Abstract
An increase in production of reactive oxygen species resulting in a decrease in nitric oxide bioavailability in the endothelium contributes to many cardiovascular diseases, and these reactive oxygen species can oxidize cellular macromolecules. Protein thiols are critical reducing equivalents that maintain cellular redox state and are primary targets for oxidative modification. We demonstrate endothelial NOS (eNOS) oxidant-induced protein thiyl radical formation from tetrahydrobiopterin-free enzyme or following exposure to exogenous superoxide using immunoblotting, immunostaining, and mass spectrometry. Spin trapping with 5,5-dimethyl-1-pyrroline N-oxide (DMPO) followed by immunoblotting using an anti-DMPO antibody demonstrated the formation of eNOS protein radicals, which were abolished by superoxide dismutase and l-NAME, indicating that protein radical formation was due to superoxide generation from the eNOS heme. With tetrahydrobiopterin-reconstituted eNOS, eNOS protein radical formation was completely inhibited. Using mass spectrometric and mutagenesis analysis, we identified Cys-908 as the residue involved in protein radical formation. Mutagenesis of this key cysteine to alanine abolished eNOS thiyl radical formation and uncoupled eNOS, leading to increased superoxide generation. Protein thiyl radical formation leads to oxidation or modification of cysteine with either disulfide bond formation or S-glutathionylation, which induces eNOS uncoupling. Furthermore, in endothelial cells treated with menadione to trigger cellular superoxide generation, eNOS protein radical formation, as visualized with confocal microscopy, was increased, and these results were confirmed by immunoprecipitation with anti-eNOS antibody, followed by immunoblotting with an anti-DMPO antibody. Thus, eNOS protein radical formation provides the basis for a mechanism of superoxide-directed regulation of eNOS, involving thiol oxidation, defining a unique pathway for the redox regulation of cardiovascular function.
Keywords: Electron Paramagnetic Resonance (EPR), Endothelium, Enzymes, Nitric Oxide, Nitric-Oxide Synthase, Oxidative Stress, Oxygen Radicals, Proteomics, Superoxide Ion, Thiol
Introduction
NOS is an important enzyme that converts l-arginine to l-citrulline and NO with the consumption of NADPH. NO, a labile free radical, promotes vascular smooth muscle relaxation and functions as an endogenous mediator of diverse effects in numerous tissues (1, 2). During ischemia and reperfusion, the burst production of superoxide (O2˙̄) and its derived oxidants, including peroxynitrite, hydrogen peroxide, and hydroxyl radical (•OH), cause NOS dysfunction (3, 4). Under this oxidative stress, depletion or damage of NOS cofactors, including tetrahydrobiopterin (BH4)2 or heme, can switch NOS from NO production to O2˙̄ generation (5, 6). Studies have shown that the imbalance of NO and O2˙̄ contributes to many cardiovascular diseases, including hypertension, atherosclerosis, and heart failure (7, 8).
Under oxidative stress, a number of mechanisms have been proposed to trigger reactive oxygen species generation, with the enzymes xanthine oxidase, cyclooxygenase, leukocyte NADPH oxidase, mitochondria and uncoupled endothelial NOS (eNOS) as putative sources (9, 10). Uncoupling of eNOS derived NO from NADPH oxidation, which increases O2˙̄ generation, is a primary factor contributing to or triggering many cardiovascular diseases, including hypertension, atherosclerosis, and ischemia/reperfusion injury. There are several molecular mechanisms proposed to cause eNOS uncoupling, including oxidation of BH4, arginine depletion, increased asymmetric dimethyl l-arginine production, and eNOS S-glutathionylation (4, 5, 10–13).
To prevent and reduce the risks of coronary disease and cardiovascular morbidity and mortality, several therapeutic approaches have been proposed to lower these risks, including supplementation of BH4 and its precursor, antioxidants such as vitamin C and E, and superoxide dismutase (SOD) or related enzyme mimetics (10, 14). However, these approaches may not completely prevent or rescue eNOS function and coupling (4). As such, it is important to fully investigate the molecular mechanism of eNOS redox regulation in response to oxidative stress, and the fundamental mechanisms leading to eNOS uncoupling.
Previously, it has been demonstrated that eNOS phosphorylation and protein-protein interaction play important roles in regulating eNOS function (1, 15). In our earlier study, we demonstrated that phosphorylation of eNOS Ser-1177 greatly increased eNOS O2˙̄ generation from uncoupled enzyme and altered eNOS Ca2+ sensitivity (16). In contrast, phosphorylation of eNOS Thr-495 increased the association with caveolin, by which O2˙̄ generation from the uncoupled eNOS was inhibited.
There is growing evidence that increasing oxidative stress alters the function of several enzymes through oxidative post-translational modification, such as S-glutathionylation, nitration, or nitrosylation (17–19). These modifications can either decrease or increase enzymatic activities to protect cells from further damage caused by the oxidative stress. Human eNOS, which is of critical importance in maintaining cardiovascular function, contains 27 cysteines that are conserved in all known mammalian eNOS. Thiol depletion in vivo greatly reduces NO production from eNOS (20–22). It has been shown that reducing agents, such as DTT or GSH, can enhance NO synthase activity (23, 24). Moreover, S-nitrosylation of eNOS has been shown to regulate its enzymatic activity (25), and proteomic analysis identified several cysteinyl residues involved in this modification.
Recently, we demonstrated that eNOS S-glutathionylation uncouples the enzyme leading to the increase in O2˙̄ generation from hypertensive vessels as well as bovine aortic endothelial cells (BAECs) treated with N,N′-bis(2-chloroethyl)-N-nitroso-urea (13). Two highly conserved cysteine residues were identified as primary sites for this eNOS S-glutathionylation. Together, these findings suggest that the sulfhydryl groups of cysteines in eNOS are of critical importance in maintaining and regulating NOS function. Thus, it is important to investigate the fundamental mechanism of eNOS thiol oxidation and the effect that this has on the function of this critical enzyme.
The recently developed immunospin trapping technique using an anti-DMPO antibody (26) was used to characterize the mechanism of eNOS thiol oxidation. We demonstrate that O2˙̄ oxidizes eNOS sulfhydryl groups to form protein radicals visualized by immunoblotting using an anti-DMPO antibody. Mass spectrometric analysis and site-directed mutagenesis are also used to identify the precise site of eNOS thiyl radical formation. This eNOS protein radical formation leads to modification of the function and coupling of the enzyme through subsequent S-glutathionylation or disulfide formation and is shown to occur in endothelial cells under oxidant stress. Thus, eNOS protein thiol-radical formation provides a unique mechanism for O2˙̄-directed regulation of eNOS and cardiovascular function.
EXPERIMENTAL PROCEDURES
Materials
DMPO was obtained from Dojindo Molecular Technologies, Inc. (Rockville, MD), 5-diethoxyphosphoryl-5-methyl-1-pyrroline N-oxide (DEPMPO) was from Radical Vision (Jerome, Marseille, France), and anti-DMPO antibody was from Alexis Biochemicals (San Diego, CA). Anti-NOS3 (C-20) HRP and anti-NOS3 (C-20) agarose conjugate antibodies were both obtained from Santa Cruz (Santa Cruz, CA); anti-eNOS (mouse IgG1) was from BD Biosciences (Sparks, MD); NADPH, l-Arg, calmodulin, Mn-SOD, and Tris were from Sigma-Aldrich. Secondary anti-rabbit Alexa fluor-568 and anti-mouse Alexa fluor-488-conjugated antibodies were purchased from Invitrogen. Rat nNOS reductase domain (695–1429 amino acids) was a gift from Dr. Dennis J. Stuehr (27).
Protein Expression and Purification
Human eNOS was purified from an Escherichia coli overexpression system in which plasmids eNOS (pCWeNOS) and calmodulin (pCaM) were coexpressed in BL21(DE3). The detailed expression and purification procedures have been described previously (16, 28, 29).
Protein and Heme Content Determination
Protein concentration of purified eNOS was determined by the Bradford assay from Bio-Rad using bovine serum albumin as the standard. The heme content of eNOS was determined by pyridine hemochromogen assay (16).
Measurement of O2˙̄ Generation by EPR Spin-Trapping
Spin-trapping measurements (16) of oxygen radical production from eNOS were performed in 50 mm Tris-HCl buffer, pH 7.4, containing 1 mm NADPH, 0.5 mm Ca2+, 10 μg/ml calmodulin, 15 μg/ml purified WT eNOS or C908A eNOS, and 25 mm DEPMPO. EPR spectra were recorded in a 50-μl capillary at room temperature with a Bruker EMX spectrometer operating at 9.86 GHz with 100 kHz modulation frequency as described (16). Spectra were measured with: center field, 3510 G; sweep width, 140 G; power, 20 milliwatt; receiver gain, 2 × 105; modulation amplitude, 0.5 G; time of conversion, 41 ms; time constant, 328 ms.
Immunoblotting of eNOS Protein Radicals
To form the eNOS DMPO adduct, BH4-free eNOS (5 μg) was used to self-generate O2˙̄ in presence of 0.5 mm Ca2+, 10 μg/ml calmodulin or nNOS reductase domain (4 μg) was used as an exogenous O2˙̄ source, which in turn can modify sulfhydryl groups of eNOS to form protein radicals. The reaction mixture (20 μl) was initiated by 1 mm NADPH. When nNOS reductase was used as an exogenous O2˙̄ source, no calmodulin/calcium was included in the reaction. These short-lived protein radicals were trapped with the spin trap DMPO (50 mm) (30) at room temperature for 30 min, and the reaction mixtures were then subjected to immunoblotting analysis. Standard procedures for SDS-PAGE and immunoblotting were as described previously (16).
Mass Spectrometry Analysis of eNOS Protein Radicals
The protein sample was subjected to SDS-PAGE using 4–20% gradient polyacrylamide. Protein bands on the gel were then stained with Coomassie Blue. The band containing the eNOS DMPO protein radical adduct, which was confirmed by immunoblotting with an anti-DMPO antibody, was cut and digested in-gel with trypsin, chymotrypsin, or both before MS measurement (30). The eNOS DMPO protein radical adduct was determined using capillary-liquid chromatography tandem mass spectrometry (Nano-LC-MS/MS), which was performed on a Micromass hybrid quadrupole time-of-flight II mass spectrometer (Micromass, Wythenshaw, UK) equipped with an orthogonal nanospray source (New Objective, Woburn, MA) operative in positive ion mode. The detailed parameters used in the MS measurements were as described previously (13, 30). Sequence information from MS/MS data were processed using the Mascot Distiller software with standard data processing parameters. Database searches were performed using MASCOT (Matrix Science, Boston, MA) and PEAKS (Bioinformatics Solutions, Waterloo, Canada) programs.
Molecular Modeling of eNOS Reductase Domain
The three-dimensional structure of human eNOS reductase domain was generated using the reductase domain of rat neuronal NOS (Protein Data Bank code 1F20) by Swiss molecular modeling as previous described (13).
Mutagenesis of Wild-type (WT) eNOS Cys-908 to Ala
The bacterial expression plasmid pCWeNOS (29, 31) was used to generate the eNOS Cys mutant, using a QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). The sequence of primers was as follows: sense, 5′-GAAGTGGTTCCGCGCCCCCACGCTGCTG-3′; and antisense, 5′-CAGCAGCGTGGGGGCGCGGAACCACTTC-3′. The sequence of the eNOS (C908A) mutant in pCWeNOS was confirmed by DNA sequencing at the Plant-Microbe Genomic Facility at the Ohio State University.
EPR Spin-Trapping Measurement of NO Production
Spin-trapping measurements of NO from purified WT eNOS or C908A eNOS were performed using a Bruker EMX spectrometer with Fe-MGD (0.025 mm Fe2+ and 0.25 mm MGD) as the spin trap, as described previously (32, 33). 0.4 μg/μl of eNOS was used in the presence of 10 μm BH4, and the reaction was initiated with the addition of 1 mm NADPH in the presence of eNOS cofactors.
Determination of Reductase Activity of eNOS Using Cytochrome c Assay
The reductase activities of WT or C908A mutant eNOS were determined using the cytochrome the c assay (34). The reaction was carried out in a total volume of 500 μl containing eNOS (10–15 μg) in 50 mm Tris-HCl, pH 7.2, 1 μΜ CaM, 0.2 mm CaCl, Mn-SOD (400 units/ml), and 100 μm cytochrome c. The reaction was initiated by the addition of NADPH to a final concentration of 0.5 mm. The heme reduction of cytochrome c was monitored at 550 nm using an Agilent 8453 UV-visible spectrophotometer. Mn-SOD (400 units/ml) was included to eliminate the cytochrome c reduction contributed by O2˙̄. The linear portion of the kinetic traces was used to calculate the rate of cytochrome c reduction and reductase activity of eNOS.
FAD/FMN Measurement Using HPLC Chromatography
The ratio of FAD/FMN of WT or C908A eNOS was determined using HPLC chromatography (35, 36). The purified eNOS (70 μg in 300 μl) was first boiled for 10 min to release FAD and FMN from the protein. Protein was removed by filtration through a Microcon-3 (Millipore). 20 μl of the FAD/FMN-containing filtrates were separated on an ESA HPLC system with a C18 reverse phase column (TSK-GEL ODS-80T, 4.6 mm × 25 cm). After injection, buffer A (5 mm ammonium acetate, pH 6.0, and 7% methanol) with a flow rate at 1 ml/min was used for 4 min. Then a linear gradient was developed to 70% methanol using buffer B (5 mm ammonium acetate, pH 6.0, 70% methanol) over 16 min. An ESA fluorometer with excitation wavelength set at 460 nm and emission wavelength set at 530 nm was used to detect FAD and FMN. FAD and FMN were completely separated with elution times of 14.2 and 15.8 min, respectively. The peak area was used to calculate the ratio of FAD/FMN compared with the FAD and FMN standards.
Measurement of O2˙̄-induced S-Glutathionylation and Disulfide Bond Formation
BH4-free heNOS (0.25 μg/μl, 1.85 μm) was incubated in the presence of 2 mm GSH in a volume of 20 μl in 50 mm Tris-HCl, pH 7.2, and the reaction was initiated by adding 1 mm NADPH in the presence or absence of 1 mm l-Arg at room temperature for 30 min. The control experiment without addition of NADPH was always performed. The reaction mixture was then subjected for 4–20% SDS-PAGE separation under nonreducing conditions and immunoblotting analysis using anti-GSH monoclonal antibody (ViroGen) or anti-eNOS polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA). The signal intensity of the immunoblots was digitized and quantified with Image J from the National Institutes of Health.
Measurement of O2˙̄ Generation from BAECs by EPR Spin-Trapping
Spin-trapping measurements of oxygen radical production from BAECs were performed in PBS in the presence of 50 mm DMPO. O2˙̄ generation from BAECs was activated by the addition of 10 μm menadione (37). EPR spectra were recorded in a 50-μl capillary at room temperature with a Bruker EMX spectrometer operating at 9.86 GHz with 100-kHz modulation frequency as described (5).
Cellular Studies and Immunofluorescence Microscopy
BAECs cultured on sterile coverslips (Harvard Apparatus, 22 mm2) in 35-mm sterile dishes at a density of 104 cells/dish were subjected to 10 μm menadione treatment in the presence of 50 mm DMPO. At the end of the experiment (16), cells attached to coverslips were washed with phosphate-buffered saline (PBS) and fixed with 3.7% paraformaldehyde for 10 min, permeabilized with 0.25% Triton X-100 in Tris-buffered saline with Tween (TBST) containing 0.01% Tween 20 for 10 min, and blocked for 1 h with 5% goat serum in TBST. For visualization of eNOS DMPO protein radical adducts, the fixed and permeabilized cells were incubated with rabbit anti-DMPO and mouse anti-eNOS primary antibodies at a dilution of 1:1000 in TBST containing 5% goat serum for 2.5 h at room temperature, followed by secondary anti-rabbit Alexa fluor-568- and anti-mouse Alexa fluor-488-conjugated antibody (1:1000 dilution) for 1 h at room temperature. The coverslips with cells were then mounted on a glass slide with antifade mounting medium and viewed with a Zeiss confocal microscope (LSM 510; Zeiss Inc., Peabody, MA) at a magnification of 60×, and data were captured digitally (16).
Immunoprecipitation
BAECs were treated with 10 μm menadione to activate cellular O2˙̄ generation. eNOS protein radicals were trapped in vivo using 50 mm DMPO. Cellular eNOS DMPO adducts were first immunoprecipitated using either anti-DMPO or anti-eNOS antibodies, followed by immunoblotting with anti-eNOS or anti-DMPO antibodies, respectively.
RESULTS
DEPMPO EPR Spin-Trapping of O2˙̄ Generation from Purified eNOS in the Absence of BH4
In the absence of BH4, eNOS is uncoupled and generates O2˙̄. Consistent with prior reports, this O2˙̄ generation from eNOS is inhibited by l-NAME and requires Ca2+/CaM (Fig. 1a) (5, 11). The addition of 10 μm or 1 mm l-Arg further increases its O2˙̄ generation by 2.5-fold. l-NAME (1 mm) or Mn-SOD (200 units/ml) fully quenched the observed O2˙̄ generation. Upon reconstitution with BH4 no O2˙̄-DEPMPO adduct was detected from eNOS. These results along with inhibition by l-NAME and Ca2+/CaM dependence demonstrate that O2˙̄ generated from BH4-depleted eNOS is derived from the heme center.
FIGURE 1.
O2˙̄ generated from uncoupled eNOS induces eNOS protein radical formation. a, EPR spin trapping of O2˙̄ generated by eNOS. The reaction system contained 50 mm Tris-HCl buffer, pH 7.4, 1 mm NADPH, 0.5 mm Ca2+, 10 μg/ml CaM, 15 μg/ml purified eNOS, and 25 mm spin trap DEPMPO. The spectra were recorded at room temperature with a microwave frequency of 9.863 GHz, 20 milliwatts of microwave power, and 1.0 G modulation amplitude. b, immunoblotting analysis of protein radical formation. Lanes 1–6, O2˙̄ is generated from BH4-free eNOS. Lane 1, BH4-free eNOS; lane 2, +10 μm Arg; lane 3, +1 mm Arg; lane 4, +1 mm l-NAME; lane 5, +Mn-SOD; lane 6, +10 μm BH4. Lanes 7 and 8, O2˙̄ is generated from nNOS reductase domain. Lane 7, BH4-free eNOS + nNOS reductase domain (nNOSR); lane 8, +Mn-SOD.
Characterization of eNOS Protein Radicals Using Uncoupled Enzyme or an Exogenous O2˙̄ Source
We have previously identified the site of protein radical formation in NADH dehydrogenase induced by O2˙̄ using immunospin trapping (30). In this study, BH4-free eNOS was used, as noted above, to self-generate O2˙̄, or a purified nNOS reductase domain was used to provide an exogenous O2˙̄ source. This O2˙̄ in turn can modify labile functional groups of eNOS to form protein radicals. We trapped these short-lived protein radicals with the spin trap DMPO. Immunoblotting with anti-DMPO antibody demonstrated that BH4-free eNOS formed the protein-centered radicals, by self-generated O2˙̄ or exogenous O2˙̄, that were abolished by Mn-SOD (Fig. 1b). The addition of the NOS substrate, l-Arg (10 μm and 1 mm) increased eNOS-mediated O2˙̄ generation (Fig. 1a) and also increased protein radical formation. Protein radical formation from BH4-free eNOS was also largely inhibited by the NOS-specific inhibitor, l-NAME (1 mm). Following the addition of BH4, which abolishes eNOS-mediated O2˙̄ generation, no eNOS-derived protein radical was observed. These results demonstrate that O2˙̄, generated from either eNOS or from an exogenous source, oxidatively modifies eNOS.
Identification of the eNOS Protein Radical by Mass Spectrometry
To begin to understand how the O2˙̄-induced eNOS protein radical formation could affect eNOS function, it is important to determine the precise sites of the DMPO-trapped eNOS protein radicals. Purified BH4-free eNOS was incubated with NADPH (1 mm) and 50 mm DMPO at room temperature for 1 h. This reaction mixture was then subjected to SDS-PAGE separation. The band corresponding to the DMPO-trapped eNOS protein radical adduct was cut and digested with trypsin, chymotrypsin, or both, and then the digested peptide fragments were determined by LC-MS/MS as described under “Experimental Procedures.”
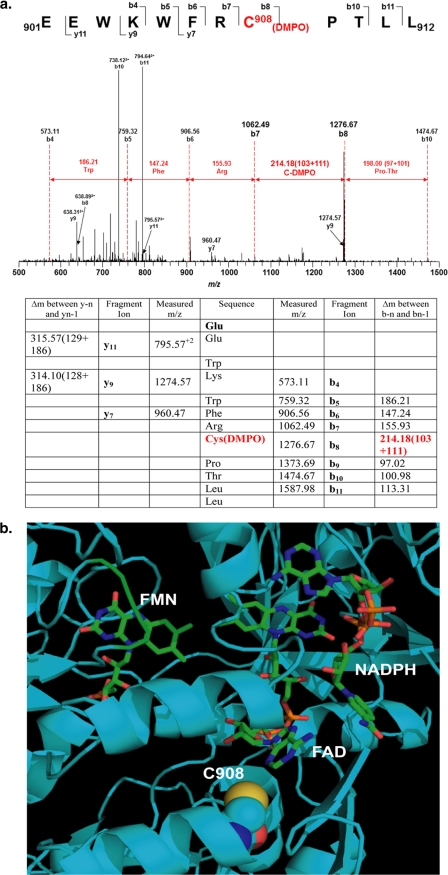
With the addition of one molecule of DMPO, the molecular mass of the DMPO-modified peptide fragment will increase by 111 Da compared with the nonmodified peptide fragment. The peptides with a mass difference of 111 Da were identified by LC/MS, and the sequence of the modified peptides was determined by MS/MS. From the mass determination, one specific modified cysteine residue, Cys-908, was identified in the tryptic/chymotryptic fragment EEWKWFRC*PTLL (amino acids 901–912; Fig. 2a).
FIGURE 2.
Mass spectrometry and molecular modeling reveal that Cys-908 is the site for eNOS protein radical formation. a, LC-MS/MS analysis of eNOS protein radical formation. Protein radical formation was demonstrated to be on Cys-908 using DMPO as a trap. The molecular mass difference between fragment ions b7 and b8 demonstrated a mass shift of 111 Da compared with the native fragment ions; this allowed unequivocal assignment of the DMPO adduct to Cys-908. b, molecular modeling of human eNOS reductase domain. The three-dimensional structure of human eNOS reductase domain was generated using the reductase domain of rat neuronal NOS (Protein Data Bank code 1F20) by Swiss molecular modeling. Cys-908 (C908) is located at the interface of FAD and FMN domain. This residue is conserved throughout all mammalian species. Structural perturbation of this domain is expected when this residue is mutated to Ala allowing oxygen to access this pocket.
Molecular Modeling of Human eNOS Reductase Domain
From the three-dimensional structure of human eNOS reductase domain, Cys-908 is located at the interface of FAD and FMN domain (Fig. 2b). This residue is conserved throughout all mammalian species. Structural perturbation on this interface will be expected when this residue is mutated to Ala, but not to Ser, allowing oxygen to access this pocket.
Mutagenesis of eNOS Cys-908 to Ala Resists Thiyl Radical Formation and Uncouples eNOS
Site-directed mutagenesis of eNOS Cys-908 to Ala was used to further confirm this assignment and to determine the importance of Cys-908 on the eNOS function and redox regulation. C908A mutant totally abolished eNOS protein radical formation in BH4-free eNOS as determined by immunoblotting (Fig. 3a). The NO production from WT and C908A eNOS was directly measured by EPR spin trapping using Fe-MGD. The mutation of this Cys residue greatly decreased measured NO production from the enzyme with more than a 10-fold decrease compared with WT eNOS (Fig. 3b). The addition of Mn-SOD (200 units/ml) significantly increased the measured NO, indicating that the C908A mutation alters the function of eNOS to produce O2˙̄, in lieu of NO, which in turn scavenges and decreases the trapped NO. With the mutation to Ala, because of a change in size and polarity of this residue, the perturbation at the FAD/FMN interface is expected, and this can lead to a decrease in NO production and electron leakage. Previously, it has been demonstrated that site-directed mutagenesis of electronegative residues at the FMN module of neuronal NOS affects electron transfer and NOS catalytic activity (38). This further supports our results of Cys-908 playing an important role in electron transfer in the reductase domain, as well as regulating eNOS activity.
FIGURE 3.
C908A mutation of eNOS prohibits thiyl radical formation and uncouples eNOS. a, immunoblotting of WT eNOS and eNOS C908A to measure protein radical formation. The upper panel is eNOS protein radical formation trapped by DMPO, when eNOS is uncoupled, and immunoblotting against anti-DMPO antibody. Lane 1 is protein radical formation from WT eNOS and trapped by DMPO. Lane 2 is the same as in lane 1 with the addition of Mn-SOD. Lane 3, as in lane 1, eNOS C908A and trapped DMPO. Lane 4 is same as in lane 3 with the addition of Mn-SOD. The lower panel shows an immunoblot for eNOS. b, EPR Fe-MGD NOS activity determination. The top spectrum is the NO generated from WT eNOS. The middle spectrum is the NO generated from eNOS C908A. The bottom spectrum is the NO generated from eNOS C908A plus Mn-SOD. c, determination of O2˙̄ generation from WT eNOS and eNOS C908A by DEPMPO EPR spin-trapping. The left panel is the O2˙̄-DEPMPO EPR spectra of WT eNOS. The right panel is the O2˙̄-DEPMPO EPR spectra of eNOS C908A. All of the experiments were performed at least in triplicate.
To further study the role of Cys-908 in eNOS function, O2˙̄ generation from WT and C908A mutant was measured in the presence of specific NOS inhibitors. Under BH4-free conditions eNOS-derived O2˙̄ is inhibited by l-NAME and imidazole and is sensitive to Ca2+/CaM (Fig. 3c). However, O2˙̄ generated from the C908A mutant is not inhibited by either l-NAME or imidazole and is not Ca2+/CaM-dependent (Fig. 3c). This demonstrates that the O2˙̄ generated from the C908A eNOS is not from the heme center but rather derived from the reductase domain.
Mutagenesis of eNOS Cys-908 to Ala Affects Reductase Activity of eNOS and FAD/FMN Binding
Cytochrome c reductase activity, which is a useful predictor of changes in the equilibrium between FAD and FMN of NOS (37), was used to determine the effect of mutation of Cys-908 to Ala on the reductase activity of eNOS. The reductase activity of WT eNOS is 473.8 ± 13.4 nmol/mg/min at room temperature. The reductase activity of C908A eNOS is 198.9 ± 12.5 nmol/mg/min. The content of FAD/FMN of eNOS was measured to determine the effect of Cys mutation on the binding of FAD/FMN to eNOS. The ratio of FAD/FMN was determined using HPLC. The ratio of FAD/FMN for WT eNOS is 0.98 ± 0.02. The ratio of FAD/FMN for C908A eNOS is 0.80 ± 0.02. The results are expressed as the means ± S.E. (n = 3).
O2˙̄ Generated from Uncoupled eNOS Induces S-Glutathionylation and Disulfide Bond Formation through Protein Radical Intermediates
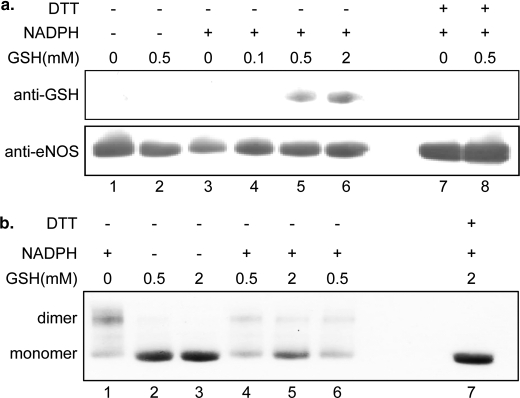
BH4-free eNOS was used to self-generate O2˙̄. This O2˙̄ in turn reacted with eNOS thiols to form protein thiyl radical intermediates. In the presence of GSH (0.1, 0.5, and 2 mm), this eNOS thiyl radical can react with GSH to generate S-glutathionylated eNOS (Fig. 4a). This protein S-glutathionylation is reversible when it is treated with 1 mm DTT. In the absence of GSH, this eNOS thiyl radical can react with a vicinal free thiol to form interdisulfide bonds. Under nonreducing eNOS immunoblotting analysis, the formation of eNOS dimer is increased when eNOS is uncoupled (Fig. 4b). This eNOS interdisulfide bond formation can be prevented with the addition of GSH and is reversed with 1 mm DTT.
FIGURE 4.
Mechanistic study of eNOS S-glutathionylation and interdisulfide bond formation. a, O2˙̄ generated from BH4-free eNOS induces protein S-glutathionylation formation. The upper panel is eNOS S-glutathionylation formation through eNOS protein thiyl radical in the presence of GSH, when eNOS is uncoupled, and immunoblotting against anti-GSH antibody. Lane 1 is eNOS only. Lane 2 is eNOS with 0.5 mm GSH. Lane 3 is the uncoupled eNOS. Lane 4 is the uncoupled eNOS with 0.1 mm GSH (20 min). Lane 5 is the uncoupled eNOS with 0.5 mm GSH (20 min). Lane 6 is the uncoupled eNOS with 2 mm GSH (20 min). Lanes 7 and 8 are samples from lanes 3 and 5, respectively, treated with 1 mm DTT. The lower panel shows immunoblotting for eNOS. b, O2˙̄ generated from BH4-free eNOS induces eNOS interdisulfide formation. Immunoblotting against anti-eNOS antibody is used to determine eNOS monomer and dimer under the nonreducing SDS-PAGE separation. Lane 1 is the uncoupled eNOS. Lanes 2 and 3 are eNOS with 0.5 or 2 mm GSH, respectively. Lane 4 is the uncoupled eNOS with 0.5 mm GSH (20 min). Lane 5 is the uncoupled eNOS with 2 mm GSH (20 min). Lane 6 is the uncoupled eNOS with 0.5 mm (60 min). Lane 7 is sample from lane 5 treated with 1 mm DTT.
Glutathione and DMPO Compete for eNOS Protein Radical
An increase in O2˙̄ production has been shown to increase eNOS DMPO adduct formation (see the prior section). We also show that the formation of eNOS protein radical can further react with GSH to form eNOS S-glutathionylation. To examine whether the eNOS protein radical trapped by DMPO, also contributes to eNOS S-glutathionylation, GSH competition for eNOS DMPO protein radical formation or DMPO competition for O2˙̄-induced eNOS S-glutathionylation experiments were performed. GSH (1–10 mm) dose-dependently inhibits eNOS DMPO protein radical formation (Fig. 5a). No eNOS DMPO protein radical is seen when concentration of GSH is 5 mm. DMPO (20–100 mm) also dose-dependently inhibits O2˙̄-induced eNOS S-glutathionylation in the presence of 2 mm GSH (Fig. 5b). Thus, this suggests that the mechanism of formation of eNOS S-glutathionylation at Cys-908 is from eNOS thiyl radical intermediates that are induced by O2˙̄.
FIGURE 5.
Glutathione and DMPO compete for O2˙̄-induced eNOS protein radical. a, GSH dose-dependently inhibits O2˙̄-induced eNOS DMPO adduct. The upper panel is a blot against anti-DMPO antibody. The lower panel is a blot against anti-eNOS antibody. Lane 1, BH4-free eNOS in the presence of all NOS cofactors and 50 mm DMPO, and the reaction is initiated with 1 mm NADPH for 30 min at room temperature. Lane 2, the reaction is the same as in lane 1 with the addition of 1 mm GSH. Lane 3, with addition of 5 mm GSH. Lane 4, with addition of 10 mm GSH. b, DMPO dose-dependently inhibits O2˙̄-induced eNOS S-glutathionylation. The upper panel is a blot against anti-GSH antibody. The lower panel is a blot against anti-eNOS antibody. Lane 1, BH4-free eNOS in the presence of all NOS cofactors and 2 mm GSH, with the reaction initiated by 1 mm NADPH for 30 min at room temperature. Lane 2, the reaction is the same as in lane 1 with the addition of 20 mm DMPO. Lane 3, with the addition of 50 mm DMPO. Lane 4, with the addition of 100 mm DMPO.
Formation and Trapping of eNOS Protein Radicals in Endothelial Cells under Oxidative Stress
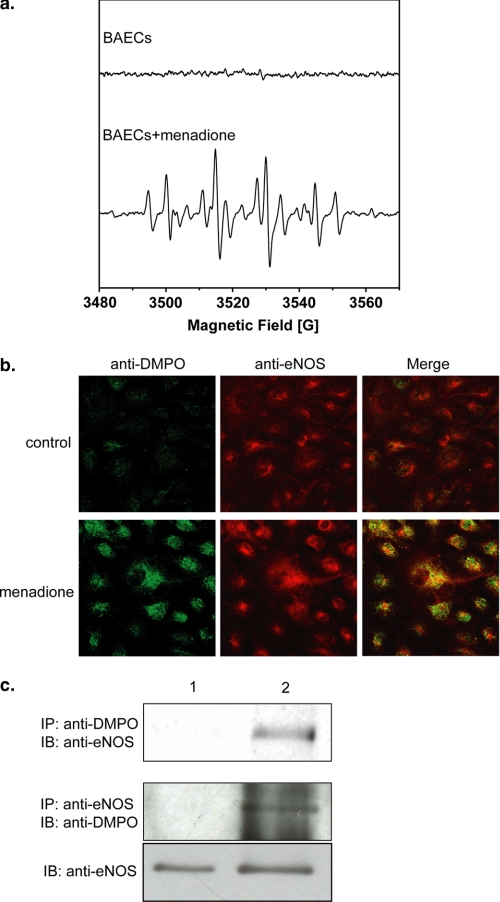
Experiments were performed to determine whether eNOS protein radicals occur in endothelial cells under oxidative stress. DMPO EPR spin-trapping was first used to determine the extent of O2˙̄ generation from BAECs induced by menadione, which uncouples flavin-containing reductases (37). Menadione-treated BAECs exhibited a prominent DMPO radical adduct signal composed mostly of DMPO-OH with a smaller DMPO-R EPR signal (Fig. 6a) (39–41). As previously reported, this indicates that menadione activates O2˙̄-derived radical generation (37). In BAECs treated with menadione, confocal microscopy with anti-DMPO and anti-eNOS antibodies demonstrated a marked increase in cellular DMPO protein radicals that colocalized with eNOS (Fig. 6b). Immunoprecipitation with anti-eNOS and anti-DMPO followed by immunoblotting with anti-DMPO or anti-eNOS antibodies confirmed that eNOS protein radical formation occurred in these cells (Fig. 6c).
FIGURE 6.
In vivo protein radical formation from endothelium under oxidative stress. a, DMPO EPR spin-trapping of O2˙̄ generated from BAECs induced by 10 μm menadione. The upper spectrum is the EPR signal of control BAECs with 50 mm DMPO spin trap. The lower spectrum is the EPR signal of BAECs with 50 mm DMPO spin trap activated by the addition of 10 μm menadione. b, immunomicroscopic detection of eNOS protein radical formation in endothelium using anti-DMPO antibody (green) and anti-eNOS antibody (red). The upper panels are control BAECs with the addition of DMPO trap, demonstrating no detectable protein radical formation. The lower panels are BAECs treated with 10 μm menadione with the addition of the DMPO trap, demonstrating marked protein radical formation (green) that colocalized with eNOS (red). c, immunoprecipitation of DMPO protein radical adducts from BAECs. Lane 1, control BAECs; lane 2, BAECs are treated with menadione. The upper panel is the immunoprecipitation (IP) with anti-DMPO antibody followed by immunoblotting (IB) against anti-eNOS. The lower panels are the immunoprecipitation with anti-eNOS antibody followed immunoblotting with anti-DMPO antibody. All of the experiments were performed at least in triplicate.
DISCUSSION
Oxidative post-translational modifications of many enzymes, such as S-glutathionylation, nitration, or nitrosylation, have been implicated in signal transduction (42–48). Recently, we have demonstrated that eNOS can be S-glutathionylated under oxidative stress in vivo and in vitro, leading to an uncoupling of the enzyme (13). Indeed, several reports have also shown that protein S-glutathionylation plays an important role in redox signaling and can be protective against irreversible oxidation of the protein thiols in cardiovascular diseases (17, 49). As such, it is important to understand the mechanism of this redox-directed enzyme regulation, opening another potential mode of therapeutic intervention to treat cardiovascular or other oxidant-derived diseases.
Under oxidative stress or disease, O2˙̄ generated from eNOS is expected to increase with the reversible depletion of cellular BH4 (4, 16, 50). The increased O2˙̄ will in turn react with functional groups of proteins, including eNOS, with the formation of labile intermediate protein radicals, leading to protein oxidation or oxidative modification. Irreversible oxidation is the primary cause of protein damage under oxidative stress leading to cell death; in contrast, reversible oxidation, such as the formation of disulfide or mixed disulfide bonds, can protect cells from oxidative damage and act as a redox sensor.
In the current report, we demonstrate that eNOS can be oxidized by eNOS-derived O2˙̄ or an exogenous O2˙̄ source leading to protein-centered radical formation. This eNOS protein radical formation is dependent upon NOS uncoupling, substrate availability, or an exogenous O2˙̄ source, whereas antioxidant enzymes, such as SOD, can protect eNOS from this thiol oxidation. As such, identification of the precise site of eNOS protein radical formation provides key insight regarding how oxidative modification of this enzyme is involved in redox regulation.
Using mass spectrometric analysis, we identified eNOS Cys-908 as the site of protein radical formation. The formation of the thiyl radical is one of the primary mechanisms for thiol oxidation. Thiyl radical formation is a one-electron oxidation, which in turn can react with a vicinal protein thiol or GSH to form a disulfide bond or a mixed disulfide bond (i.e. S-glutathionylation) (45, 46). Protein S-glutathionylation occurs through the reversible formation of a mixed disulfide bond between protein cysteine thiols and GSH, the most abundant low molecular mass thiol in cells. The identification of Cys-908 as the site of protein radical formation provides direct mechanistic evidence for an oxidant-induced process of eNOS regulation that can lead to S-glutathionylation (13).
There are several proposed mechanisms of S-glutathionylation (45, 46). Thiol-disulfide exchange with oxidized glutathione (GSSG) is one of the mechanisms for S-glutathionylation. In addition, reactive oxygen species and reactive nitrogen species-derived thiyl radicals can in turn react with GSH and protein thiols to generate protein S-glutathionylation. It has been suggested that thiol-disulfide exchange with GSSG can only occur when cellular GSSG/GSH ratio is high (42, 43, 46). It is believed that formation of thiol intermediates, such as the thiyl radical, sulfenic acid, or S-nitrosyl, is a more rapid and efficient mechanism for protein S-glutathionylation in vivo and that these mechanisms could play a role in signal transduction.
In our current study, we demonstrate that eNOS thiols can be oxidized by O2˙̄ generated from uncoupled eNOS, an exogenous source, or cellular oxidative stress through the formation of a thiyl radical in vitro or in vivo. The specific site of protein radical formation at Cys-908 provides a novel mechanism of eNOS thiol modification that can lead to further redox modifications, such as S-glutathionylation, which has been reported to regulate eNOS function in response to oxidative stress (13, 51, 52). We demonstrate that protein thiyl radicals generated from uncoupled eNOS do indeed react with reduced glutathione to form eNOS S-glutathionylation. Thus, the formation of an eNOS protein radical on Cys-908 provides a post-translational mechanism that can regulate eNOS function through oxidative modification to form eNOS S-glutathionylation or disulfide bond formation as demonstrated in this study. Furthermore, an increase in GSH can inhibit eNOS DMPO radical adduct formation, whereas increasing DMPO concentrations inhibit eNOS S-glutathionylation, suggesting that Cys-908 is the residue for eNOS thiyl radical formation, which in turn can react with GSH to form S-glutathionylated eNOS.
Site-directed mutagenesis of eNOS Cys-908 to Ala confirmed that Cys-908 is of critical importance for the regulation of eNOS function in response to oxidative stress. Mutagenesis of this residue demonstrated that Cys-908 is the primary site for the O2˙̄-induced formation of the eNOS protein radical in response to oxidative stress. Moreover, this mutation greatly decreases the NOS activity and cytochrome c reductase activity of the C908A. The cytochrome c reductase activity of NOS has been used to measure electron efflux from the reductase domain of NOS and is a useful predictor of changes in the equilibrium between FAD and FMN of NOS (53). Thus, the decrease in the cytochrome c reductase activity of C908A mutant suggests that this residue is involved in regulating electron transfer in the reductase domain of eNOS by altering the electron transfer between FAD and FMN, leading to possible electron leakage from this site and a decrease in NO production from the enzyme. In our previous report (13), when Cys-908 was mutated to Ser, a conservative mutation, there was no effect on NO production from the enzyme. However, when mutated to Ala, there is a change in the size and polarity of this residue, the perturbation at the FAD/FMN interface is expected, and this can lead to a decrease in NO production and electron leakage from the site.
O2˙̄ generated from the C908A mutant is not inhibited by either l-NAME, a NOS competitive inhibitor, or imidazole, a heme-binding ligand, and is also independent of Ca2+/CaM, which is necessary for transfer of electrons from the flavins in the reductase domain to the heme center. The mutation of Cys-908 to Ala slightly alters the binding of FAD and FMN to the enzyme. Moreover, from the predicted three-dimensional structure of the eNOS reductase domain, we found that Cys-908 is located in the eNOS reductase domain near the FAD-binding site (Fig. 2a). As such, we hypothesize that mutagenesis of eNOS Cys-908 to alanine could alter the structure of this FAD and FMN interface and increase oxygen accessibility to the flavins contributing to electron leakage from this domain leading to the observed increase in O2˙̄ generation and a decrease in electron transfer from the reductase domain to the heme center decreasing NO production. Thus, the observed O2˙̄-induced eNOS thiol oxidation at Cys-908 and subsequent S-glutathionylation could play an important role in regulating eNOS function by allowing for the regulation of electron flow from the reductase domain to the oxygenase domain, and given the known reactions of protein thiol-radicals, this residue could be involved in a redox-signaling based regulation of eNOS and, ultimately, cardiovascular function.
The oxidative stress produced during ischemia/reperfusion injury has been found to lead to the S-glutathionylation of several important proteins, such as aconitase and triose phosphate isomerase (54). eNOS is of critical importance in maintaining proper cardiovascular function. The detailed understanding of how oxidants alter its structure/function with increasing oxidative stress as occurs during ischemia/reperfusion will facilitate the development of strategies to prevent these injuries and alleviate the underlying processes of oxidant induced disease. We have demonstrated that O2˙̄ generation in endothelium leads to increases in eNOS protein radical formation, and this observation is consistent with our results on purified eNOS, which demonstrated eNOS protein radical formation via self-generated O2˙̄ or an exogenous O2˙̄-generating source. Thus, in vivo, eNOS cysteine residues are potential targets for both reversible and irreversible oxidative modifications and are potentially involved in redox signaling during oxidative stress.
In conclusion, the increase in O2˙̄ generation found under oxidative stress can induce eNOS protein radical formation contributing to oxidative modification. The formation of the eNOS protein radical is dependent on the decreased availability of cofactors, and substrates of eNOS, or the presence of an exogenous O2˙̄ source. These eNOS protein radical intermediates can in turn react with vicinal protein thiol or GSH to form a disulfide bond or S-glutathionylation. The identification of Cys-908 as a primary target for radical formation provides a novel mechanism by which eNOS is regulated in response to oxidative stress. This site-specific thiol redox modification can modulate cardiovascular function as demonstrated here and in our prior study (13) by altering eNOS function switching the enzyme from NO production to O2˙̄ generation. Thus, the molecular mechanism of eNOS thiol oxidation delineated in this work serves as a critical step toward understanding the redox regulation of eNOS and endothelial-mediated vascular function as well as the alterations that occur in diseases associated with oxidative stress.
Acknowledgments
We thank Dr. Liwen Zhang and Dr. Kari Green-Church at Ohio State University Campus Chemical Instrument Center proteomics center for support with mass spectrometric analysis. We thank Dr. Dennis J. Stuehr (Cleveland Clinic) for providing us with rat nNOS reductase domain.
This work was supported, in whole or in part, by National Institutes of Health Grants HL63744, HL65608, and HL38324 (to J. L. Z.); HL081734 (to L. J. D.); HL83237 (to Y.-R. C.); and K99HL103846 (to C.-A. C.).
- BH4
- tetrahydrobiopterin
- BAEC
- bovine aortic endothelial cell
- CaM
- calmodulin
- DEPMPO
- 5-diethoxyphosphoryl-5-methyl-1-pyrroline N-oxide
- MGD
- N-methyl-d-glucamine dithiocarbamate
- l-NAME
- l-NG-nitroarginine methyl ester hydrochloride
- eNOS
- endothelial NOS
- Mn-SOD
- superoxide dismutase
- DMPO
- 5,5-dimethyl-1-pyrroline N-oxide.
REFERENCES
- 1. Sessa W. C. (2004) J. Cell Sci. 117, 2427–2429 [DOI] [PubMed] [Google Scholar]
- 2. Kone B. C., Kuncewicz T., Zhang W., Yu Z. Y. (2003) Am. J. Physiol. Renal Physiol. 285, F178–F190 [DOI] [PubMed] [Google Scholar]
- 3. Wang P., Zweier J. L. (1996) J. Biol. Chem. 271, 29223–29230 [DOI] [PubMed] [Google Scholar]
- 4. Dumitrescu C., Biondi R., Xia Y., Cardounel A. J., Druhan L. J., Ambrosio G., Zweier J. L. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 15081–15086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Xia Y., Tsai A. L., Berka V., Zweier J. L. (1998) J. Biol. Chem. 273, 25804–25808 [DOI] [PubMed] [Google Scholar]
- 6. Vásquez-Vivar J., Kalyanaraman B., Martásek P., Hogg N., Masters B. S., Karoui H., Tordo P., Pritchard K. A., Jr. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 9220–9225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kojda G., Harrison D. (1999) Cardiovasc. Res. 43, 562–571 [DOI] [PubMed] [Google Scholar]
- 8. Cave A. C., Brewer A. C., Narayanapanicker A., Ray R., Grieve D. J., Walker S., Shah A. M. (2006) Antioxid. Redox. Signal. 8, 691–728 [DOI] [PubMed] [Google Scholar]
- 9. Zweier J. L., Talukder M. A. (2006) Cardiovasc. Res. 70, 181–190 [DOI] [PubMed] [Google Scholar]
- 10. Förstermann U. (2006) Biol. Chem. 387, 1521–1533 [DOI] [PubMed] [Google Scholar]
- 11. Druhan L. J., Forbes S. P., Pope A. J., Chen C. A., Zweier J. L., Cardounel A. J. (2008) Biochemistry 47, 7256–7263 [DOI] [PubMed] [Google Scholar]
- 12. Pou S., Pou W. S., Bredt D. S., Snyder S. H., Rosen G. M. (1992) J. Biol. Chem. 267, 24173–24176 [PubMed] [Google Scholar]
- 13. Chen C. A., Wang T. Y., Varadharaj S., Reyes L. A., Hemann C., Talukder M. A., Chen Y. R., Druhan L. J., Zweier J. L. (2010) Nature 468, 1115–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Muscoli C., Cuzzocrea S., Riley D. P., Zweier J. L., Thiemermann C., Wang Z. Q., Salvemini D. (2003) Br. J. Pharmacol. 140, 445–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Stuehr D. J. (1999) Biochim. Biophys. Acta 1411, 217–230 [DOI] [PubMed] [Google Scholar]
- 16. Chen C. A., Druhan L. J., Varadharaj S., Chen Y. R., Zweier J. L. (2008) J. Biol. Chem. 283, 27038–27047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martínez-Ruiz A., Lamas S. (2007) Cardiovasc. Res. 75, 220–228 [DOI] [PubMed] [Google Scholar]
- 18. Parodi O., De Maria R., Roubina E. (2007) J. Cardiovasc. Med. 8, 765–774 [DOI] [PubMed] [Google Scholar]
- 19. Peluffo G., Radi R. (2007) Cardiovasc. Res. 75, 291–302 [DOI] [PubMed] [Google Scholar]
- 20. Hothersall J. S., Cunha F. Q., Neild G. H., Norohna-Dutra A. A. (1997) Biochem. J. 322, 477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Harbrecht B. G., Di Silvio M., Chough V., Kim Y. M., Simmons R. L., Billiar T. R. (1997) Ann. Surg. 225, 76–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cuzzocrea S., Zingarelli B., O'Connor M., Salzman A. L., Szabó C. (1998) Br. J. Pharmacol. 123, 525–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hofmann H., Schmidt H. H. (1995) Biochemistry 34, 13443–13452 [DOI] [PubMed] [Google Scholar]
- 24. Huang A., Xiao H., Samii J. M., Vita J. A., Keaney J. F., Jr. (2001) Am. J. Physiol. Cell Physiol. 281, C719–C725 [DOI] [PubMed] [Google Scholar]
- 25. Tummala M., Ryzhov V., Ravi K., Black S. M. (2008) DNA Cell Biol. 27, 25–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mason R. P. (2004) Free Radic. Biol. Med. 36, 1214–1223 [DOI] [PubMed] [Google Scholar]
- 27. Ilagan R. P., Tejero J., Aulak K. S., Ray S. S., Hemann C., Wang Z. Q., Gangoda M., Zweier J. L., Stuehr D. J. (2009) Biochemistry 48, 3864–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Martasek P., Liu Q., Liu J., Roman L. J., Gross S. S., Sessa W. C., Masters B. S. (1996) Biochem. Biophys. Res. Commun. 219, 359–365 [DOI] [PubMed] [Google Scholar]
- 29. Rodríguez-Crespo I., Ortiz de Montellano P. R. (1996) Arch. Biochem. Biophys. 336, 151–156 [DOI] [PubMed] [Google Scholar]
- 30. Chen Y. R., Chen C. L., Zhang L., Green-Church K. B., Zweier J. L. (2005) J. Biol. Chem. 280, 37339–37348 [DOI] [PubMed] [Google Scholar]
- 31. Gerber N. C., Ortiz de Montellano P. R. (1995) J. Biol. Chem. 270, 17791–17796 [DOI] [PubMed] [Google Scholar]
- 32. Xia Y., Cardounel A. J., Vanin A. F., Zweier J. L. (2000) Free Radic. Biol. Med. 29, 793–797 [DOI] [PubMed] [Google Scholar]
- 33. Vanin A. F., Liu X., Samouilov A., Stukan R. A., Zweier J. L. (2000) Biochim. Biophys. Acta 1474, 365–377 [DOI] [PubMed] [Google Scholar]
- 34. Abu-Soud H. M., Loftus M., Stuehr D. J. (1995) Biochemistry 34, 11167–11175 [DOI] [PubMed] [Google Scholar]
- 35. Gnanaiah W., Omdahl J. L. (1986) J. Biol. Chem. 261, 12649–12654 [PubMed] [Google Scholar]
- 36. Stuehr D. J., Cho H. J., Kwon N. S., Weise M. F., Nathan C. F. (1991) Proc. Natl. Acad. Sci. U.S.A. 88, 7773–7777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rosen G. M., Freeman B. A. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 7269–7273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Panda K., Haque M. M., Garcin-Hosfield E. D., Durra D., Getzoff E. D., Stuehr D. J. (2006) J. Biol. Chem. 281, 36819–36827 [DOI] [PubMed] [Google Scholar]
- 39. Zweier J. L. (1988) J. Biol. Chem. 263, 1353–1357 [PubMed] [Google Scholar]
- 40. Zweier J. L., Kuppusamy P., Williams R., Rayburn B. K., Smith D., Weisfeldt M. L., Flaherty J. T. (1989) J. Biol. Chem. 264, 18890–18895 [PubMed] [Google Scholar]
- 41. Buettner G. R. (1987) Free Radic. Biol. Med. 3, 259–303 [DOI] [PubMed] [Google Scholar]
- 42. Winterbourn C. C., Hampton M. B. (2008) Free Radic. Biol. Med. 45, 549–561 [DOI] [PubMed] [Google Scholar]
- 43. Gallogly M. M., Mieyal J. J. (2007) Curr. Opin. Pharmacol. 7, 381–391 [DOI] [PubMed] [Google Scholar]
- 44. Foster M. W., Hess D. T., Stamler J. S. (2009) Trends Mol. Med. 15, 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Adachi T., Schoneich C., Cohen R. A. (2005) Drug Discov. Today Dis. Mech. 2, 39–46 [Google Scholar]
- 46. Biswas S., Chida A. S., Rahman I. (2006) Biochem. Pharmacol. 71, 551–564 [DOI] [PubMed] [Google Scholar]
- 47. Turko I. V., Murad F. (2002) Pharmacol. Rev. 54, 619–634 [DOI] [PubMed] [Google Scholar]
- 48. Zhang H., Xu Y., Joseph J., Kalyanaraman B. (2008) Methods Enzymol. 440, 65–94 [DOI] [PubMed] [Google Scholar]
- 49. Mieyal J. J., Gallogly M. M., Qanungo S., Sabens E. A., Shelton M. D. (2008) Antioxid. Redox. Signal. 10, 1941–1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zweier J. L., Flaherty J. T., Weisfeldt M. L. (1987) Proc. Natl. Acad. Sci. U.S.A. 84, 1404–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Chen C. A., Druhan L. J., Wang T. Y., Chen Y. R., Zweier J. L. (2008) Circulation 118, S274 [Google Scholar]
- 52. Chen C. A., Varadharaj S., Reyes L., Talukder A. H., Chen Y. R., Druhan L. J., Zweier J. L. (2009) Circulation 120, S1070 [Google Scholar]
- 53. Stuehr D. J., Tejero J., Haque M. M. (2009) FEBS J. 276, 3959–3974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Eaton P., Byers H. L., Leeds N., Ward M. A., Shattock M. J. (2002) J. Biol. Chem. 277, 9806–9811 [DOI] [PubMed] [Google Scholar]