Abstract
The aspartate:alanine antiporter (AspT) of the lactic acid bacterium Tetragenococcus halophilus is a member of the aspartate:alanine exchanger (AAEx) transporter family. T. halophilus AspT catalyzes the electrogenic exchange of l-aspartate1− with l-alanine0. Although physiological functions of AspT were well studied, l-aspartate1−:l-alanine0 antiport mechanisms are still unsolved. Here we report that the binding sites of l-aspartate and l-alanine are independently present in AspT by means of the kinetic studies. We purified His6-tagged T. halophilus AspT and characterized its kinetic properties when reconstituted in liposomes (Km = 0.35 ± 0.03 mm for l-aspartate, Km = 0.098 ± 0 mm for d-aspartate, Km = 26 ± 2 mm for l-alanine, Km = 3.3 ± 0.2 mm for d-alanine). Competitive inhibition by various amino acids of l-aspartate or l-alanine in self-exchange reactions revealed that l-cysteine selectively inhibited l-aspartate self-exchange but only weakly inhibited l-alanine self-exchange. Additionally, l-serine selectively inhibited l-alanine self-exchange but barely inhibited l-aspartate self-exchange. The aspartate analogs l-cysteine sulfinic acid, l-cysteic acid, and d-cysteic acid competitively and strongly inhibited l-aspartate self-exchange compared with l-alanine self-exchange. Taken together, these kinetic data suggest that the putative binding sites of l-aspartate and l-alanine are independently located in the substrate translocation pathway of AspT.
Keywords: Amino Acids Transport, Bacterial Metabolism, Kinetics, Liposomes, Membrane Energetics
Introduction
In some strains of the lactic acid bacterium Tetragenococcus halophilus, a proton-motive force is generated by the combined action of an intracellular l-aspartate decarboxylation reaction, catalyzed by an l-aspartate-4-decarboxylase (AspD, EC 4.1.1.12), and an electrogenic aspartate1−:alanine0 exchange reaction, catalyzed by an aspartate:alanine antiporter (AspT, TC number 2.A.81.1.1): l-aspartate (out) + l-alanine (in) → l-aspartate (in) + l-alanine (out). The proton-motive force generated is sufficiently high to drive ATP synthesis via the bacterial F0F1-ATPase. This combination of proton-motive force and ATP synthesis has been proposed as a proton-motive metabolic cycle, the prototype model of which is found in Oxalobacter formigenes (1–3). Such decarboxylation reactions are thought to be advantageous for cells because they generate metabolic energy and regulate intracellular pH (4, 5).
In previous work with proteoliposomes, we found that the aspartate:alanine exchange catalyzed by AspT is electrogenic (4, 6). AspT is classified as a conventional secondary transport protein and belongs to the newly classified aspartate:alanine exchanger family (TC number 2.A.81) of transporters in the system developed by Saier et al. (25, 26). Recently, the results of a BLAST (7) search of the nucleotide sequence of the aspT gene and the amino acid sequence of the AspT protein against current nucleotide and protein data bases, respectively, suggested that the aspartate:alanine exchanger transporters are conserved in many bacterial species (8, 9). Very recently, Fukui et al. (10) found SucE1, an aspartate:alanine exchanger transporter from Corynebacterium glutamicum, plays a role in the export of succinate generated during fermentation. The putative broad distribution of AspT orthologs and paralogs in bacteria suggests that further biochemical study of AspT could provide valuable insights into membrane transport.
AspT is a membrane protein containing 543 amino acids (57.2 kDa). Its membrane topology has been studied by using alkaline phosphatase and β-lactamase fusion methods (11); AspT has also been studied by using the cysteine-substituted accessibility method (12, 13), which uses the impermeant, fluorescent thiol-specific probe Oregon Green 488 maleimide and the impermeant, nonfluorescent thiol-specific probe [2-(trimethylammonium)ethyl]methanethiosulfonate bromide. These analyses revealed that AspT has a unique topology and that transmembrane domain 3 participates in the formation of a hydrophilic cleft in the membrane, implicating the transmembrane domain in the ligand-induced conformational changes (14). In previous work, we developed a solubilization and purification scheme by using n-dodecyl-β-d-maltoside (DDM),3 and we studied the oligomerization of AspT by means of a glutaraldehyde cross-linking assay (14). Here, we sought to characterize, in detail, the kinetic properties of AspT by analyzing the transport kinetics of reconstituted AspT under various conditions. In addition, to reveal the substrate recognition mechanism of AspT, we performed competition analysis with a series of amino acids and the aspartate analogs. The competition analysis suggested that the putative binding sites are independently located in the substrate translocation pathway of AspT.
EXPERIMENTAL PROCEDURES
Chemicals, Cells, and Expression Plasmids
l-[2,3-3H]Aspartic acid (1.07 GBq/mmol) was purchased from GE Healthcare. 1-O-n-Octyl-β-d-glucopyranoside, DDM, and l-cysteic acid (l-CA) were obtained from Nacalai Tesque (Kyoto, Japan). Escherichia coli phospholipids were provided by Avanti Polar Lipids (Alabaster, AL) (15). l-Cysteine sulfinic acid (l-CSA) was purchased from Sigma. E. coli strain XL1 blue harboring pMS421 (Specr LacIq), and referred to as strain XL3 (1), was used to express histidine-tagged AspT with pTrc99A (Amersham Biosciences).
Expression, Solubilization, and Purification of AspT(WT)-His
Expression of histidine-tagged AspT was performed as described previously (14). In brief, a preculture of E. coli XL3 carrying pTrcAsp-His was diluted 100-fold in fresh Luria-Bertani (LB) medium containing 30 mm d-glucose, 30 μg/ml of carbenicillin, and 30 μg/ml of spectinomycin. These cells were grown for 2.5 h at 37 °C with shaking and then diluted 2-fold in fresh Luria broth containing 30 mm d-glucose, 60 mm l-aspartate, and 1 mm pyridoxal 5′-phosphate. The cell suspension was incubated statically for 13 h at 37 °C. At 12 h prior to cell harvest, 200 μm isopropyl-β-d-thiogalactoside was added to the cultures. Isopropyl-β-d-thiogalactoside-induced cells were harvested by using centrifugation at 7,000 × g for 15 min and washed with 100 mm potassium phosphate (pH 7); membrane vesicles were prepared by using high-pressure lysis in the presence of 100 mm potassium phosphate (pH 7), as previously described (1). Membrane vesicles were solubilized (15) at 4 °C for 6 h with 1.5% (w/v) DDM in the presence of 50 mm l-aspartate, 20 mm potassium phosphate (pH 7), and 20% glycerol. After centrifugation at 150,000 × g for 30 min, the supernatant was incubated with a nickel-nitrilotriacetic acid affinity resin (400-μl bed volume for a 2-liter culture) at 4 °C for 7 h. The column was washed on ice with a total of 15 ml/2-liter of culture wash buffer (50 mm l-aspartate, 20 mm potassium phosphate, pH 7, 20% glycerol, 0.01% DDM, and 25 mm imidazole). AspT(WT)-His was then eluted by using brief centrifugation in the cold with elution buffer (50 mm l-aspartate, 20 mm potassium phosphate, pH 7, 20% glycerol, 0.01% DDM, and 0.25 m imidazole) at 1 ml/2-liter culture. The elution fractions were stored at −80 °C as concentrated stocks.
Reconstitution and Transport of Purified AspT(WT)-His
Solubilized membrane proteins were reconstituted in a final volume of 1 ml with 800 μl of detergent extracts (10 to 20 μg of protein) (or control lipid extract), 130 μl of bath-sonicated liposomes (5.9 mg of E. coli phospholipid), and 18 μl of 15% 1-O-n-octyl-β-d-glucopyranoside, with the balance made up by 50 mm potassium phosphate (pH 7). After incubation for 20 min on ice, proteoliposomes (or control liposomes) were formed at room temperature (RT) by rapid injection of the mixtures into 20 ml of loading buffer containing buffer solution and suitable countersubstrate such as 100 mm l-aspartate as the potassium salt for l-aspartate exchange reaction. Because buffer components were changed in each experiment, components of loading buffer were described under supplemental “Methods”. The substrate-loaded proteoliposomes (or liposomes) were kept at RT for 20 min. To assess l-aspartate transport by l-[2,3-3H]aspartate-loaded particles, proteoliposomes were diluted 20-fold from the concentrated stock into an appropriate volume of assay buffer (50 mm potassium phosphate, pH 7, 100 mm K2SO4). After 1 to 3 min of preincubation at 25 °C, l-[2,3-3H]aspartate was added to a final concentration of 100 μm; at various times, 50–100-μl aliquots were removed for membrane filtration with a 0.22-μm pore size GSTF Millipore filter (Millipore Co., Billerica, MA). The membrane filters were washed twice with 5 ml of assay buffer (15).
Filtration Assay of Substrate Transport
Unless otherwise noted, initial rates of l-[2,3-3H]aspartate entry were measured in duplicate at 25 °C by means of a filtration assay (16, 17). Proteoliposomes were applied directly to the center of a 0.22-μm pore size GSTF Millipore filter (Millipore Co.) and washed twice with 5 ml of chilled assay buffer (50 mm potassium phosphate, pH 7, 100 mm K2SO4). Upon release of the vacuum, the proteoliposomes were covered with assay buffer that was preincubated at 25 °C with 0.1 mm l-[2,3-3H]aspartate, and the reaction was terminated after 1 min by use of filtration and washing.
Synthesis of the Aspartate Analog d-Cysteic Acid
To study the inhibitory effect of an aspartate analog, we synthesized d-cysteic acid (d-CA), as previously described (18). Hydrochloric acid (2.1 ml, 12 m) and bromine (1.0 ml, 19.4 mmol) were added to a solution of d-cystine (1.00 g, 4.16 mmol) in deionized water (6.3 ml) at RT; the mixture was then heated to about 65 °C and stirred without incubation. After 40 min, the mixture was evaporated in a vacuum to remove unreduced bromine and solvent. The resulting residue was dissolved in deionized water (4.0 ml), and insoluble matter was removed by use of filtration. The filtrate was concentrated by evaporation to 2.7 ml and allowed to crystallize by standing overnight in a refrigerator. The crystals were filtered with suction, washed with cold deionized water, and dried under reduced pressure over 6 h to give 262 mg (34%) of pure d-CA monohydrate as a white crystalline solid; IR νmax 3168 (m), 1766 (s), 1505 (s); 1H NMR (solvent: D2O, reference: DSS) δ 3.39 (1H, dd, J = 15.0, 9.5 Hz), 3.56 (1H, dd, J = 15.0, 3.0 Hz), 4.34 (1H, m); HRMS (FAB) m/z calculated for C3H8O5NS ([M + H]+) 170.0123, found 170.0123.
RESULTS
Transport Properties of Purified AspT
In previous studies using crude membrane extracts it was unclear whether AspT alone catalyzes the electrogenic l-aspartate:l-alanine exchange reaction (6). Nanatani et al. (14) established AspT purification and reconstitution methods and confirmed l-aspartate self-exchange properties. Here, we clearly demonstrated that purified AspT alone is sufficient for the electrogenic l-aspartate1−:l-alanine0 exchange reaction (Figs. 1 and 2, Table 1). Purified AspT exhibited all of the antiporter properties anticipated from previous studies with crude membrane extracts (6).
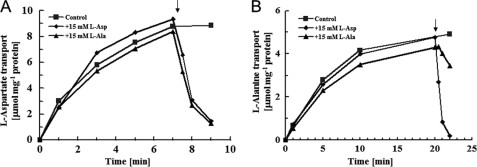
FIGURE 1.
Reactions of l-aspartate and l-alanine self-exchange. Proteoliposomes were loaded with 100 mm l-aspartate (A) or l-alanine (B) plus 50 mm potassium phosphate (pH 7) and then washed and resuspended as described under “Experimental Procedures.” Proteoliposomes (♦, ■, ▴) were placed in 100 mm (A) or 50 mm (B) K2SO4 plus 50 mm potassium phosphate (pH 7) at 3.3 μg of protein/ml, at which point 100 μm l-[3H]aspartate (A) or 100 μm l-[3H]alanine (B) were added. To estimate substrate transport, aliquots were taken for filtration and washing at the times indicated. The arrow denotes the addition of 15 mm unlabeled l-aspartate (♦) or 15 mm unlabeled l-alanine (▴).
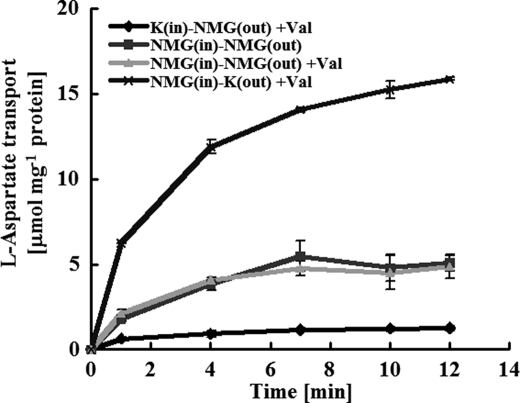
FIGURE 2.
The electrogenic nature of l-aspartate:l-alanine exchange. Proteoliposomes or liposomes were loaded with 100 mm l-alanine plus 50 mm phosphate (pH 7) as either the N-methyl-d-glucamine (■, ▴, ×) or potassium (♦) salt. To start the experiment, proteoliposomes, suspended at 1.9 μg of protein/ml in their respective assay buffers, were diluted 133-fold in assay medium containing 100 μm l-[3H]aspartate along with 100 mm SO4 plus 50 mm phosphate as the potassium (×) or NMG (♦, ▴, ■) salt; with one exception (■), 1 μm valinomycin (Val) was also present.
TABLE 1.
Kinetic parameters of AspT
| Km | Vmax | Vmax/Km | |||
|---|---|---|---|---|---|
| mm | μmol min−1mg−1of protein | ||||
| l-Asp | 0.35 | ±0.03 | 175 | ±21 | 500 |
| l-Ala | 26 | ±2 | 155 | ±9 | 6.0 |
| d-Asp | 0.098 | ±0 | 3.8 | ±0.3 | 39 |
| d-Ala | 3.3 | ±0.2 | 3.3 | ±0.4 | 1.0 |
| l-Ser | 11 | ±1 | 103 | ±24 | 9.5 |
| l-Cys | 8.2 | ±0.1 | 11 | ±1 | 1.3 |
The steady-state incorporation of l-[3H]aspartate and l-[3H]alanine was ∼8 μmol/mg of protein and 4.5 μmol/mg of protein, respectively (Fig. 1). The incorporated material was readily expelled after the addition of excess unlabeled l-aspartate or l-alanine, as if l-[3H]aspartate or l-[3H]alanine had been taken up by an exchange reaction and alanine had been the countersubstrate for aspartate. Because accumulated l-[3H]aspartate and l-[3H]alanine were released by l-aspartate or l-alanine (Fig. 1), it seems feasible that both compounds served as substrates.
In our previous studies, we examined aspartate transport by using l-alanine-loaded proteoliposomes formed with crude membrane extracts and using valinomycin along with external or internal potassium to generate a membrane potential that would accelerate or retard such an electrogenic reaction (4). Here, we confirmed the electrogenic nature of AspT by using proteoliposomes and purified protein. Control proteoliposomes were prepared and assayed by using N-methyl-d-glucamine as the internal and external cation. These proteoliposomes were largely unaffected by valinomycin and took up l-[3H]aspartate at an initial rate of about 2.2–2.8 nmol mg −1 of protein in 15 s (Fig. 2). However, when the proteoliposomes were made with internal potassium, such that an internally negative potential was established in the presence of valinomycin, there was profound inhibition of l-[3H]aspartate transport (from 2.7 to 0.1 nmol to 15 s−1 mg−1 of protein), whereas in the reciprocal experiment, when an internally positive potential was present, l-[3H]aspartate movement was stimulated (from 7.9 to 15.1 nmol/15 s). These experiments strongly suggest that l-aspartate:l-alanine exchange is an electrogenic reaction in which a net negative charge moves in parallel with l-aspartate.
Temperature and pH Dependence of AspT
To investigate the optimal temperature for AspT, we measured initial rates of uptake over a temperature range of 25 to 65 °C by use of a filtration assay. Proteoliposomes were prepared as described under “Experimental Procedures” and supplemental “Methods”. The reaction was stopped after 10 s. Uptake activity was lowest at the high end of the temperature range, with a roughly linear increase between 20 and 45 °C. Over 45 °C, the activity of AspT decreased, and it was abolished at 65 °C (supplemental Fig. S1A).
To study the thermal stability of AspT, we incubated solubilized AspT at 30–60 °C. After the incubation, the solubilized AspT was reconstituted into liposomes. Incubation at 30, 40, 50, and 60 °C gave half-lives of AspT activity of 66, 2.1, 0.37, and 0.18 min, respectively (supplemental Fig. S1B). Solubilized AspT was unstable over 40 °C and its activity was lost at temperatures above 40 °C.
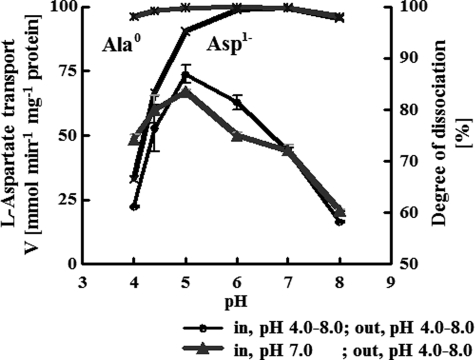
The pH dependence of AspT transport was assessed with uptake experiments (Fig. 3). The purified AspT was incubated at either acidic or neutral pH, and then reconstituted into proteolipsomes at a lipid-to-protein ratio of 5000:1 (molar ratio) before the initiation of the assay. Uptake activity of l-[3H]aspartate tended to increase as the external pH was lowered from neutral values to a maximum of around pH 5.0. The uptake activity subsequently decreased at pH values lower than 5.0.
FIGURE 3.
pH dependence of AspT transport. The pH dependence of AspT transport was studied in uptake experiments. l-Aspartate:l-alanine exchange experiments were carried out with AspT-reconstituted proteoliposomes loaded with 10 mm l-alanine and citrate-phosphate buffer (pH 4.0 to 8.0), the ionic strength of which was adjusted to 75 mm with KCl, whereas the external pH was changed from 4.0 to 8.0 by using citrate-phosphate buffer, the ionic strength of which was adjusted to 80 mm with KCl. Initial rates of l-[3H]aspartate transport were plotted as a function of the pH of the outside buffer; the inside buffer was pH 7 under all conditions (▴) or the pH of the inside buffer was changed to match that of the outside buffer (●). Rates of dissociation into l-aspartate1− (×) and l-alamine0 (*) at the indicated pH values are shown. Data are from three independent experiments and presented as the average with S.D.
Substrate Protection of Solubilized AspT
We also directly measured the dissociation constant, KD(l-aspartate) and KD(l-alanine), by monitoring the kinetics with which l-aspartate or l-alanine stabilized solubilized AspT. The solubilized AspT was incubated with l-aspartate (5, 10, and 20 mm) or l-alanine (50, 100, and 200 mm) at 37 °C for several minutes. After the reconstitution, the remaining transport activities of AspT reconstituted into proteoliposomes loaded with 200 mm l-alanine and 50 mm N-methyl-d-glucamine-phosphate (pH 7.0) were examined by using a simple filtration method (14).
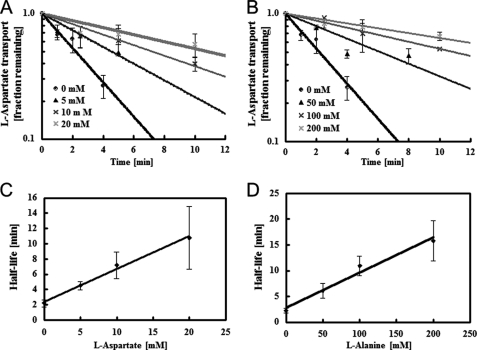
When a detergent extract was incubated at 37 °C, the AspT activity decreased in an exponential fashion, with a half-life of about 2.2 min (Fig. 4). Added substrate was clearly protective and increased the AspT lifetime. l-Aspartate at 5, 10, and 20 mm gave half-lives of 4.5, 7.2, and 11 min, respectively (Fig. 4A), and l-alanine at 50, 100, and 200 mm gave half-lives of 6.1, 11, and 16 min, respectively (Fig. 4B). This stabilization presumably reflects substrate binding by AspT and the generation of ligand complexes more resistant to denaturation, as noted for OxlT (19). Assuming that liganded AspT is much more stable than unliganded carrier, a reasonable assumption given the observed response to excess substrate (l-aspartate and l-alanine), then the stabilization achieved by substrate binding can be analyzed quantitatively to derive the dissociation constant of the AspT-substrate complex (19). In such cases, the fold-increase in AspT lifetime (R) is related to KD(substrate) by the following expression: r = 1 + (S)/KD(substrate) (Eq. 1), where (S) represents the substrate concentration used for stabilization. For the l-aspartate levels (5 to 20 mm) and l-alanine levels (50 to 200 mm) tested here, KD(l-aspartate) and KD(l-alanine) were 4.6 ± 0.4 and 29 ± 4 mm, respectively (Fig. 4, C and D).
FIGURE 4.
Substrate protection of solubilized AspT. A, solubilized AspT (27 μg of protein/ml) was incubated at 37 °C in the absence (●) or presence of aspartate at 5 (▴), 10 (×), or 20 mm (*). To follow the decay of AspT, aliquots were removed at the indicated times and placed in chilled quench tubes containing 20 mm l-aspartate and the other components required for reconstitution. After reconstitution, proteoliposomes were tested for residual AspT activity by use of an abbreviated assay (see “Experimental Procedures”). In the absence of added substrate, recoverable AspT activity disappeared with a half-life of 0.15 min. Aspartate concentrations of 5, 10, and 20 mm gave half-lives of 4.5, 7.2, and 11 min, respectively; from Equation 1 (see text), the predicted corresponding KD(l-aspartate) value was 4.6 ± 0.4 mm. B, solubilized AspT was incubated in the absence (●) or presence of l-alanine (50 mm, ▴; 100 mm, ×; 200 mm, *) and residual AspT activity was measured by use of an abbreviated assay. In the absence of added substrate, recoverable AspT activity disappeared with a half-life of 0.15 min. Alanine concentrations of 50, 100, and 200 mm gave half-lives of 6.1, 11, and 16 min, respectively; from Equation 1, the predicted corresponding KD(l-alanine) value was 29 ± 4 mm. C and D, half-life of solubilized AspT in the presence of l-aspartate (C) and l-alanine (D). Data are from three independent experiments and presented as the average with S.D.
Kinetic Analysis of AspT Transport
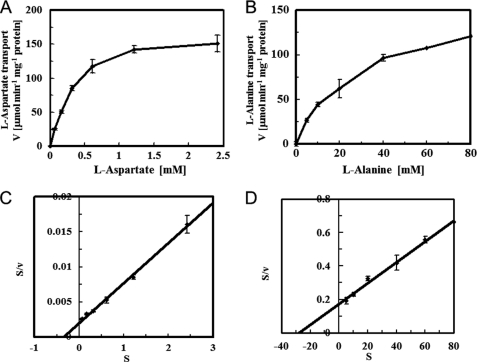
We undertook a kinetic study, relying on samples filtered after a 10-s reaction, to estimate the initial velocities of the l-aspartate and l-alanine self-exchange reactions. We used a Hanes-Woolf plot to determine the Michaelis constant (Km) and the maximum reaction (Vmax) (Fig. 5). We found that the l-aspartate self-exchange reaction had a Michaelis constant (Km) of 0.35 ± 0.03 mm and a maximum velocity of 175 ± 21 μmol min−1 mg−1 of protein and that the l-alanine self-exchange reaction had a Km of 26 ± 2 mm and a maximum velocity of 155 ± 9 μmol min−1 mg−1 of protein (mean ± S.E.). The Km of the l-aspartate self-exchange reaction was therefore 74-fold lower than that of the l-alanine self-exchange reaction. These results are consistent with the tendency of KD values reported for l-aspartate and l-alanine binding to Lactobacillus sp. AspT (4). l-Aspartate had a higher affinity than l-alanine for AspT, but the l-aspartate and l-alanine self-exchange reactions had almost the same maximal velocities (Vmax). Thus, the l-alanine self-exchange reaction showed almost the same transport rate as that of the l-aspartate self-exchange reaction in the presence of sufficiently high concentrations of l-alanine (>50 mm). We also undertook a kinetic study of d-aspartate and d-alanine self-exchange reactions; the results are summarized in Table 1. d-Aspartate and d-alanine showed a higher affinity than l-aspartate and l-alanine, respectively, for AspT. On the other hand, the l-isomers had higher Vmax values than those of the d-isomers. These results suggest that the d-isomers form a tight and stable complex with AspT, with slow dissociation rates and low Vmax values. AspT thus recognizes not only l-aspartate and l-alanine, but also d-aspartate and d-alanine, as substrates.
FIGURE 5.
Kinetic analysis of l-aspartate and l-alanine self-exchange. The kinetic parameters of l-[3H]aspartate (A) and l-[3H]alanine (B) transport were determined by sampling after a 15-s reaction to estimate initial velocities. Substrate concentrations were corrected for the residual external aspartate carried into the assay from the proteoliposome stock. A linear transform was used to calculate the Michaelis constant (Km) and maximum velocity (Vmax) of the l-[3H]aspartate (C) and l-[3H]alanine (D) transport reactions. Data are from three independent experiments and presented as the average with S.D.
Substrate Screening of AspT
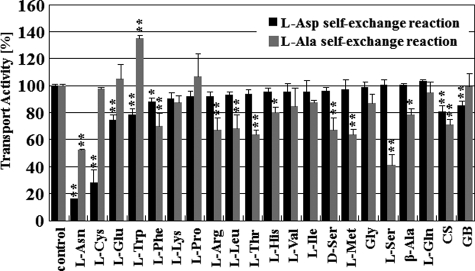
To investigate the substrate specificity of AspT, we performed substrate-competition experiments by measuring the uptake of l-[3H]aspartate and l-[3H]alanine in self-exchange reactions in the presence of 15 mm of various unlabeled amino acids (18 l-amino acids, glycine, three d-amino acids, β-alanine, d-cycloserine, and glycine betaine) (Fig. 6). l-Asparagine and l-cysteine strongly inhibited l-[3H]aspartate uptake (relative accumulation of l-[3H]aspartate: 10 to 30%), whereas l-glutamate and l-tryptophan moderately inhibited l-[3H]aspartate uptake (relative accumulation of l-[3H]aspartate: 70 to 80%). None of the amino acids completely inhibited l-[3H]alanine uptake. l-Asparagine and l-serine medially inhibited l-[3H]alanine uptake (relative accumulation of l-[3H]aspartate: 40 to 60%). l-Phenylalanine, l-arginine, l-leucine, l-threonine, d-serine, l-methionine, β-alanine, and d-cycloserine moderately inhibited l-[3H]alanine uptake (relative accumulation of l-[3H]aspartate: 60 to 80%). These results suggest that AspT recognizes as substrates not only l-aspartate, l-alanine, d-aspartate, and d-alanine, but also many other d- and l-amino acids, such as l-serine and l-cysteine. l-Cysteine and l-serine had unique inhibitory effects. l-[3H]Aspartate uptake was strongly inhibited by l-cysteine; however, l-[3H]alanine uptake was not inhibited. On the other hand, l-serine moderately inhibited l-[3H]alanine uptake and barely inhibited l-[3H]aspartate uptake. d-Cycloserine, an analog of d-alanine, and glycine betaine, a natural osmolyte, did not strongly inhibit the l-aspartate and l-alanine self-exchange reactions.
FIGURE 6.
Competitive inhibition of AspT counterflow with amino acids and analogs. Amino acid inhibition pattern of AspT transport activity. Transport of 39 μm radiolabeled l-aspartate was measured for 10 s in AspT-reconstituted proteoliposomes containing 100 mm l-aspartate. Transport was measured in the absence (no amino acids) or presence of the indicated amino acids or d-cycloserine (CS) or glycine betaine (GB) at 15 mm in the external medium (black bars). Transport of 2.9 mm radiolabeled l-alanine was measured for 10 s in AspT-reconstituted proteoliposomes containing 100 mm l-alanine. Transport was measured in the absence (no amino acids) or presence of the indicated amino acids at 15 mm in the external medium (gray bars). Transport activities are expressed as a percentage of the transport in AspT proteoliposomes containing 100 mm l-aspartate (black bars) or l-alanine (gray bars) in the absence of inhibitors (no amino acids). Data are from two or three independent experiments with three replicates per condition and presented as the average with S.D. (*, p < 0.05; **, p < 0.01, Dunnett's test).
Kinetic Analysis of l-Cysteine and l-Serine Self-exchange
Because l-serine and l-cysteine were found to be substrates, we undertook a simple kinetic study in which they served as self-exchange substrates. We relied on samples that were filtered after a 10-s reaction to estimate initial velocities (Table 1). We found that the l-serine self-exchange reaction had a Michaelis constant (Km) of 11 ± 1 mm and a maximum velocity of 103 ± 24 μmol min−1 mg−1 of protein and that the l-cysteine self-exchange reaction had a Km of 8.2 ± 0.1 mm and a maximum velocity of 11 ± 1 μmol min−1 mg−1 of protein (mean ± S.E.). These results indicate that AspT catalyzes self-exchange reactions with these two substrates.
Inhibitory Effect of l-Cysteine and l-Serine
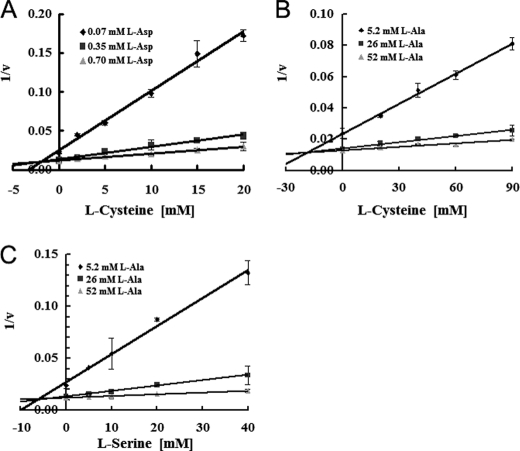
To evaluate the inhibitory effects of l-cysteine and l-serine, we performed a Dixon-Webb plot analysis of data from samples that were filtered after a 10-s reaction to estimate initial velocities (20, 21). External substrate concentrations were adjusted to 0.07, 0.35 (=Km value), and 0.7 mm for l-aspartate self-exchange, and 5.8, 26 (=Km value), and 58 mm for l-alanine self-exchange. The three lines in the Dixon plot crossed at one point (Fig. 7, A–C), demonstrating competitive inhibition of l-aspartate and l-alanine by l-cysteine and l-serine, respectively. The calculated inhibition constant values (Ki) were 2.0 ± 0.2 mm (l-aspartate exchange inhibited by cysteine), 20 ± 2.7 mm (l-alanine exchange inhibited by cysteine), and 5.3 ± 1.7 mm (l-alanine exchange inhibited by serine) (Table 2). The Ki of l-serine for l-aspartate self-exchange was too high to measure, suggesting that serine is a specific inhibitor of alanine self-exchange but not of aspartate self-exchange. These analyses reveal that the mechanism of inhibition of l-aspartate self-exchange and l-alanine self-exchange by l-cysteine and l-serine is competitive inhibition.
FIGURE 7.
l-Cysteine and l-serine inhibit l-aspartate and l-alanine uptake. Dixon-Webb plot analysis, relying on samples filtered after a 10-s reaction to estimate the initial velocities. The initial rates of uptake in AspT-reconstituted proteoliposomes were measured in the presence of (A) 0.07 (♦), 0.35 (■), or 0.70 mm (▴) l-aspartate or (B and C) 5.2 (♦), 26 (■), or 52 mm (▴) l-alanine. The reciprocal value of the rate was plotted versus l-cysteine (A and B) and l-serine (C). The inhibition constants (Ki) were calculated to be 2.0 ± 0.2 (l-aspartate exchange inhibited by cysteine), 20 ± 2.7 (l-alanine exchange inhibited by serine), and 5.3 ± 1.7 mm (l-alanine exchange inhibited by serine).
TABLE 2.
Calculated Ki values for compounds that competed for l-Asp or l-Ala uptake in proteoliposomes
|
l-Asp self-exchange |
l-Ala self-exchange |
|||
|---|---|---|---|---|
| Ki | IC50 | Ki | IC50 | |
| mm | mm | |||
| l-Cysteine | 2.0 ± 0.2 | 8.9 | 20 ± 2.7 | 65 |
| l-Serine | NDa | ND | 5.3 ± 1.7 | 28 |
| l-CSA | 0.59 ± 0.08 | 0.83 | 28 ± 0 | 28 |
| l-CA | 0.29 ± 0.05 | 0.47 | 14 ± 0 | 16 |
| d-CA | 0.14 ± 0.02 | 0.19 | 5.0 ± 0.2 | 9.0 |
a ND, not determined. The Ki of serine for aspartate self-exchange was too high to measure, suggesting that serine is a specific inhibitor of l-alanine self-exchange but not l-aspartate self-exchange.
Because the Km of l-serine to AspT was larger than the Km of l-aspartate to AspT but smaller than the Km of l-alanine to AspT, the apparent selective inhibitory effect of l-serine on l-alanine self-exchange can be explained by the difference in the Km values of l-serine and l-alanine. However, the Km of l-cysteine was larger than that of l-aspartate; this cannot explain the apparent selective inhibition of l-aspartate self-exchange with l-cysteine.
Inhibitory Effect of l-Aspartate Analogs
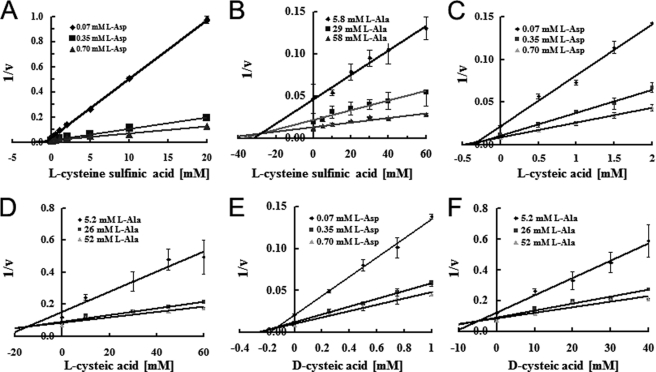
To further investigate the selective inhibition of l-aspartate self-exchange with l-cysteine, we used substrate analogs for both cysteine and aspartate, namely l-CSA, l-CA, and d-CA (supplemental Fig. 2, we synthesized the d-CA as described under “Experimental Procedures”). We measured l-aspartate and l-alanine self-exchange in their presence. l-CSA, d-CA, and l-CA inhibited both the l-aspartate and l-alanine self-exchange reactions via AspT (Fig. 8 and Table 2). The three plot lines crossed at one point in the Dixon plot (Fig. 8, A–F), revealing the competitive inhibition of l-aspartate and l-alanine by these analogs. The inhibition constants (Ki) were calculated and are summarized in Table 2. d-CA was the strongest inhibitor of both the l-aspartate and l-alanine self-exchange reactions. To determine whether these inhibitors were transportable via AspT, we applied a counter-flow and exchange reaction assay. The inhibitors induced the export of internal l-[3H]aspartate and l-[3H]alanine from proteoliposomes, indicating that they were competitive inhibitors (supplemental Fig. S3). All three analogs markedly inhibited l-aspartate self-exchange but only weakly inhibited l-alanine self-exchange.
FIGURE 8.
l-Cysteine sulfinic acid, l-cysteic acid, and d-cysteic acid inhibit l-aspartate and l-alanine uptake. Dixon-Webb plot analysis, relying on samples filtered after a 10-s reaction to estimate initial velocities. The initial rates of uptake in AspT-reconstituted proteoliposomes were measured in the presence of (A, C, and E) 0.07 mm (♦), 0.35 mm (■), or 0.70 mm (▴) l-aspartate or (B, D, and F) 5.2 mm (♦), 26 mm (■), or 52 mm (▴) l-alanine. The reciprocal value of the rate was plotted versus the concentration of l-CSA (A and B), l-CA (C and D), or d-CA (E and F). For l-aspartate and l-alanine, straight-line plots were obtained that intersected at a common point (-Ki), indicating that these compounds competitively inhibited l-aspartate and l-alanine uptake. Ki values for the other inhibitors were similarly obtained.
DISCUSSION
Kinetic Properties of AspT
Purified His6 AspT reconstituted in proteoliposomes catalyzed l-aspartate1−:l-alanine0 exchange (Fig. 1), and internal positive and negative membrane potentials accelerated and inhibited the exchange reaction, respectively (Fig. 2). These results clearly demonstrate that the l-aspartate:l-alanine exchange reaction is catalyzed solely by AspT and that the reaction is electrogenic. Although the optimum temperature for aspartate self-exchange by reconstituted AspT was 45 °C (supplemental Fig. S1A), solubilized AspT was unstable over 40 °C and its half-life at 40 °C was 2.1 min (supplemental Fig. S1B). The optimum pH for l-aspartate:l-alanine exchange by reconstituted AspT was pH 5.0 (Fig. 3, gray line). Because more than 95% of l-aspartate molecules release protons from their β-carboxyl group at pH 5.0, l-aspartate:l-alanine exchange seems to be electrogenic at pH 5.0.
The KD values calculated from the stabilization of solubilized AspT with l-aspartate and l-alanine were 4.6 ± 0.4 and 29 ± 4 mm, respectively (Fig. 4). The Km values for the l-aspartate and l-alanine self-exchange reactions were 0.35 ± 0.03 and 26 ± 2 mm, respectively (Table 1). Both the Km and KD for l-aspartate were lower than those for l-alanine, suggesting that these values reflect the physiological conditions for AspT function. In the natural habitat of T. halophilus, intracellular levels of l-alanine are high (>50 mm) and the extracellular levels of l-aspartate substrate for AspT import are relatively low (<20 mm).
The Km values for the self-exchange reactions were 0.35 (l-aspartate), 26 (l-alanine), 0.098 (d-aspartate), and 3.3 mm (d-alanine) (Fig. 5 and Table 1). The Vmax values were 175 μmol min−1 mg−1 protein (l-aspartate), 155 μmol min−1 mg−1 protein (l-alanine), 3.8 μmol min−1 mg−1 protein (d-aspartate), and 3.3 μmol min−1 mg−1 protein (d-alanine). Because AspT in intact T. halophilus cells generally exchanges external l-aspartate with internal l-alanine, the similar Vmax values of AspT for self-exchange of l-aspartate and l-alanine are physiologically reasonable. The Vmax/Km values were 500 (l-aspartate), 39 (d-aspartate), 6.0 (l-alanine), and 1.0 (d-alanine). The Km and Vmax values of AspT to d-aspartate and d-alanine were much lower than those to the l-forms (Table 1). AspT may, therefore, form a more stable complex with d-aspartate and d-alanine than with the l-forms, and the release of the d-amino acids from AspT would be a rate-limiting step.
Binding Sites for l-Aspartate and l-Alanine Are Independently Located in AspT?
In our competitive inhibition experiments, 15 mm of each competitive substrate or analog was added to the counter-flow reaction mixture. Kinetics analysis of the self-exchange reactions revealed Km values of 8.2 mm for l-cysteine and 11 mm for l-serine, and Vmax values of 11 μmol min−1 mg−1 of protein for l-cysteine and 103 μmol min−1 mg−1 of protein for l-serine. l-Cysteine and l-serine are, therefore, good substrates for AspT (Table 1). In the competitive inhibition experiments, l-serine strongly inhibited l-alanine self-exchange but barely affected l-aspartate self-exchange (Fig. 6 and Table 2). This selective inhibitory effect of l-serine on l-alanine self-exchange may be explained by the affinity of l-serine for AspT (Km = 11 mm), which is higher than that of l-aspartate but lower than that of l-alanine (Table 1). On the other hand, l-cysteine strongly inhibited l-aspartate self-exchange but barely affected l-alanine self-exchange (Fig. 6 and Table 2). This selective inhibitory effect cannot be explained by the difference in putative affinities of l-cysteine, l-aspartate, and l-alanine for AspT, because the order of AspT affinity for these three amino acids is l-aspartate (Km = 0.35 mm) < l-cysteine (Km = 8.2 mm) < l-alanine (Km = 26 mm). Although l-cysteine has a lower affinity for AspT than l-aspartate, l-cysteine inhibited l-aspartate self-exchange. Moreover, l-cysteine had a higher affinity for AspT than did l-alanine, whereas l-cysteine barely inhibited l-alanine self-exchange.
To examine whether the selective inhibitory effect of l-cysteine on l-aspartate self-exchange was dependent on the structural similarity between the two amino acids, we examined three aspartate analogs, l-CSA, l-CA, and d-CA. l-CSA, l-CA, and the d-aspartate analog d-CA strongly inhibited l-aspartate self-exchange but not l-alanine self-exchange (Fig. 8 and Table 2). Dixon plot analysis of the inhibitors clearly demonstrated competitive inhibition by these analogs.
The selective inhibitory effects of l-cysteine and aspartate analogs on aspartate self-exchange (but not l-alanine self-exchange) were explained by the following hypothesis. The l-serine binding site overlaps with the putative l-alanine binding site, which differs from the putative l-aspartate binding site. The thiol residue in l-cysteine has a pKa of 8.3 and can release protons akin to the β-carboxyl group of l-aspartate (pKa = 3.86). At pH 7, 6.6% of thiol residues in l-cysteine release protons. Compared with l-cysteine, the hydroxyl residue in l-serine has a higher pKa of 13 and the OH-group in the side residue barely releases protons at neutral or even alkaline pH. In addition, l-cysteine sulfinic acid (oxidized cysteine to a sulfonate, pKa 1.5) showed smaller IC50 than l-cysteine (Table 1). The state of proton dissociation in the side residues would thus be involved in substrate recognition by AspT. Another example of the two independent substrate binding site model was proposed in the arginine:agmatine antiporter AdiC. In E. coli, the acid-resistance system exports intracellular protons by the combined action of an intracellular arginine decarboxylation reaction and an electrogenic arginine:agmatine exchange reaction catalyzed by the AdiC (5, 22). Gao et al. (23, 24) solved the crystal structure of E. coli AdiC and proposed a model for arginine:agmatine exchange. These crystal structures and ITC analyses revealed the existence of two independent substrate binding sites in the substrate translocation pathway and identified key residues that are involved in binding to arginine and agmatine (23).
The selective inhibitory effects suggest the presence of two independent substrate binding sites in the substrate translocation pathway of AspT. However, in a kinetic sense, it would be necessary for alanine binding to be coupled with aspartate binding. If binding of the two substrates were fully independent, the exchange would be decoupled and the transporter could not fulfill its physiological role. In fact, l-cysteine and l-aspartate analogs slightly inhibited l-alanine self-exchange. Therefore, the translocation pathways for l-aspartate and l-alanine appeared to overlap. For complete understanding of substrate binding and recognition mechanisms of AspT, further biochemical and structural studies of AspT are needed.
Supplementary Material
Acknowledgments
We thank the Kikkoman Corporation for their gift of the T. halophilus asp operon. We also thank Dr. Peter C. Maloney (Department of Physiology, Johns Hopkins Medical School) and Dr. Hiroshi Yoneyama (Graduate School of Agricultural Science, Tohoku University) for critical reading of this article.
This work was supported by a Grant-in-aid for challenging exploratory research from Japan Society for the Promotion of Science (JSPS) Grant 2265108 (to K. A.), the Kato Memorial Bioscience Foundation (to K. N.), and a NISR (Noda Institute for Scientific Research) Research grant (to K. A.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3 and “Methods.”
- DDM
- n-dodecyl-β-d-maltoside.
REFERENCES
- 1. Abe K., Ruan Z. S., Maloney P. C. (1996) J. Biol. Chem. 271, 6789–6793 [DOI] [PubMed] [Google Scholar]
- 2. Anantharam V., Allison M. J., Maloney P. C. (1989) J. Biol. Chem. 264, 7244–7250 [PubMed] [Google Scholar]
- 3. Maloney P. C., Yan R. T., Abe K. (1994) J. Exp. Biol. 196, 471–482 [DOI] [PubMed] [Google Scholar]
- 4. Abe K., Hayashi H., Maloney P. C., Malone P. C. (1996) J. Biol. Chem. 271, 3079–3084 [DOI] [PubMed] [Google Scholar]
- 5. Gong S., Richard H., Foster J. W. (2003) J. Bacteriol. 185, 4402–4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Abe K., Ohnishi F., Yagi K., Nakajima T., Higuchi T., Sano M., Machida M., Sarker R. I., Maloney P. C. (2002) J. Bacteriol. 184, 2906–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. (1990) J. Mol. Biol. 215, 403–410 [DOI] [PubMed] [Google Scholar]
- 8. Barabote R. D., Tamang D. G., Abeywardena S. N., Fallah N. S., Fu J. Y., Lio J. K., Mirhosseini P., Pezeshk R., Podell S., Salampessy M. L., Thever M. D., Saier M. H., Jr. (2006) Biochim. Biophys. Acta 1758, 1557–1579 [DOI] [PubMed] [Google Scholar]
- 9. Fujiki T., Nanatani K., Nishitani K., Yagi K., Ohnishi F., Yoneyama H., Uchida T., Nakajima T., Abea K. (2007) J. Biochem. 141, 85–91 [DOI] [PubMed] [Google Scholar]
- 10. Fukui K., Koseki C., Yamamoto Y., Nakamura J., Sasahara A., Yuji R., Hashiguchi K., Usuda Y., Matsui K., Kojima H., Abe K. (2011) J. Biotechnol. 154, 25–34 [DOI] [PubMed] [Google Scholar]
- 11. Nanatani K., Ohonishi F., Yoneyama H., Nakajima T., Abe K. (2005) Biochem. Biophys. Res. Commun. 328, 20–26 [DOI] [PubMed] [Google Scholar]
- 12. Deleted in proof.
- 13. Nanatani K., Fujiki T., Kanou K., Takeda-Shitaka M., Umeyama H., Ye L., Wang X., Nakajima T., Uchida T., Maloney P. C., Abe K. (2007) J. Bacteriol. 189, 7089–7097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nanatani K., Maloney P. C., Abe K. (2009) J. Bacteriol. 191, 2122–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ambudkar S. V., Maloney P. C. (1986) J. Biol. Chem. 261, 10079–10086 [PubMed] [Google Scholar]
- 16. Wang X., Sarker R. I., Maloney P. C. (2006) Biochemistry 45, 10344–10350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ye L., Maloney P. C. (2002) J. Biol. Chem. 277, 20372–20378 [DOI] [PubMed] [Google Scholar]
- 18. Clarke H. T. (1955) Organic Syntheses, Collective Volumes (Horning E. C., Allen C. F. H., Shriner R. L., Drake N. L., Smith L. I., Hamilton C. S., Snyder H. R. eds) p. 226, John Wiley & Sons, Inc., New York [Google Scholar]
- 19. Maloney P. C., Anantharam V., Allison M. J. (1992) J. Biol. Chem. 267, 10531–10536 [PubMed] [Google Scholar]
- 20. Dixon M. (1953) Biochem. J. 55, 170–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cornish-Bowden A., Eisenthal R. (1974) Biochem. J. 139, 721–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Iyer R., Williams C., Miller C. (2003) J. Bacteriol. 185, 6556–6561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gao X., Lu F., Zhou L., Dang S., Sun L., Li X., Wang J., Shi Y. (2009) Science 324, 1565–1568 [DOI] [PubMed] [Google Scholar]
- 24. Gao X., Zhou L., Jiao X., Lu F., Yan C., Zeng X., Wang J., Shi Y. (2010) Nature 463, 828–832 [DOI] [PubMed] [Google Scholar]
- 25. Saier M. H., Jr., Tran C. V., Barabote R. D. (2006) Nucleic Acids Res. 34, D181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saier M. H., Jr., Yen M. R., Noto K., Tamang D. G., Elkan C. (2009) Nucleic Acids Res. 37, D274–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.