Abstract
The pig is important for agriculture and as an animal model in human and veterinary medicine, yet despite over 20 years of effort, there has been a failure to generate pluripotent stem cells analogous to those derived from mouse embryos. Here we report the production of leukemia inhibitory factor-dependent, so-called naive type, pluripotent stem cells from the inner cell mass of porcine blastocysts by up-regulating expression of KLF4 and POU5F1. The alkaline phosphatase-positive colonies resulting from reprogramming resemble mouse embryonic stem cells in colony morphology, cell cycle interval, transcriptome profile, and expression of pluripotent markers, such as POU5F1, SOX2, and surface marker SSEA1. They are dependent on leukemia inhibitory factor signaling for maintenance of pluripotency, can be cultured over extended passage, and have the ability to form teratomas. These cells derived from the inner cell mass of pig blastocysts are clearly distinct from the FGF2-dependent “primed” induced pluripotent stem cells described recently from porcine mesenchymal cells. The data are consistent with the hypothesis that the up-regulation of KLF4, as well as POU5F1, is required to create and stabilize the naive pluripotent state and may explain why the derivation of embryonic stem cells from pigs and other ungulates has proved so difficult.
Keywords: Cell Differentiation; Embryo; Embryonic Stem Cell; Stem Cells; Transcription Factors; Embryonic Stem Cells; Induced Pluripotent Stem Cells; KLF4, POU5F1; Naive; Primed
Introduction
Pluripotent stem cell lines from the inner cell mass (ICM)3 of the embryo, the so-called embryonic stem cells (ESC), were first established over 30 years ago from day 3.5 mouse (m) blastocysts (1, 2) and more recently from totipotent blastomeres (3, 4). These authentic mESC lines are dependent on LIF/STAT3 signaling for maintenance of pluripotency (5), tolerate complete enzymatic dispersal, and fulfill the stringent criteria of pluripotency, such as the lack of senescence when cultured in vitro, the ability to differentiate into multiple cell types representing the three germ layers both in vitro and in vivo, and finally, contribution to the germ line in chimeric offspring (6). These attributes of mESC formed the basis for the transformation of the field of mammalian genetics and developmental biology and propelled the mouse as the prime biomedical model for studying the genetic basis of disease. Until now, attempts to isolate equivalent kinds of ESC from the blastocysts of all but a few “permissive” mouse strains without a major pharmacological intervention have met with failure. ESC established from the human (7), monkey (8), and more recently from the pig (9) and those lines established from the epiblast of non-permissive mouse strains, the epiblast stem cells (10, 11), demonstrate a stark contrast in phenotype and gene expression profile relative to the mESC. These ESC were characterized by their flattened morphology, dependence on FGF2 and TGFB/activin A signaling for maintenance of their pluripotency (12), inactivation of one of the X chromosomes in female cell lines (13), intolerance to passage as single cells, and lack of competence for producing germ line chimeras (10, 11). Also, they are more susceptible to spontaneous differentiation, making the standard practice of culture and manipulation much more demanding. Nichols and Smith (5) have suggested that the two types of ESC differ fundamentally in the gene networks that maintain their pluripotency and named the LIF-dependent ESC “naive” and the FGF2-dependent type “primed.”
Efforts to establish naive ESC from pig (p) embryos began over two decades ago, soon after the first studies describing mESC from blastocysts (14, 15), but the resulting lines bore only a limited resemblance to mESC and failed to meet the full criteria for pluripotency. Four recent studies (16–19) have reported the derivation of porcine induced pluripotent stem cells (piPSC) from fibroblasts by employing the classic combination of reprogramming factors POU5F1 (OCT4), SOX2, KLF4, and c-MYC (OSKM) originally developed to reprogram fibroblasts from the mouse (20). The resulting pluripotent cell lines resembled human cells rather than mESC in terms of their general morphological features and in their requirement for FGF2 rather than LIF (21). Naive ESC have recently been generated successfully from the embryos of rat, but only with major modifications to standard culture conditions (22, 23). In particular, protein kinase inhibitors, CHIR99021 (CH) and PD0325901, were added to the medium to activate the WNT signaling pathway (CH) and to inhibit the ERK-mediated differentiation pathway (PD0325901), respectively. A slightly different strategy was employed to generate naive pluripotent stem cells from the NOD strain of mouse, which previously had failed to yield ESC (24). Here two approaches proved successful. One was to transduce the founder cells with lentiviral vectors designed to overexpress KLF4 and c-MYC, whereas the other was to use pharmacological inhibitors, namely CH to bypass c-MYC function (25) and kenpaullone (KP) to substitute for KLF4 (26). We rationalized that the previous failures to establish ESC from porcine embryos might have been due to low endogenous levels of c-MYC and KLF4 in ICM cells (27). In addition, we had noted that concentrations of mRNA for endogenous c-MYC and KLF4 in porcine piPSC reprogrammed from fetal fibroblasts were extremely low, possibly accounting for the primed phenotype of these cells (17). Accordingly, we explored strategies to ectopically overexpress KLF4 in porcine ICM and culture in medium containing KP and CH medium during the reprogramming steps (27).
EXPERIMENTAL PROCEDURES
Lentiviral Transduction of Reprogramming Factors and Derivation of pESC
The Tet-inducible human (h) KLF4 vector and FUW-M2rtTA described earlier (28) were obtained from Addgene (plasmid numbers 20727 and 20342, respectively). Pseudovirus was produced in human 293FT cells (Invitrogen) by co-transfecting each lentiviral vector with the Vesicular Stomatitis Virus-G envelope (pMD2.G) and packaging vector (psPAX2) (17) with PolyJet (SignaGen, Gaithersburg, MD). For single-factor pESC (pESK) derivation, late day 5 in vitro-produced porcine blastocysts were stripped of trophectoderm by immunosurgery (Experiment-1, 31 blastocysts; Experiment-2, 32 blastocysts) (29) to expose the ICM for lentiviral transduction with hKLF4 and M2rtTA virus. Two days following transduction, the blastomeres were plated onto irradiated mouse embryonic fibroblast feeders at 4 × 104/cm2 in standard LIF medium with the two KP/CH inhibitors (DMEM-F12, 20% knock-out serum replacement, KP (1 μm final; Sigma), CH (3 μm final; Stemgent, San Diego, CA), 250 units/ml hLIF (GenScript, Piscataway, NJ), and 0.5 μg/ml doxycycline (DOX) (Stemgent) at 38.5 °C in a 4% O2, 5% CO2, 91% N2 atmosphere. For the derivation of two-factor (pESOK)stem cell lines, lentiviral vectors expressing hPOU5F1 and hKLF4 under the regulation of constitutive promoters, as described by Ezashi et al. (17), were used to transduce the pESK lines. The cells were cultured continuously under identical conditions to those described above. The two-factor derived stem cell lines (pESO2K) were also established by transducing pESK lines with a Tet-inducible lentiviral vector bearing the open reading frames of hPOU5F1 and hKLF4 transgenes separated by an IRES2 site (O2K) to facilitate bicistronic expression from a single vector. The pESO2K cells were adopted for culture on a laminin-coated substratum (equivalent of 20 μg/35-mm dish; Stemgent) in the presence of commercially available GS2-M medium (Stem Cell Sciences, Cambridge, UK) supplemented with 2 μg/ml DOX and 1000 units/ml hLIF. All cell types were routinely passaged 1:10 every 2–3 days by using Accutase® (Millipore, Billerica, MA).
Population Doubling Time
Population doubling times for pESK I were estimated as reported before (17). Approximately 0.5 × 105 pESK were seeded in each well of 12-well plates precoated with Matrigel under standard culture conditions(17). The number of cells in each well (triplicates for each time point) was counted at 24, 48, 72, and 96 h with daily exchange of fresh medium.
Transcriptional Profiling by Microarray
RNA from two separate pESK, piPSC, and porcine fetal fibroblast (PFF) lines was extracted by using STAT-60 (Tel-Test Inc., Friendswood, TX). Microarray analysis of the samples was performed with porcine Affymetrix arrays as described previously (17).
Alkaline Phosphatase Staining, Immunofluorescence, and Western Blotting
Alkaline phosphatase staining was performed by the nitro blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate method (Promega, Madison, WI). For immunofluorescence, cells grown on coverslips with feeders were fixed in 4% paraformaldehyde in PBS for 15 min at room temperature, washed, and exposed to either 5% goat serum or 5% donkey serum (Sigma), 1% BSA (Jackson ImmunoResearch Laboratories, West Grove, PA), and 0.1% Triton X-100 (Fisher) in PBS for 30 min. Colonies were permeabilized with methanol for 10 min at −20 C prior to pSTAT3 staining. The cells were then incubated with primary antibody for 2 h at room temperature. After washing, the cells were incubated with secondary antibody. For Cy5 tyramide signal amplification, a secondary biotin antibody and then HRP-conjugated streptavidin were utilized. Primary antibodies were: POU5F1 (1:200) (30), SOX2 (1:1,000; Millipore), SSEA-1 (1:50; Millipore), SSEA-4 (1:50; Millipore), phospho-STAT3 (1:200; Cell Signaling, Beverly, MA), TUJ-1 (1:100, Millipore), and SOX17 (1:100; R&D Systems). Secondary antibodies were Alexa Fluor 546-conjugated goat anti-rabbit IgG (POU5F1) and Alexa Fluor 546-conjugated goat anti-mouse IgG (SOX2) (all from Invitrogen), anti-mouse biotin (SSEA-1 and -4) (1:2000; Sigma B7151), and anti-rabbit biotin (pSTAT3) (1:2000 Sigma B3275). Bound biotin was detected with streptavidin HRP (1:2000 PerkinElmer Life Sciences NEL750001EA), and staining was performed with a Cy5 tyramide signal amplification kit (PerkinElmer Life Sciences). The images from Cy5 far-red staining were pseudocolored (green) for better visualization. VECTASHIELD mounting medium with DAPI (Vector Laboratories, Burlingame, CA, catalog number H-1200) was utilized to mount the coverslips. For Western blotting, 30 μg of total cell lysate from pESOK and PFF were first resolved on 12% SDS-PAGE gels. Protein bands were transferred electrophoretically onto PVDF membranes (Millipore) overnight, which were incubated in blocking buffer (10 mm Tris-HCl, 150 mm NaCl, 0.0.05% Tween, pH 7.5 (TBST)) containing 5% nonfat milk and 1% BSA (Sigma) for 1 h. Membranes were then exposed to respective primary antibodies (1:2000) in blocking buffer for 2 h at room temperature, washed (three times) with TBST, and incubated with HRP-conjugated anti-rabbit or anti-mouse secondary antibodies (1:5000). After further washing (three times) in TBST, the blots were developed by using the Phototope-HRP Western blot detection system (Cell Signaling).
Directed Differentiation of pESO2K Cells under Chemically Defined Conditions
The two-factor pESO2K cells were tested for their potential to differentiate into defined lineages in the absence of DOX support and under chemically defined medium conditions. They were differentiated into an endodermal lineage following 7 days of culture in the presence of a small molecule IDE1 (Stemgent) on a Matrigel substratum (31) and into neuronal lineage in the presence of 20 ng/ml FGF2, 100 ng/ml FGF8, and 400 ng/ml Sonic hedgehog (R&D Systems) (32) on a laminin substratum.
Teratoma Formation
pESOK II cells (5 × 106) were injected in 0.25-ml volume with 30% Matrigel (BD Biosciences) solution subcutaneously into two 6-month-old CD1 nude mice (CD1-Foxn1nu, Charles River Laboratories). The derived tumors were dissected out and fixed in 10% (v/v) neutral buffered formalin. Paraffin-embedded tissue was sectioned and then stained with hematoxylin and eosin. All animal experiments were approved by the University of Missouri Institutional Animal Care and Use Committee under Protocol 4467. Additionally, teratoma injections underneath the kidney capsule of CD1 nude mice were performed successfully as a service by Applied StemCell Inc. (Menlo Park, CA) (data not shown).
Reverse Transcription PCR (RT-PCR) and Real Time RT-PCR Analysis
RNA was extracted in STAT-60 reagent (Tel-Test), treated with Turbo-DNase I (Ambion, Austin, TX), and reverse transcribed by using oligo(dT) and SuperScript III reverse transcriptase (Invitrogen). PCR was performed with KOD Hot Start polymerase mix (Novagen, Darmstadt, Germany) under recommended cycling conditions: 95 °C for 2 min followed by 35 amplification cycles (95 °C, 15 s; specific annealing temperature, 15 s; 70 °C, 10 s) with a final extension cycle at 70 °C for 10 s. The relevant primers and annealing temperatures have been reported elsewhere (17). Real time RT-PCR analysis was also performed as described previously (17).
Karyotyping
Standard G-banding chromosome analysis was performed as a service by Cell Line Genetics (Madison, WI). Cytogenetic analysis was performed on 20 G-banded metaphase spreads for each cell line.
RESULTS
Putative pESC Generated by Single-factor Transduction
ICMs from in vitro-produced day 5.5 porcine blastocysts were freed of trophectoderm by immunosurgery (29). Each ICM provided an average of about eight cells, which were immediately exposed to a tet-inducible lentivirus carrying the open reading frame of the hKLF4 gene and a second, similar vector incorporating the tetracycline transactivator (M2rtTA) (28). After 2 weeks of culture on irradiated mouse embryonic fibroblasts, two putative pESC colonies (named pESK I & II) emerged from two independent experiments (Fig. 1A). Although colony numbers were few, they were each obtained from only about 250 initiating cells, i.e. ∼0.4% efficiency. A SNP-CHIP 60K porcine array (a service provided by GeneSeek, Lincoln, NE) established that the two lines were of porcine origin and that there was a total of 27,636 discordant genotype mismatches between the two cell lines, thereby verifying that the two cell lines were distinct from one another (GEO accession number GSE26436). Once the colonies reached a diameter of ∼50 μm, they were picked and dispersed by using Accutase. All subsequent cultures were performed in this manner. The colonies exhibited a morphology similar to mESC (Fig. 1B) that was distinct from the flatter colonies of porcine iPSC derived by the four-factor (OSKM) procedure (17). The cells had a high nuclear-to-cytoplasmic ratio and a short cell cycle interval (∼9.5 h). They showed no signs of senescence, altered morphology, or tendency to differentiate over 50 passages (∼300 cell doublings).
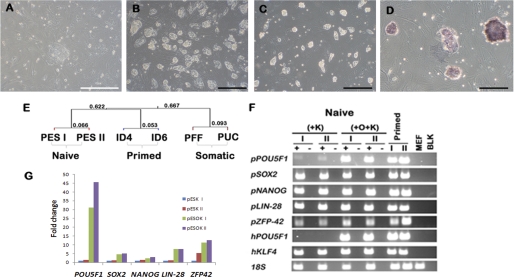
FIGURE 1.
Light micrograph images and transcriptional profile of colonies derived from the ICM of pig blastocysts. A–D, light micrographs of colonies generated by single-factor (KLF4) induction from the ICM of pig blastocysts. A, the single colony (pESK I line) appearing 2 weeks after KLF4 transduction and culture in modified LIF-KP/CH medium (scale bar, 100 μm). B, colonies of pESK I on day 3 of culture at passage 8. C, two-factor pESOK II (hPOU5F1 and hKLF4) on day 2 of culture following four passages after induction. D, alkaline phosphatase staining of pESOK II cells after 4 days in culture (scale bar in B–D, 200 μm). E, hierarchical clustering of the microarray data obtained from two of the single-factor induced pESK lines (pESK I and II (PES I and PES II)), four-factor piPSC reported in Ezashi et al. (17) (ID4 and ID6), and the porcine somatic cells, PFF, and porcine umbilical cord-derived explant cells (PUC). Hybridizations were performed on the Porcine Gene Chip® array (Affymetrix) and the microarray data analyzed by using the GeneSpring GX (Agilent) software. F, RT-PCR analysis of the single- (+K) and two-factor (O+K) pES lines (I and II), and the piPSC (ID4 and ID6) generated by Ezashi et al. (17). Mouse embryonic fibroblast cells (MEF) and no-template blank (BLK) were used as negative controls. The symbols + and − represent amplifications in which reverse transcriptase was either included or not included in the RT reactions. G, real time RT-PCR analysis of pluripotent genes in the two pESK and pESOK lines. Note the relative up-regulation of pluripotent factors in the two-factor derived cells when compared with the single-factor cell lines. The relative expression of the genes was represented as -fold changes when compared with the pESK I line.
Transcriptome Profiling of Single-factor Derived pESC
Transcriptome profiling on Affymetrix porcine microarrays (GEO accession number GSE26369, pESK I & II, passage numbers 14 and 5 in Fig. 1E) indicated that the reprogramming had been successful and that the two cell lines had a similar mRNA composition. The pESK lines clustered separately from PFF, primary cultures of porcine umbilical cord cells, and two FGF2-dependent, piPSC lines derived from PFF by four-factor reprogramming, which have properties resembling primed ESC (17) (Fig. 1E). The pESK I and II lines were, however, only weakly alkaline phosphatase-positive (not shown), and although they clearly expressed many of the endogenous (porcine) genes indicative of pluripotency (Fig. 1, F and G, and supplemental Fig. S1), the patterns were clearly anomalous. In particular, they exhibited unexpected low amounts of POU5F1, KLF4, and STAT3 transcripts, although mRNA for the LIF receptor gene, LIFR, appeared elevated (supplemental Fig. S1). The transcript concentrations for many genes, including SOX2 and NANOG, could not be assessed from the porcine arrays, which continue to remain poorly annotated.
Two-factor Transduction Is Required for the Establishment of ICM-derived piPSC
The two ICM-derived single-factor lines, although exhibiting many characteristics of naive pluripotency, were clearly anomalous in gene expression profile and also failed to form teratomas when injected subcutaneously into immunodeficient mice, suggesting that they had not achieved ground state pluripotency. To counter the low expression of pPOU5F1 and pKLF4 in the pESK I and II lines, the cells were transduced with lentiviral vectors (17) expressing hPOU5F1 and hKLF4, a tactic recently used with human cells (13). The emerging new colonies (pESOK), although little changed in growth rate, were less flat and had the compact, “glistening” appearance typical of mESC colonies (Fig. 1C). They also exhibited high alkaline phosphatase activity (Fig. 1D) and elevated mRNA levels for endogenous, i.e. porcine, genes consistent with a pluripotent phenotype, e.g. pPOU5F1, pSOX2, pNANOG, pLIN-28, and pZFP42 (pREX1) (Fig. 1G). Immunocytochemical analysis confirmed that cells within the colonies expressed POU5F1 and SOX2 in their nuclei (Fig. 2, A and B) and the surface markers SSEA-1, which stained strongly, and SSEA-4, which stained relatively weakly (Fig. 2, D and E; staining controls, supplemental Fig. S2). Western blotting analysis further verified the expression of pluripotent markers POU5F1, SOX2, and NANOG (Fig. 2F). LIF dependence was confirmed by two parameters, (i) the detection of phosphorylated STAT3 by immunofluorescence (Fig. 2C) and Western blotting (Fig. 2F) and (ii) the rapid induction of differentiation in the absence of LIF and in the presence of an inhibitor for STAT3 signaling (1 μm JAK inhibitor 1, Calbiochem) (supplemental Fig. S3) (33). The two cell lines, pESOK I and II, that were characterized in most detail were karyotypically normal (supplemental Fig. S4), and unlike pESK I and II, were capable of forming teratomas in immune-compromised mice (Fig. 3A). The tumors contained cell types representing all the three germ layers (Fig. 3B), suggesting that robust expression of POU5F1 is required to provide pluripotency.
FIGURE 2.
Pluripotent phenotype of the two-factor pESOK II colonies. A–E, epifluorescence images of colonies immunostained with antibodies directed against the pluripotency markers POU5F1 (A) and SOX2 (B), phosphorylated STAT3 (pSTAT3) (C), and carbohydrate antigens SSEA-1 (D) and SSEA-4 (E). Colonies shown in A, B, and C were at day 3 after passage; those in D and E had been cultured for 5 days. The upper panels show specific staining reactions, and the lower panels show nuclear staining by DAPI. Note that the staining of POU5F1, SOX2, and phosphorylated STAT3 in these raised colonies is consistent with a nuclear localization, whereas the robust surface staining for SSEA1 and the weaker SSEA4 fluorescence are consistent with their expected surface localization. F, Western blotting analysis confirming the expression of pluripotent genes in the pESOK line (II). Lysate from PFF serves as a negative control.
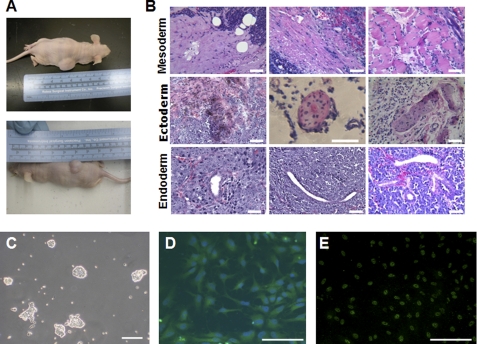
FIGURE 3.
In vivo and in vitro differentiation potential of the two-factor derived pES lines. A, gross images of the two mice showing robust subcutaneous teratomas in the dorsal flank regions. B, mesoderm, ectoderm, and endoderm present in teratomas derived from the pESOK II cell line. The images are H&E-stained sections showing representative mesoderm (left to right: connective tissue, connective tissue and smooth muscle, and striated muscle), ectoderm (left to right: pigmented cells, stratified epithelium, and epidermis), and endoderm lineage (left to right: cuboidal epithelium lined gland, duct, and branched glands). C, representative light micrograph of pESO2K colonies cultured on laminin substratum in the presence of GS2-M medium with DOX and LIF supplementation. Note the small, compact three-dimensional colonies with an undifferentiated morphology. D, fluorescent micrograph of pESO2K colonies undergoing directed differentiation into TUJ1-positive (green staining) neuronal precursors. Nuclei are stained blue with DAPI. E, fluorescent micrograph of pESO2K colonies undergoing directed differentiation into SOX17-positive (green nuclear staining) endodermal precursors. In all panels, the scale bar represents 50 μm.
Feeder-independent Culture and Differentiation of Two-factor pESO2K Cells under Chemically Defined Conditions
The two-factor pESO2K cells were able to adapt to culture on a laminin-coated surface in the presence of a commercially available chemically defined medium GS2-M in the presence of LIF and DOX (Fig. 3C). They survived extended passages (>20 passages) under these conditions and showed no signs of either senescence or change in morphology. The cells could also be induced to undergo successful differentiation under defined conditions. They differentiated into SOX17-positive endodermal precursors (Fig. 3E) (31) and into βIII-tubulin (TUJ1)-positive neuroectodermal precursors (Fig. 3D) (32). Additionally, when membrane-labeled (1 μm CM-diI, Molecular Probes) cells were injected into precompacted day 3 porcine in vitro embryos, they were able to proliferate and colonize the resultant blastocysts (supplemental Fig. S5).
DISCUSSION
Here we describe the generation of a putative naive class of porcine pluripotent cells using ICM of blastocysts as the founder population. The rationale for the experiments was that such cells would likely have many of the desirable features of mESC, including the rapid growth rate, resistance to spontaneous differentiation, ease of genetic manipulation, and ability to incorporate into chimeras that have made mESC such a valuable experimental tool for studies on the mouse model. We also hypothesized that if such porcine cells could be derived from a pluripotent source, namely the ICM, they would lack the epigenetic memory that might be carried from more differentiated somatic cells (34).
The experiments clearly demonstrate that it is possible to derive pluripotent stem cells, seemingly of the naive class, directly and relatively efficiently from the ICM of the pig, an important advance because swine are valuable alternative models to the mouse in biomedical research (21), and naive cells are much more amenable to physical and genetic manipulation than pluripotent cells of the primed type. The studies also demonstrate an evolutionarily shared requirement for KLF4, a key transcription factor that is downstream of the LIF signaling cascade, in achieving pluripotency and LIF dependence. Low expression of KLF4 and c-MYC probably underpins the long history of failure to establish naive pluripotent stem cells from pig and other domestic species, as well as several strains of mouse (24). An examination of deep sequencing data obtained on cDNA from porcine blastocysts confirmed a relatively low abundance of these two crucial transcripts, although POU5F1 mRNA was relatively abundant (35). Although POU5F1 was represented by 344 reads, KLF4 had 49 and c-MYC only had three. Our choice of culture in a 4% O2 environment, conditions that favor glycolytic metabolism in human embryonic stem cells (36), may have facilitated the derivation of pluripotent lines by compensating for low levels of endogenous c-MYC expression. Among its many pleiotropic roles, c-MYC promotes glycolysis (37). Additionally, our laboratory has previously shown that maintenance of hESC in an undifferentiated state is favored by culture in low oxygen environment (30), whereas others have demonstrated that such physiological O2 conditions enhance reprogramming efficiency (38, 39) and maintain the active chromatin state of pluripotent cells (40), again functions ascribed to c-MYC. On the other hand, although culturing in low O2 and transducing the pig ICM cells with KLF4 generated colonies bearing a superficial resemblance to naive cells, they were not pluripotent by conventional standards. As creation of LIF-dependent human ESC required additional, supplementary expression of either POU5F1 or KLF2 to achieve a “true” pluripotent state (13), we added such an extra step and showed that the requirement for an additional factor also held true for pigs. Our choice of POU5F1 over KLF2 was based on the fact that KLF2 mRNA levels seem to increase concomitantly with POU5F1 expression (41). Taken together, we conclude that culturing under physiological O2 conditions is ideal for derivation and propagation of pig pluripotent stem cells, KLF4 is a major player distinguishing naive and primed pluripotent stemness states, and ectopic POU5F1 overexpression is required in the pig and potentially other domestic animals to achieve and maintain that state.
Supplementary Material
Acknowledgment
We thank Dr. Kirk of the Department of Biology for providing the nucleoside solution.
This work was supported, in whole or in part, by National Institutes of Health Grant HD-21896 (to R. M. R.). This work was also supported by MO Life Sciences Board Grant 00022147 (to T. E.) and an Addgene Challenge Award (Innovation award) (to B. P. V. L. T. and R. M. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- ICM
- inner cell mass
- ESC
- embryonic stem cells
- iPSC
- induced pluripotent stem cells
- LIF
- leukemia inhibitory factor
- CH
- CHIR99021
- KP
- kenpaullone
- DOX
- doxycycline
- PFF
- porcine fetal fibroblast(s)
- h
- human
- p
- pig
- m
- mouse.
REFERENCES
- 1. Evans M. J., Kaufman M. H. (1981) Nature 292, 154–156 [DOI] [PubMed] [Google Scholar]
- 2. Martin G. R. (1981) Proc. Natl. Acad. Sci. U.S.A. 78, 7634–7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eistetter H. R. (1989) Dev. Growth Differ. 31, 275–282 [DOI] [PubMed] [Google Scholar]
- 4. Delhaise F., Bralion V., Schuurbiers N., Dessy F. (1996) Eur. J. Morphol. 34, 237–243 [DOI] [PubMed] [Google Scholar]
- 5. Nichols J., Smith A. (2009) Cell Stem Cell 4, 487–492 [DOI] [PubMed] [Google Scholar]
- 6. Wobus A. M., Boheler K. R. (2005) Physiol. Rev. 85, 635–678 [DOI] [PubMed] [Google Scholar]
- 7. Thomson J. A., Itskovitz-Eldor J., Shapiro S. S., Waknitz M. A., Swiergiel J. J., Marshall V. S., Jones J. M. (1998) Science 282, 1145–1147 [DOI] [PubMed] [Google Scholar]
- 8. Thomson J. A., Kalishman J., Golos T. G., Durning M., Harris C. P., Becker R. A., Hearn J. P. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 7844–7848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Alberio R., Croxall N., Allegrucci C. (2010) Stem Cells Dev. 19, 1627–1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tesar P. J., Chenoweth J. G., Brook F. A., Davies T. J., Evans E. P., Mack D. L., Gardner R. L., McKay R. D. (2007) Nature 448, 196–199 [DOI] [PubMed] [Google Scholar]
- 11. Brons I. G., Smithers L. E., Trotter M. W., Rugg-Gunn P., Sun B., Chuva de Sousa Lopes S. M., Howlett S. K., Clarkson A., Ahrlund-Richter L., Pedersen R. A., Vallier L. (2007) Nature 448, 191–195 [DOI] [PubMed] [Google Scholar]
- 12. Xu R. H., Peck R. M., Li D. S., Feng X., Ludwig T., Thomson J. A. (2005) Nat. Methods 2, 185–190 [DOI] [PubMed] [Google Scholar]
- 13. Hanna J., Cheng A. W., Saha K., Kim J., Lengner C. J., Soldner F., Cassady J. P., Muffat J., Carey B. W., Jaenisch R. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 9222–9227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Notarianni E., Laurie S., Moor R. M., Evans M. J. (1990) J. Reprod. Fertil. Suppl. 41, 51–56 [PubMed] [Google Scholar]
- 15. Piedrahita J. A., Anderson G. B., Bondurant R. H. (1990) Theriogenology 34, 879–901 [DOI] [PubMed] [Google Scholar]
- 16. Wu Z., Chen J., Ren J., Bao L., Liao J., Cui C., Rao L., Li H., Gu Y., Dai H., Zhu H., Teng X., Cheng L., Xiao L. (2009) J. Mol. Cell Biol. 1, 46–54 [DOI] [PubMed] [Google Scholar]
- 17. Ezashi T., Telugu B. P., Alexenko A. P., Sachdev S., Sinha S., Roberts R. M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 10993–10998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Esteban M. A., Xu J., Yang J., Peng M., Qin D., Li W., Jiang Z., Chen J., Deng K., Zhong M., Cai J., Lai L., Pei D. (2009) J. Biol. Chem. 284, 17634–17640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. West F. D., Terlouw S. L., Kwon D. J., Mumaw J. L., Dhara S. K., Hasneen K., Dobrinsky J. R., Stice S. L. (2010) Stem Cells Dev. 19, 1211–1220 [DOI] [PubMed] [Google Scholar]
- 20. Takahashi K., Yamanaka S. (2006) Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 21. Roberts R. M., Telugu B. P., Ezashi T. (2009) Cell Cycle 8, 3078–3081 [DOI] [PubMed] [Google Scholar]
- 22. Buehr M., Meek S., Blair K., Yang J., Ure J., Silva J., McLay R., Hall J., Ying Q. L., Smith A. (2008) Cell 135, 1287–1298 [DOI] [PubMed] [Google Scholar]
- 23. Li P., Tong C., Mehrian-Shai R., Jia L., Wu N., Yan Y., Maxson R. E., Schulze E. N., Song H., Hsieh C. L., Pera M. F., Ying Q. L. (2008) Cell 135, 1299–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hanna J., Markoulaki S., Mitalipova M., Cheng A. W., Cassady J. P., Staerk J., Carey B. W., Lengner C. J., Foreman R., Love J., Gao Q., Kim J., Jaenisch R. (2009) Cell Stem Cell 4, 513–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marson A., Foreman R., Chevalier B., Bilodeau S., Kahn M., Young R. A., Jaenisch R. (2008) Cell Stem Cell 3, 132–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lyssiotis C. A., Foreman R. K., Staerk J., Garcia M., Mathur D., Markoulaki S., Hanna J., Lairson L. L., Charette B. D., Bouchez L. C., Bollong M., Kunick C., Brinker A., Cho C. Y., Schultz P. G., Jaenisch R. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 8912–8917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Telugu B. P., Ezashi T., Roberts R. M. (2010) Stem Cell Rev. 6, 31–41 [DOI] [PubMed] [Google Scholar]
- 28. Soldner F., Hockemeyer D., Beard C., Gao Q., Bell G. W., Cook E. G., Hargus G., Blak A., Cooper O., Mitalipova M., Isacson O., Jaenisch R. (2009) Cell 136, 964–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Han Y. M., Abeydeera L. R., Kim J. H., Moon H. B., Cabot R. A., Day B. N., Prather R. S. (1999) Biol. Reprod. 60, 1110–1113 [DOI] [PubMed] [Google Scholar]
- 30. Ezashi T., Das P., Roberts R. M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4783–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Borowiak M., Maehr R., Chen S., Chen A. E., Tang W., Fox J. L., Schreiber S. L., Melton D. A. (2009) Cell Stem Cell 4, 348–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ying Q. L., Stavridis M., Griffiths D., Li M., Smith A. (2003) Nat. Biotechnol. 21, 183–186 [DOI] [PubMed] [Google Scholar]
- 33. Telugu B. P., Ezashi T., Roberts R. M. (2010) Int. J. Dev. Biol. 54, 1703–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim K., Doi A., Wen B., Ng K., Zhao R., Cahan P., Kim J., Aryee M. J., Ji H., Ehrlich L. I., Yabuuchi A., Takeuchi A., Cunniff K. C., Hongguang H., McKinney-Freeman S., Naveiras O., Yoon T. J., Irizarry R. A., Jung N., Seita J., Hanna J., Murakami P., Jaenisch R., Weissleder R., Orkin S. H., Weissman I. L., Feinberg A. P., Daley G. Q. (2010) Nature 467, 285–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bauer B. K., Isom S. C., Spate L. D., Whitworth K. M., Spollen W. G., Blake S. M., Springer G. K., Murphy C. N., Prather R. S. (2010) Biol. Reprod. 83, 791–798 [DOI] [PubMed] [Google Scholar]
- 36. Westfall S. D., Sachdev S., Das P., Hearne L. B., Hannink M., Roberts R. M., Ezashi T. (2008) Stem Cells Dev. 17, 869–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vander Heiden M. G., Cantley L. C., Thompson C. B. (2009) Science 324, 1029–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yoshida Y., Takahashi K., Okita K., Ichisaka T., Yamanaka S. (2009) Cell Stem Cell 5, 237–241 [DOI] [PubMed] [Google Scholar]
- 39. Zhu S., Li W., Zhou H., Wei W., Ambasudhan R., Lin T., Kim J., Zhang K., Ding S. (2010) Cell Stem Cell 7, 651–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lengner C. J., Gimelbrant A. A., Erwin J. A., Cheng A. W., Guenther M. G., Welstead G. G., Alagappan R., Frampton G. M., Xu P., Muffat J., Santagata S., Powers D., Barrett C. B., Young R. A., Lee J. T., Jaenisch R., Mitalipova M. (2010) Cell 141, 872–883 [DOI] [PubMed] [Google Scholar]
- 41. Hall J., Guo G., Wray J., Eyres I., Nichols J., Grotewold L., Morfopoulou S., Humphreys P., Mansfield W., Walker R., Tomlinson S., Smith A. (2009) Cell Stem Cell 5, 597–609 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.