropriate autophagic response to changing environmental conditions. Despite the importance of this kinase, relatively little is known about the molecular details of Atg1 activation. In this study we show that Atg13, an evolutionarily conserved regulator of Atg1, promotes the formation of a specific Atg1 self-interaction in the budding yeast, Saccharomyces cerevisiae. The appearance of this Atg1-Atg1 complex is correlated with the induction of autophagy, and conditions that disrupt this complex result in diminished levels of both autophagy and Atg1 kinase activity. Moreover, the addition of a heterologous dimerization domain to Atg1 resulted in elevated kinase activity both in vivo and in vitro. The formation of this complex appears to be an important prerequisite for the subsequent autophosphorylation of Thr-226 in the Atg1 activation loop. Previous work indicates that this modification is necessary and perhaps sufficient for Atg1 kinase activity. Interestingly, this Atg1 self-association does not require Atg17, suggesting that this second conserved regulator might activate Atg1 in a manner mechanistically distinct from that of Atg13. In all, this work suggests a model whereby this self-association stimulates the autophosphorylation of Atg1 within its activation loop.
Keywords: Autophagy, Protein Degradation, Protein Kinases, Protein-Protein Interactions, TOR Complex (TORC), Atg1 Protein Kinase, Atg17, Macroautophagy, ULK1
Introduction
The term autophagy refers to a collection of membrane-trafficking pathways in eukaryotic cells that are responsible for the degradation of cytoplasmic protein and organelles (1–3). During this transport, these cytoplasmic materials are encapsulated within a membrane-bound intermediate and targeted to the lysosome for degradation (4). Defects in these transport pathways have been linked to a variety of human ailments including specific cancers, Crohn disease, and neurological disorders such as Huntington disease (5–8). In many of these conditions autophagy is being considered as a potential point of therapeutic intervention (9–12). It is, therefore, critical that we develop a thorough understanding of the proteins involved in these transport pathways and the manner in which these degradative activities are regulated.
The basic machinery of autophagy is evolutionarily conserved and has been found in all eukaryotic cells (13). The best understood of these degradative pathways is the nonspecific macroautophagy that was initially characterized at the molecular level in the budding yeast, Saccharomyces cerevisiae (2, 14, 15). This pathway, like most autophagy-based transport, is highly regulated, and the Atg1 protein kinase appears to be a key element of this control (16–18). This enzyme and its associated partners are targeted by multiple signaling pathways to ensure that macroautophagy activity is appropriately coordinated with the cellular environment. For example, this kinase is directly phosphorylated in mammals by both the TORC1 and AMP-activated protein kinases (19–23). The former phosphorylation appears to inhibit autophagy, whereas the latter stimulates this degradative process. The S. cerevisiae Atg1 complex is also targeted by multiple pathways, and Atg1 itself has been shown to be a direct substrate for the cAMP-dependent protein kinase (24, 25). In addition, Atg1 kinase activity has been shown to require a specific autophosphorylation that occurs within the activation loop of this enzyme (26, 27). This phosphorylation site is conserved in Atg1 orthologs and is likely to be an important site of regulation for all eukaryotes. Finally, recent work has identified a site of phosphorylation within a glycine-rich loop that appears to be inhibitory for kinase activity (28). A major challenge for the field is to develop an understanding of how these different regulatory events are properly coordinated within the eukaryotic cell.
The Atg1 protein kinase exists in a complex with a number of proteins, including Atg13 and Atg17 (29, 30). These latter two proteins were originally identified in S. cerevisiae and have been shown to be required for Atg1 kinase activity (2, 31). Functional counterparts of both of these proteins have been found in other eukaryotes, including humans (20, 32–34). In S. cerevisiae, Atg13 directly associates with Atg1, and the presence of this interaction is correlated with Atg1 kinase activity (31). This situation differs somewhat in animals, where Atg13 appears to be constitutively complexed with Atg1 (20, 34). However, it is not yet clear in any case how Atg13 or Atg17 stimulates Atg1 kinase activity. In this study we show that Atg13 promotes an Atg1-Atg1 self-association that appears to be important for both Atg1 kinase activity and the induction of autophagy. Atg17 is not required for this interaction, suggesting that these regulators stimulate Atg1 in distinct ways. Based on our data here, we propose that the interaction with Atg13 allows for the formation of Atg1 dimers (or higher order structures) that are required for Atg1 autophosphorylation within the activation loop.
EXPERIMENTAL PROCEDURES
Strains and Growth Media
Standard Escherichia coli growth conditions and media were used in this study. The yeast-rich growth medium, YPAD, consists of 2% glucose, 1% yeast extract, 2% Bacto-peptone, and 500 mg/liter adenine-HCl. The SC-glucose minimum growth medium has been described (35, 36). The yeast strains used were TN125 (MATa ade2 his3 leu2 lys2 trp1 ura3 pho8::pho8Δ60), YYK126 (TN125 atg1Δ::LEU2), and BY4741 (MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0) (31, 37, 38).
Plasmid Construction
The plasmids used in this study are listed in Table 1. Plasmid pPHY1115 was originally named pRS316-3×mycATG1 and was generously provided by Dr. Yoshinori Ohsumi. Plasmid pPHY2376 is a pRS423-based construct where Atg1 has three copies of the HA epitope at its N terminus and was provided by Dr. Daniel Klionsky. To construct the ATG1 deletions, the appropriate fragments were PCR-amplified from plasmid pPHY1115 and cloned into pRS424 with a C-terminal 3×Myc epitope tag. For the Atg1-ZIP construct, an XmaI site was introduced by a site-directed mutagenesis into the ATG1 locus of pPHY2376 just before the stop codon. The leucine zipper, or ZIP, domain of GCN4 (encoding residues 250–281) was subsequently cloned as a PCR fragment into this newly introduced XmaI site to generate the ATG1-ZIP construct; yeast genomic DNA was used as template in this PCR reaction. For the GAL1-ATG13 construct, the GAL1/10 promoter region and a 6× His tag were PCR-amplified from plasmid pEGH. This PCR fragment was then cloned into pRS316-ATG13 to generate a plasmid encoding a His6-tagged Atg13 under the control of the galactose-inducible promoter from the GAL1 gene. The plasmids pEGH and pRS316-ATG13 were provided by Drs. Michael Snyder and Takeshi Noda, respectively. The ATG13 plasmid pPHY2426 and ATG17 plasmid pPHY2806 have been described (39).
TABLE 1.
Plasmids used in this study
aa, amino acids.
| Plasmid | Description | Source |
|---|---|---|
| pPHY1115 | Atg1, endogenous promoter, C-terminal 3× Myc tag, pRS316 | (52) |
| pPHY2728 | Atg1, endogenous promoter, C-terminal 3× Myc tag, pRS413 | This study |
| pPHY3395 | Atg1, endogenous promoter, C-terminal 3× Myc tag, pRS426 | (27) |
| pPHY2869 | Atg1, endogenous promoter, N-terminal 3× HA tag, pRS416 | This study |
| pPHY2376 | Atg1, endogenous promoter, N-terminal 3× HA tag, pRS423 | (53) |
| pPHY2920 | Atg1(K54A), endogenous promoter, N-terminal 3× HA tag, pRS423 | (27) |
| pPHY2925 | Atg1, aa 1–330, endogenous promoter, N-terminal 3× HA tag, pRS423 | This study |
| pPHY3368 | Atg1, aa 331–897, copper-inducible promoter, N-terminal 3× HA tag, pRS426 | This study |
| pPHY2462 | Atg1, aa 1–529, endogenous promoter, C-terminal 3× Myc tag, pRS424 | This study |
| pPHY2466 | Atg1, aa 1–330, endogenous promoter, C-terminal 3× Myc tag, pRS424 | This study |
| pPHY2935 | Atg1, aa 331–897, endogenous promoter, C-terminal 3× Myc tag, pRS424 | This study |
| pPHY2864 | Atg1, aa 448–897, endogenous promoter, C-terminal 3× Myc tag, pRS424 | This study |
| pPHY2962 | Atg1, aa 550–897, endogenous promoter, C-terminal 3× Myc tag, pRS424 | This study |
| pPHY2965 | Atg1, aa 600–897, endogenous promoter, C-terminal 3× Myc tag, pRS424 | This study |
| pPHY2968 | Atg1, aa 650–897, endogenous promoter, C-terminal 3× Myc tag, pRS424 | This study |
| pPHY2887 | Atg1, aa 731–897, endogenous promoter, C-terminal 3× Myc tag, pRS424 | This study |
| pPHY3381 | Atg1-ZIP, endogenous promoter, N-terminal 3× HA tag, pRS423 | This study |
| pPHY3531 | Atg13, GAL1/10 promoter, N-terminal 6× His tag, pRS426 | This study |
| pPHY2426 | Atg13, CUP1 promoter, N-terminal 3× HA tag, pRS426 | (39) |
| pPHY2806 | Atg17, endogenous promoter, N-terminal 6× Myc tag, pRS424 | (39) |
Autophagy Assays
Autophagy was induced by treating mid-log phase cells with 200 ng/ml rapamycin for 4 h unless otherwise specified. Autophagy activity was assessed with the alkaline phosphatase-based assay that measures the delivery and subsequent activation in the vacuole of an altered form of the Pho8 phosphatase, known as Pho8Δ60 (38). Pho8Δ60 lacks the N-terminal 60 amino acids that include a membrane-spanning region needed to target this protein into the secretion pathway and ultimately to the vacuole (40, 41). Because Pho8 is activated by a proteolytic event that occurs in the vacuole, the Pho8Δ60 protein is a catalytically inactive resident of the cytoplasm. However, Pho8Δ60 can be delivered to the vacuole and subsequently activated by autophagy-mediated transport. The level of autophagy activity was equivalent to the difference in alkaline phosphatase activity detected between extracts prepared from log phase and rapamycin-treated cells. The data presented here are the average of at least three independent experiments.
Western Blotting and Immunoprecipitation
Protein samples for Western blotting were prepared as described (39). The proteins were separated on SDS-polyacrylamide gels, transferred to a nitrocellulose membrane, and then probed with the appropriate primary and secondary antibodies (GE Healthcare). The primary antibodies used were the anti-HA (Roche Applied Science) and anti-myc (Cell Signaling). A chemiluminescent substrate (Thermo Fisher Scientific Inc.) or LI-COR Biosciences Odyssey infrared imaging system was then used to detect the reactive bands or fluorescence intensities, respectively. Immunoprecipitations were performed as described (24, 42). The Atg1-Atg1 analyses were all carried out in the atg1Δ strain, YYK126. HA-tagged proteins were precipitated on an anti-HA affinity matrix (Roche Applied Science), whereas myc-tagged proteins were immunoprecipitated with a monoclonal anti-myc antibody and subsequently collected on Protein A-Sepharose beads. All of the co-immunoprecipitation experiments were performed with cells that had been treated with rapamycin for 60 min unless otherwise indicated. For the cross-linking experiments, atg1Δ cells in mid-log phase that had or had not been treated with rapamycin for 1 h were collected by centrifugation, washed twice with PBS, and then treated with the reversible cross-linker, dithiobis[succinimidylpropionate] (Thermo Scientific), at a final concentration of 1 mm for 30 min at 37 °C. The cross-linking reaction was terminated by adding Tris-HCl, pH 7.5, to a final concentration of 20 mm and incubating for an additional 15 min at 25 °C. Cell lysates were prepared by glass-beading, and the co-immunoprecipitations were carried out as above. To reverse the cross-links, the samples were then incubated at 100 °C for 5 min in a gel-loading buffer containing 5% β-mercaptoethanol. The proteins in the immunoprecipitates were subsequently separated by SDS-polyacrylamide gel electrophoresis, and the relative level of the associated protein was assessed by Western blotting with the appropriate antibody. The generation and characterization of the anti-Thr(P)-226 antiserum has been described (27). Briefly, this antiserum was generated in rabbits immunized with a phosphorylated peptide, FLPNTSLAEpTLCGSPLY, which corresponds to the sequence of the Atg1 activation loop.
In Vitro Kinase Assays
After immunoprecipitation, Atg1 proteins were collected on beads and incubated with 10 μCi of [γ-32P]ATP for 30 min in kinase buffer (25 mm MOPS, 1 mm EGTA, 100 μm Na3VO4, 15 mm MgCl2, and 15 mm p-nitrophenyl phosphate) (31). The reaction products were subsequently separated by SDS-polyacrylamide gel electrophoresis, and the relative level of radioactivity incorporated into the Atg1 proteins was assessed by with a Typhoon Trio PhosphorImager (GE Healthcare). The input level of each protein was assessed by Western blotting with the appropriate antibody.
RESULTS
Atg1 Self-interaction Was Detected Specifically in Cells Undergoing Autophagy
To better understand the regulation of Atg1 kinase activity, we have been examining Atg1 interactions with other proteins in yeast. During the course of this work we found that the full-length Atg1 exhibited a robust interaction with itself in wild-type cells (Fig. 1, A and B). This self-association was detected in extracts prepared from rapamycin-treated cells that contained two Atg1 proteins with different epitope tags. For the experiments here, the Atg1 proteins had either the HA or c-myc epitope. The self-interaction was demonstrated by precipitating one of the tagged proteins and then determining the relative level of the other version of Atg1 in the immunoprecipitates by Western blotting. The presence of Atg1 in different macromolecular complexes has been well documented, but the biological relevance of an Atg1-Atg1 interaction has not been addressed (17, 18, 43).
FIGURE 1.
Atg1 participates in a self-interaction in vivo. A, a schematic shows the different Atg1 constructs used in these binding studies. Atg1-FL refers to the full-length Atg1 protein, and the shaded region indicates the N-terminal protein kinase domain. B, the full-length Atg1 exhibited a robust self-interaction. Extracts were prepared from cells expressing the indicated myc epitope-tagged Atg1 fragments together with an HA-tagged full-length Atg1. The HA3-Atg1 was precipitated on an α-HA affinity resin, and the relative amounts of the associated Atg1 fragments were assessed by Western blotting with an α-myc antibody. The protein bands indicated by the closed circles are degradation products present in the samples. Vec, vector. C, the protein kinase domain of Atg1 exhibited a weak interaction with the full-length Atg1. The interaction between two full-length Atg1 proteins or the 1–330 fragment and a full-length Atg1 was assessed with a co-immunoprecipitation assay as described above. The full-length HA-tagged Atg1 was precipitated in each case, and the relative amount of the associated full-length Atg1 or 1–330 fragment was assessed by Western blotting with an α-myc antibody. To assess the relative strengths of the interactions, 1/20× and 1/50× amounts of the immunoprecipitation mixture from the extracts containing two full-length Atg1 proteins were loaded onto the gel. FL refers to the full-length Atg1. The positions of the full-length Atg1 and the 1–330 fragment of Atg1 are indicated. D, an interaction was observed between two 1–330 fragments of Atg1. The HA-tagged fragment was precipitated on an α-HA affinity resin, and the amount of the associated myc-tagged fragment was assessed by Western blotting with an α-myc antibody.
To map the domains responsible for this Atg1 self-interaction, we tested whether different Atg1 fragments were able to interact with a full-length protein. However, none of the Atg1 fragments examined exhibited a strong interaction in this assay (see Fig. 1, A and B). A weak but detectable interaction was observed between the full-length protein and an N-terminal fragment that contained the kinase domain (Fig. 1, B and C). A similarly weak interaction was also found with two differentially tagged kinase domain fragments (Fig. 1D). One potential explanation for these data is that a strong self-interaction might require domains from both the N- and C-terminal halves of the Atg1 protein.
To assess the physiological relevance of this self-association, we asked whether the amount of this Atg1-Atg1 complex was elevated in cells undergoing macroautophagy (referred to as autophagy for the remainder of this report). For this analysis we used cells expressing the differentially tagged Atg1 proteins at endogenous levels. The cells were treated briefly with a membrane-permeable cross-linking agent, and one of the Atg1 proteins was immunoprecipitated from the resulting extracts. The relative level of the second Atg1 protein in this complex was then determined by Western blotting with the appropriate antibody. Interestingly, we found that the extent of Atg1 self-association was significantly elevated after rapamycin treatment, a condition that induces autophagy (Fig. 2). In contrast, little if any of the Atg1-Atg1 complex was detected in log phase cells where autophagy activity is low (Fig. 2). This correlation with autophagy activity was consistent with the Atg1 self-association having a functional role in this degradative process.
FIGURE 2.
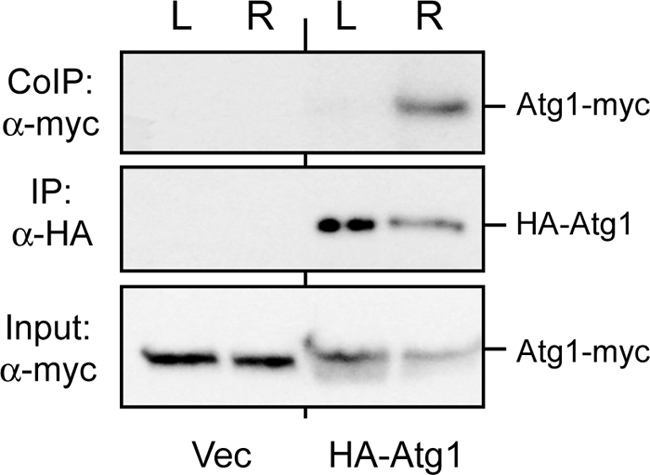
The level of the Atg1-Atg1 complex was elevated in cells undergoing autophagy. Log phase (L) or rapamycin-treated (R) cells were incubated with the cross-linking agent, dithiobis[succinimidylpropionate], for 30 min as described under “Experimental Procedures.” The cells expressed the full-length Atg1-myc3 with or without the full-length HA3-Atg1, as indicated. Cell extracts were prepared, and the HA3-Atg1 was precipitated on an α-HA affinity resin. The relative amount of associated Atg1-myc3 was then assessed by Western blotting as described under “Experimental Procedures.”
The Atg1-Atg1 Interaction Required Atg13 but Not Atg17
Atg13 and Atg17 are required for Atg1 kinase activity, and we tested here whether these regulators were needed for the Atg1 self-interaction. We found that the Atg1 self-association was strongly attenuated in cells lacking Atg13 but was essentially normal in atg17Δ mutants (Fig. 3A). Consistent with these observations, the overexpression of Atg13 led to an increased level of the Atg1-Atg1 complex (Fig. 3B). In contrast, the over-expression of Atg17 had no significant effect on this self-interaction. The results with the atg17Δ mutant suggested that Atg1 kinase activity was not needed for the self-association. This prediction was confirmed when a normal self-interaction was observed with the kinase-defective Atg1 variant, Atg1K54A (Fig. 3C). Altogether, these data suggested that Atg13 and Atg17 might function in different ways to stimulate Atg1 kinase activity. In support of this assertion, we found that the overexpression of Atg17 inhibited both Atg1 kinase activity and the autophagy levels induced by rapamycin (Fig. 4, A and B). In contrast, both of these activities were elevated in the presence of excess Atg13 (Fig. 4, A and B) (27). For this analysis, autophagy activity was measured with a standard assay that assesses the vacuolar delivery of an altered form of the Pho8 alkaline phosphatase known as Pho8Δ60 (38, 41). Atg1 kinase activity was assessed by determining the relative level of autophosphorylation at Thr-226 within the activation loop (27). In all, these studies suggested that Atg13, but not Atg17, promotes an Atg1 self-association that could be important for Atg1 kinase activity.
FIGURE 3.
Atg1 self-association required the presence of the Atg13 protein. A, the level of the Atg1-Atg1 complex was greatly diminished in atg13Δ cell extracts. The relative level of the Atg1 self-interaction was assessed in the indicated strains expressing the full-length Atg1-myc3 with or without the full-length HA3-Atg1. The HA3-Atg1 was precipitated on an α-HA affinity resin, and the relative amount of the associated Atg1-myc3 was assessed by Western blotting with an α-myc antibody. B, the relative level of the Atg1 self-association was elevated in cells overexpressing (OE) Atg13. The values shown below the top panel indicate the relative level of Atg1-myc protein present in the co-immunoprecipitation reaction. The Western blot signal was quantified with the ImageJ program and normalized to that present in the vector (Vec) control lane. C, the kinase-defective Atg1K54A variant exhibited a normal self-interaction. The Atg1 self-association was examined in cells expressing myc- and HA-tagged versions of either the wild-type Atg1 or the kinase-defective variant, Atg1K54A, as indicated.
FIGURE 4.
The overexpression of Atg17 resulted in diminished levels of both autophagy and Atg1 kinase activity. A, autophagy levels were reduced in cells overexpressing Atg17. Autophagy was assessed with the Pho8Δ60-based alkaline phosphatase assay as described under “Experimental Procedures.” The graph shows the relative autophagy activity present in the indicated strains; the wild type is assigned the value of 100%. B, the levels of Atg1 autophosphorylation at Thr-226 were reduced in cells overexpressing (OE) Atg17. The relative level of Thr-226 phosphorylation in Atg1 was assessed in cells overexpressing either Atg13 or Atg17 by Western blotting with an α-Thr(P)-226 phosphospecific antibody (27). The levels of Thr-226 phosphorylation and total Atg1 protein were determined with a LICOR Biosciences Odyssey infrared imaging system as described under “Experimental Procedures.” The bottom panel shows the merged image for the α-Thr(P)-226 and α-HA panels where the former is false-colored green, and the latter is red. Note that different exposure times were used to illustrate the relevant effects of Atg13 and Atg17 overexpression. Atg1-P indicates the position of an autophosphorylated form of Atg1 that migrates anomalously on SDS-polyacrylamide gels (24, 28).
Disruption of the Atg1-Atg13 Interaction Resulted in a Reduced Level of Atg1 Self-association
The above data showed that the Atg1 self-association was diminished in cells that lacked Atg13. However, it was formally possible that Atg13 did not need to bind to Atg1 to influence this self-association. To test this latter possibility, we set out to examine the Atg1 self-association in cells where the Atg1-Atg13 interaction was disrupted. The experimental plan was to identify the Atg1 domain responsible for the interaction with Atg13 and to assess the effects of over-expressing this protein element. Using a series of deletion constructs, we found that a C-terminal region of Atg1 was sufficient for an interaction with Atg13 (Fig. 5, A and B). A strong interaction was observed with the Atg1550-C fragment, but detectable binding was still present with the smaller Atg1600-C fragment (Fig. 5C). These results are consistent with previous work on both the mammalian and yeast Atg1 that implicated a C-terminal element in the binding to Atg13 (19, 26, 32, 43). Unfortunately, we were unable to map the C-terminal end point of this binding domain because of the instability exhibited by fragments lacking the C terminus of Atg1. For the remainder of this report we will refer to the C-terminal region of Atg1 as the thirteen interaction domain (TID).2 The full TID is likely contained within the 550-C fragment of Atg1, although the 600-C fragment also exhibited detectable binding to Atg13.
FIGURE 5.
A C-terminal domain of Atg1 was necessary and sufficient for the interaction with Atg13. A, a schematic depicts the Atg1 fragments used for the mapping analysis described here. B and C, Atg13 interacted specifically with C-terminal fragments of Atg1. HA-Atg13 was immunoprecipitated from yeast cells that also expressed the indicated myc-tagged fragments of Atg1. The relative levels of the Atg1 fragments present in the immunoprecipitates were assessed by Western blotting with an α-myc antibody. The closed circles indicate degradation products present in the samples. FL, full-length. Vec, vector.
To disrupt the Atg1-Atg13 interaction, we overexpressed Atg1 fragments that contained the TID. The expectation was that these fragments would bind to Atg13 and thereby sequester this protein away from the full-length Atg1. Consistent with this prediction, we found that overexpression of the Atg1331-C fragment resulted in a diminished Atg1-Atg13 interaction (Fig. 6A). These effects were specific, as this overexpression had no significant effect on Atg13 binding to Atg17 (Fig. 6B). Interestingly, Atg17 interacted specifically with a C-terminal fragment of Atg1, and this latter interaction was also disrupted by the overexpression of Atg1331-C (Fig. 6, B and C). These data are consistent with previous reports indicating that the Atg17 interaction with Atg1 is mediated at least in part by Atg13 (29, 30). More germane to our discussion here, we found that the overexpression of the TID-containing fragment, Atg1331-C, also disrupted the Atg1 self-association (Fig. 7A). In contrast, the overexpression of the N-terminal Atg11–330 region did not have an effect upon this Atg1-Atg1 interaction. Finally, we found that the observed effects of the Atg1331-C fragment were dose-dependent, and only fragments that were capable of binding to Atg13 disrupted the Atg1 self-association (Fig. 7, B and C). For these latter two analyses, we employed a mixing strategy and added the different Atg1 fragments back to a cell extract that contained the already-formed Atg1-Atg1 complex. In all, the results here suggested that Atg13 binding to Atg1 was a prerequisite for the Atg1 self-interaction.
FIGURE 6.
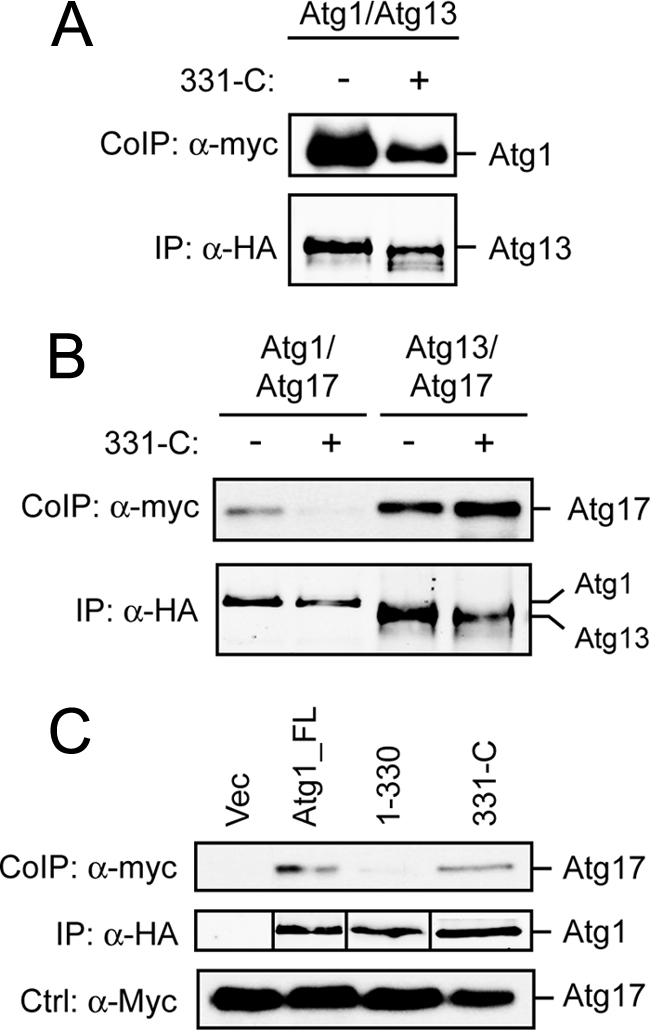
The overexpression of the 331-C fragment of Atg1 specifically disrupted the interaction between Atg13 and Atg1. A, overexpression of the 331-C fragment of Atg1 disrupted the interaction between Atg13 and Atg1. The 331-C fragment of Atg1 was overexpressed in cells containing an HA-tagged Atg13 and a myc-tagged Atg1. Atg13 was collected on an α-HA affinity resin, and the amount of the associated Atg1 was assessed by Western blotting with an α-myc antibody. B, overexpression of the 331-C fragment of Atg1 did not affect the interaction between Atg13 and Atg17. The 331-C fragment of Atg1 was overexpressed in cells containing a myc-tagged Atg17 and an HA-tagged version of either Atg1 or Atg13. The latter HA-tagged proteins were collected on an α-HA affinity resin, and the amount of the associated Atg17 was assessed by Western blotting with an α-myc antibody. C, Atg17 exhibited an interaction with the C-terminal 331-C domain of Atg1. HA-tagged versions of the indicated fragments of Atg1 (full-length, 1–330, and 331-C) were co-expressed in cells with a myc-tagged Atg17. The HA-tagged fragments were collected on an α-HA affinity resin, and the amount of the associated Atg17 was assessed by Western blotting with an α-myc antibody. Note that the Western blot bands corresponding to the different HA-tagged fragments of Atg1 were spliced together to facilitate the comparison of protein levels.
FIGURE 7.
Overexpression of TID-containing fragments of Atg1 led to a disruption of the Atg1 self-interaction. A, overexpression of the 331-C fragment of Atg1 resulted in a diminished level of the Atg1-Atg1 complex. The 1–330 and 331-C fragments of Atg1 were overexpressed in cells that were also expressing HA- and myc-tagged versions of the full-length Atg1. The Atg1 self-interaction was detected by Western blotting for the presence of the Atg1-myc protein in immunoprecipitates that had been collected on an α-HA affinity resin. B, increasing amounts of the 331-C fragment of Atg1 led to a correspondingly greater disruption of the Atg1 self-interaction. C, the 600-C fragment was the minimal Atg1 domain capable of disrupting the Atg1 self-interaction. The effects of overexpressing the indicated fragments of Atg1 on the Atg1 self-interaction were assessed as described above in A.
The Overexpression of a TID-containing Fragment of Atg1 Resulted in Diminished Levels of Both Atg1 Kinase Activity and Autophagy
We also assessed whether the above conditions that disrupted the Atg1-Atg1 interaction would have an effect upon the autophagy process. Indeed, we found that the overexpression of Atg1 fragments containing the TID led to a significant decrease in both Atg1 kinase activity and autophagy (Fig. 8, A and B). Moreover, these inhibitory effects on autophagy were reversed by the simultaneous overexpression of Atg13 (Fig. 8C). In all, these data suggested that both Atg13 binding to Atg1 and the Atg1 self-association are required in vivo for autophagy and Atg1 kinase activity.
FIGURE 8.

Overexpression of TID-containing fragments of Atg1 resulted in diminished levels of both autophagy and Atg1 protein kinase activity. A, overexpression of Atg1 fragments that contained the TID led to an inhibition of autophagy activity. Autophagy activity was assessed in the wild-type strain, TN125, overexpressing the indicated fragments of Atg1 with the Pho8Δ60-based alkaline phosphatase assay. The graph shows the relative autophagy activity present in the indicated strains after normalization to the wild type. B, the levels of Atg1 autophosphorylation at Thr-226 were reduced in cells overexpressing TID-containing fragments of Atg1. The relative level of Thr-226 phosphorylation was assessed by Western blotting with an α-Thr(P)-226 phosphospecific antibody (27). The levels of Thr-226 phosphorylation and total Atg1 protein were determined with a LICOR Biosciences Odyssey infrared imaging system as described under “Experimental Procedures.” Atg1-P indicates the position of an autophosphorylated form of Atg1 that migrates anomalously on SDS-polyacrylamide gels (24, 28). C, the inhibition of autophagy associated with overexpression of the 550-C fragment of Atg1 was suppressed by the presence of excess Atg13. Autophagy activity was assessed in the indicated cells with the Pho8Δ60-based alkaline phosphatase assay. The graph shows the relative autophagy activity present in the indicated strains after normalization to the wild type. Wt refers to the wild-type yeast strain, TN125.
The Presence of a Heterologous Dimerization Domain Resulted in Elevated Atg1 Kinase Activity
The above data suggested that the formation of an Atg1 dimer (or higher order structure) might be an important step in the activation of Atg1. To address this possibility further, we assessed the effects of adding a heterologous dimerization domain to the C terminus of Atg1. For this experiment we used the leucine zipper segment of the Gcn4 transcription factor (44). This domain has been extensively studied and has been shown to be sufficient to mediate the dimerization of an attached heterologous protein (45). Both the wild-type Atg1 and the Atg1-ZIP fusion protein were expressed in an atg13Δ mutant, and Atg1 kinase activity was assessed. For the in vitro assays we immunoprecipitated the Atg1 proteins from both log phase and rapamycin-treated cells and then carried out an autophosphorylation assay with [γ-32P]ATP. In both conditions the Atg1-ZIP protein exhibited a 2-fold higher level of autophosphorylation activity than the wild-type Atg1 (Fig. 9A). For the in vivo determinations, the level of Thr-226 autophosphorylation was assessed in rapamycin-treated cells. Here, we found that the presence of the leucine zipper motif resulted in a more than 3-fold increase in Atg1 activity (Fig. 9B). However, it should be noted that the presence of Atg1-ZIP at endogenous levels was not sufficient to induce autophagy in an atg13Δ mutant. In all, these data indicated that Atg1 kinase activity was stimulated by the presence of a heterologous dimerization domain and were consistent with models suggesting that the formation of an Atg1-Atg1 complex was an important prerequisite for Atg1 activation in vivo.
FIGURE 9.

The addition of a heterologous dimerization domain resulted in elevated Atg1 protein kinase activity in an atg13Δ strain. A, the presence of a leucine zipper dimerization motif led to increased Atg1 autophosphorylation in vitro. Atg1 and Atg1-ZIP were immunoprecipitated from either log phase (gray bars) or rapamycin-treated (black bars) cells, and an in vitro autophosphorylation assay was carried out with [γ-32P] ATP as described under “Experimental Procedures.” The reaction products were then run out on an SDS-polyacrylamide gel, and the amount of radiolabel incorporated into Atg1 was assessed by phosphorimaging. The lower panel indicates the relative Atg1 protein amount present in the kinase reactions. L, log phase; R, rapamycin-treated. B, the presence of the dimerization domain resulted in an elevated level of Atg1 autophosphorylation at Thr-226 in vivo. The relative level of autophosphorylation at Thr-226 on the indicated proteins was assessed by Western blotting with the α- Thr(P)-226 phosphospecific antibody. The cells were treated for 1 h with 200 ng/ml rapamycin before analysis. The levels of Thr-226 phosphorylation and total Atg1 protein were determined with a LICOR Biosciences Odyssey infrared imaging system. The Thr(P)-226 Index refers to the α-Thr(P)-226 signal after normalization for the amount of Atg1 protein present. C, shown is a model for the activation of Atg1. Atg13 binds to a C-terminal domain of Atg1 and to Atg17. This interaction with Atg13 promotes an Atg1 self-association that results in the subsequent autophosphorylation of Thr-226 in the activation loop; an intermolecular, or in trans, reaction is shown. Note that the relative stoichiometry of the different proteins in this complex has not yet been determined. See “Discussion” for further details.
DISCUSSION
Autophagy-mediated degradation is highly regulated in eukaryotic cells, and the Atg1 protein kinase appears to be a key target of this control. Atg1 kinase activity is required for autophagy and is elevated in response to conditions known to induce this degradative pathway (18, 31). However, the molecular events required for the activation of Atg1 remain poorly understood. The work here examined this activation process and provides insight into the role of Atg13, an evolutionarily conserved regulator of Atg1 kinase activity. In particular, we find that Atg13 promotes the formation of a specific Atg1-Atg1 complex whose appearance is correlated with increased autophagy activity. Conditions that disrupt this Atg1 self-association result in diminished autophagy and Atg1 kinase activity. Finally, the addition of a heterologous dimerization domain to Atg1 resulted in elevated kinase activity in both in vivo and in vitro assays. In all, these data suggest that the formation of a dimer or higher order structure is an important intermediate event during the activation of Atg1.
The data here also suggest that Atg13 and Atg17 function in distinct ways to activate Atg1. In particular, neither the absence nor overexpression of Atg17 has any significant effect on the formation of the Atg1 self-interaction. In addition, we found that the overexpression of Atg17 results in diminished autophagy and Atg1 kinase activity; both of these activities are stimulated in the presence of excess Atg13 (27). These results suggest that the relative stoichiometry of Atg17 and Atg1 might be important for the normal activation of Atg1. However, further work will be needed to determine precisely how this conserved regulator influences Atg1 activity in eukaryotic cells.
Based upon this study, we propose the following sequence of events for the activation of Atg1. Atg13 initially binds to the C terminus of Atg1 and thereby promotes the formation of a specific Atg1-Atg1 complex (Fig. 9C). The presence of this Atg1 self-association in turn leads to the autophosphorylation of Thr-226 within the activation loop. Previous work has indicated that this modification is required and is perhaps sufficient for Atg1 kinase activity (27). This autophosphorylation could occur as a result of either an in trans or in cis reaction mechanism. In the in trans reaction each Atg1 chain in the complex would be recognized and phosphorylated by a partner enzyme. An intermolecular mechanism is thought to be responsible for most instances where protein kinases are known to be autophosphorylated within the activation loop (46). Of particular interest is a model suggested by recent work with a number of protein kinases, including the Chk2 enzyme (47, 48). For these enzymes, crystallographic studies have demonstrated that the inactive enzyme is capable of forming a dimer where each partner exchanges its activation loop with the other. This exchange appears to generate an active configuration of the enzyme and allows for the trans-phosphorylation within the activation loop. Therefore, one of the roles for Atg13 could be to act as a scaffold that brings Atg1 molecules together to allow for the subsequent exchange of the activation loops. This model is consistent with the observation that both the C-terminal TID and N-terminal sequences within the kinase domain appear to be required for the Atg1 self-interaction (see Fig. 1). In addition, for the kinases that undergo this exchange, it has been noted that the phosphorylation site within the activation loop does not conform to the consensus that is recognized subsequently by the activated enzyme (46, 48). This may also be true for Atg1, as the sequence around Thr-226 differs substantially from that surrounding a second site of autophosphorylation, Ser-390 (28).
Although the autophosphorylation at Thr-226 could instead be the result of an intramolecular, or in cis phosphorylation reaction, the architecture of the kinase domain suggests that this sort of a reaction is less likely (49). However, there are a few instances where autophosphorylation within the activation loop does occur by an in cis mechanism. In these cases the kinases appear to carry out this autophosphorylation either as a nascent chain that is still associated with the ribosome or in the presence of specific chaperone proteins (50, 51). Because we have found that the autophosphorylation at Thr-226 can occur in vitro with the fully formed protein (27), we currently favor an in trans model for Atg1 activation loop phosphorylation. However, we have not yet been able to detect the in trans phosphorylation of a kinase-inactive version of Atg1 in vivo.3 One explanation for this result is that the alterations that inactivate Atg1 might also affect the structure of the activation loop and, thus, preclude its phosphorylation by the wild-type enzyme present. In any case it is clear that a complete understanding of Atg1 function will require a more detailed picture of the active form of this enzyme and that this will likely involve additional structural studies.
Acknowledgments
We thank Daniel Klionsky, Takeshi Noda, Yoshinori Ohsumi, and Michael Snyder for generously providing strains and plasmids used during the course of this work.
This work was supported, in whole or in part, by National Institutes of Health Grant GM65227 (to P. K. H.).
Y-Y. Yeh, K. H. Shah, and P. K. Herman, unpublished observations.
- TID
- thirteen interaction domain
- IP
- immunoprecipitate.
REFERENCES
- 1. Klionsky D. J. (2007) Nat. Rev. Mol. Cell Biol. 8, 931–937 [DOI] [PubMed] [Google Scholar]
- 2. Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y. (2009) Nat. Rev. Mol. Cell Biol. 10, 458–467 [DOI] [PubMed] [Google Scholar]
- 3. Todde V., Veenhuis M., van der Klei I. J. (2009) Biochim. Biophys. Acta 1792, 3–13 [DOI] [PubMed] [Google Scholar]
- 4. Mizushima N. (2007) Genes Dev. 21, 2861–2873 [DOI] [PubMed] [Google Scholar]
- 5. Mizushima N., Klionsky D. J. (2007) Annu. Rev. Nutr. 27, 19–40 [DOI] [PubMed] [Google Scholar]
- 6. Winslow A. R., Rubinsztein D. C. (2008) Biochim. Biophys. Acta 1782, 723–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Levine B., Kroemer G. (2008) Cell 132, 27–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Deretic V. (2010) Curr. Opin. Cell Biol. 22, 252–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. (2008) Nature 451, 1069–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rubinsztein D. C., Gestwicki J. E., Murphy L. O., Klionsky D. J. (2007) Nat. Rev. Drug Discov. 6, 304–312 [DOI] [PubMed] [Google Scholar]
- 11. Orvedahl A., Levine B. (2009) Cell Death Differ. 16, 57–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. White E., DiPaola R. S. (2009) Clin. Cancer Res. 15, 5308–5316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang Z., Klionsky D. J. (2010) Nat. Cell Biol. 12, 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Baba M., Osumi M., Ohsumi Y. (1995) Cell Struct. Funct. 20, 465–471 [DOI] [PubMed] [Google Scholar]
- 15. Xie Z., Klionsky D. J. (2007) Nat. Cell Biol. 9, 1102–1109 [DOI] [PubMed] [Google Scholar]
- 16. Stephan J. S., Herman P. K. (2006) Autophagy 2, 146–148 [DOI] [PubMed] [Google Scholar]
- 17. Chan E. Y., Tooze S. A. (2009) Autophagy 5, 758–765 [DOI] [PubMed] [Google Scholar]
- 18. Mizushima N. (2010) Curr. Opin. Cell Biol. 22, 132–139 [DOI] [PubMed] [Google Scholar]
- 19. Jung C. H., Jun C. B., Ro S. H., Kim Y. M., Otto N. M., Cao J., Kundu M., Kim D. H. (2009) Mol. Biol. Cell 20, 1992–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hosokawa N., Hara T., Kaizuka T., Kishi C., Takamura A., Miura Y., Iemura S., Natsume T., Takehana K., Yamada N., Guan J. L., Oshiro N., Mizushima N. (2009) Mol. Biol. Cell 20, 1981–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ganley I. G., Lam du H., Wang J., Ding X., Chen S., Jiang X. (2009) J. Biol. Chem. 284, 12297–12305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim J., Kundu M., Viollet B., Guan K. L. (2011) Nat. Cell Biol. 13, 132–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Egan D. F., Shackelford D. B., Mihaylova M. M., Gelino S., Kohnz R. A., Mair W., Vasquez D. S., Joshi A., Gwinn D. M., Taylor R., Asara J. M., Fitzpatrick J., Dillin A., Viollet B., Kundu M., Hansen M., Shaw R. J. (2011) Science 331, 456–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Budovskaya Y. V., Stephan J. S., Deminoff S. J., Herman P. K. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 13933–13938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cebollero E., Reggiori F. (2009) Biochim. Biophys. Acta 1793, 1413–1421 [DOI] [PubMed] [Google Scholar]
- 26. Kijanska M., Dohnal I., Reiter W., Kaspar S., Stoffel I., Ammerer G., Kraft C., Peter M. (2010) Autophagy 6, 1168–1178 [DOI] [PubMed] [Google Scholar]
- 27. Yeh Y. Y., Wrasman K., Herman P. K. (2010) Genetics 185, 871–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yeh Y. Y., Shah K. H., Chou C. C., Hsiao H. H., Wrasman K. M., Stephan J. S., Stamatakos D., Khoo K. H., Herman P. K. (2011) Autophagy 7, 716–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kabeya Y., Kamada Y., Baba M., Takikawa H., Sasaki M., Ohsumi Y. (2005) Mol. Biol. Cell 16, 2544–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cheong H., Yorimitsu T., Reggiori F., Legakis J. E., Wang C. W., Klionsky D. J. (2005) Mol. Biol. Cell 16, 3438–3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kamada Y., Funakoshi T., Shintani T., Nagano K., Ohsumi M., Ohsumi Y. (2000) J. Cell Biol. 150, 1507–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chan E. Y., Longatti A., McKnight N. C., Tooze S. A. (2009) Mol. Cell. Biol. 29, 157–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hara T., Takamura A., Kishi C., Iemura S., Natsume T., Guan J. L., Mizushima N. (2008) J. Cell Biol. 181, 497–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chang Y. Y., Neufeld T. P. (2009) Mol. Biol. Cell 20, 2004–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chang Y. W., Howard S. C., Budovskaya Y. V., Rine J., Herman P. K. (2001) Genetics 157, 17–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kaiser C., Michaelis S., Mitchell A. (1994) Methods in Yeast Genetics, pp. 207–210, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 37. Giaever G., Chu A. M., Ni L., Connelly C., Riles L., Véronneau S., Dow S., Lucau-Danila A., Anderson K., André B., Arkin A. P., Astromoff A., El-Bakkoury M., Bangham R., Benito R., Brachat S., Campanaro S., Curtiss M., Davis K., Deutschbauer A., Entian K. D., Flaherty P., Foury F., Garfinkel D. J., Gerstein M., Gotte D., Güldener U., Hegemann J. H., Hempel S., Herman Z., Jaramillo D. F., Kelly D. E., Kelly S. L., Kötter P., LaBonte D., Lamb D. C., Lan N., Liang H., Liao H., Liu L., Luo C., Lussier M., Mao R., Menard P., Ooi S. L., Revuelta J. L., Roberts C. J., Rose M., Ross-Macdonald P., Scherens B., Schimmack G., Shafer B., Shoemaker D. D., Sookhai-Mahadeo S., Storms R. K., Strathern J. N., Valle G., Voet M., Volckaert G., Wang C. Y., Ward T. R., Wilhelmy J., Winzeler E. A., Yang Y., Yen G., Youngman E., Yu K., Bussey H., Boeke J. D., Snyder M., Philippsen P., Davis R. W., Johnston M. (2002) Nature 418, 387–391 [DOI] [PubMed] [Google Scholar]
- 38. Noda T., Matsuura A., Wada Y., Ohsumi Y. (1995) Biochem. Biophys. Res. Commun. 210, 126–132 [DOI] [PubMed] [Google Scholar]
- 39. Stephan J. S., Yeh Y. Y., Ramachandran V., Deminoff S. J., Herman P. K. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 17049–17054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Scott S. V., Hefner-Gravink A., Morano K. A., Noda T., Ohsumi Y., Klionsky D. J. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 12304–12308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Noda T., Klionsky D. J. (2008) Methods Enzymol. 451, 33–42 [DOI] [PubMed] [Google Scholar]
- 42. Deminoff S. J., Ramachandran V., Herman P. K. (2009) Genetics 182, 529–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Abeliovich H., Zhang C., Dunn W. A., Jr., Shokat K. M., Klionsky D. J. (2003) Mol. Biol. Cell 14, 477–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Landschulz W. H., Johnson P. F., McKnight S. L. (1988) Science 240, 1759–1764 [DOI] [PubMed] [Google Scholar]
- 45. Rieker J. D., Hu J. C. (2000) Methods Enzymol. 328, 282–296 [DOI] [PubMed] [Google Scholar]
- 46. Oliver A. W., Knapp S., Pearl L. H. (2007) Trends Biochem. Sci. 32, 351–356 [DOI] [PubMed] [Google Scholar]
- 47. Oliver A. W., Paul A., Boxall K. J., Barrie S. E., Aherne G. W., Garrett M. D., Mittnacht S., Pearl L. H. (2006) EMBO J. 25, 3179–3190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Pike A. C., Rellos P., Niesen F. H., Turnbull A., Oliver A. W., Parker S. A., Turk B. E., Pearl L. H., Knapp S. (2008) EMBO J. 27, 704–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pellicena P., Kuriyan J. (2006) Curr. Opin. Struct. Biol. 16, 702–709 [DOI] [PubMed] [Google Scholar]
- 50. Lochhead P. A., Kinstrie R., Sibbet G., Rawjee T., Morrice N., Cleghon V. (2006) Mol. Cell 24, 627–633 [DOI] [PubMed] [Google Scholar]
- 51. Lochhead P. A., Sibbet G., Morrice N., Cleghon V. (2005) Cell 121, 925–936 [DOI] [PubMed] [Google Scholar]
- 52. Matsuura A., Tsukada M., Wada Y., Ohsumi Y. (1997) Gene 192, 245–250 [DOI] [PubMed] [Google Scholar]
- 53. Kim J., Huang W. P., Stromhaug P. E., Klionsky D. J. (2002) J. Biol. Chem. 277, 763–773 [DOI] [PMC free article] [PubMed] [Google Scholar]







