Abstract
Protein-arginine methyltransferases aid in the regulation of many biological processes by methylating specific arginyl groups within targeted proteins. The varied nature of the response to methylation is due in part to the diverse product specificity displayed by the protein-arginine methyltransferases. In addition to site location within a protein, biological response is also determined by the degree (mono-/dimethylation) and type of arginine dimethylation (asymmetric/symmetric). Here, we have identified two strictly conserved methionine residues in the PRMT1 active site that are not only important for activity but also control substrate specificity. Mutation of Met-155 or Met-48 results in a loss in activity and a change in distribution of mono- and dimethylated products. The altered substrate specificity of M155A and M48L mutants is also evidenced by automethylation. Investigation into the mechanistic basis of altered substrate recognition led us to consider each methyl transfer step separately. Single turnover experiments reveal that the rate of transfer of the second methyl group is much slower than transfer of the first methyl group in M48L, especially for arginine residues located in the center of the peptide substrate where turnover of the monomethylated species is negligible. Thus, altered product specificity in M48L originates from the differential effect of the mutation on the two rates. Characterization of the two active-site methionines provides the first insight into how the PRMT1 active site is engineered to control product specificity.
Keywords: Enzyme Mechanisms, Histone Methylation, Post-translational Modification, Protein Methylation, S-Adenosylmethionine, ADMA, PRMT, Arginine Methylation, Methyltransferase, Product Specificity
Introduction
Protein methylation is a significant post-translational modification in eukaryotic organisms. Protein arginine residues can be methylated on the guanidino nitrogens by protein-arginine methyltransferases (PRMTs),2 which use S-adenosyl-l-methionine (AdoMet) as a methyl group donor. This post-translational modification is important in a wide variety of fundamental biological processes, including transcription, RNA splicing, signal transduction, DNA repair, viral replication (reviewed in Ref. 1), and chromatin remodeling (2). In recent years, the significance of PRMTs in human diseases has been increasingly studied, especially in cardiovascular disease (3) and cancer (4). In all, PRMTs play a crucial role in many biological processes.
Although the biological importance of PRMTs has become well accepted, the current knowledge of the fundamental biochemistry of these enzymes is limited, due in part to the complexity of the system. So far, 11 PRMT isoforms have been identified. In mammalian cells, nine PRMTs catalyze monomethyl arginine (MMA) formation, and they can be categorized into two major types as follows: PRMT1, -2–4, -6, and -8 additionally catalyze asymmetric dimethyl arginine (ADMA) formation, demonstrating type I activity; and PRMT5, -7, and -9 catalyze symmetric dimethyl arginine (SDMA) formation, demonstrating type II activity (Fig. 1A). PRMT10 and -11 were identified as putative PRMT genes with no methylation activity shown as yet (5). As with other enzyme families that post-translationally modify protein substrates, accurate substrate recognition by the PRMTs is critical for the proper transmission of biochemical information. However, very little information is available to explain how or why any of the PRMT isoforms target their cognate protein substrates, and more so, what determines which arginine within a protein should be methylated, i.e. a consensus sequence has not been identified.
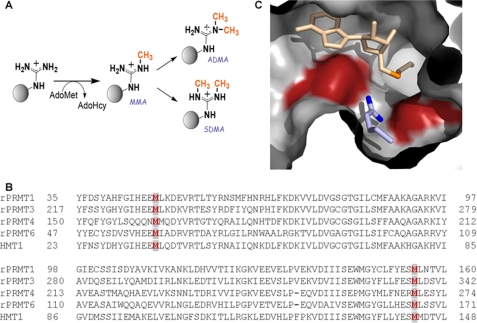
FIGURE 1.
A, methylation reactions catalyzed by PRMTs. Type I and type II PRMTs make MMA. Type I PRMTs may then go on to make ADMA, while type II PRMTs produce SDMA. The second molecules of AdoMet and AdoHcy have been omitted for clarity. B, sequence alignment for rat PRMT1, rat PRMT3, rat PRMT4, yeast RMT1/Hmt1, and rat PRMT6. The conserved methionine pair is highlighted in dark red in light gray boxes. C, rat PRMT1-AdoHcy-peptide substrate complex shows the positioning of two strictly conserved methionine residues. The active site of rat PRMT1 (Protein Data Bank code 1OR8) is shown with AdoHcy in light orange (sulfur in dark orange), methionines 155 (left) and 48 (right) in dark red, and the substrate arginine group of the R3 peptide (ac-GGRGGFGGRGGFGGRGGFGG) in blue (terminal guanidino nitrogens are shown in dark blue).
Adding to the complexity of the PRMT field, different methylation statuses (MMA, ADMA, or SDMA state) of the same substrate can lead to distinct biological outputs. One of the most striking examples is Arg-3 of histone H4 (H4R3), which can be either asymmetrically dimethylated by PRMT1 or symmetrically dimethylated by PRMT5, resulting in antagonistic effects on gene regulation (reviewed in Ref. 6). Moreover, the two methylation products of type I PRMTs, MMA and ADMA, can also have distinct biological functions. In yeast, MMA present in histone H3 at Arg-2 correlates to active transcription, whereas the ADMA state contributes to transcriptional repression (7). In contrast to the idea that MMA is simply an intermediate for ADMA generation, the current data point to a bona fide signaling role for MMA residues in much the same way that mono-, di-, and trimethylation of lysine residues differentially affect signaling (8, 9). A key to understanding the biological function of the PRMTs is to understand how product specificity may be regulated, in terms of governing which arginyl residues are modified and which state of methylation is achieved.
To gain mechanistic insight regarding the product specificity of the PRMTs, we have initiated mutational studies of rat PRMT1, a ubiquitously expressed member of the PRMT family that has been estimated to perform ∼85% of arginine methylations in mammals (10). Structural studies of PRMT1 (11) and PRMT3 (12), both type I methyltransferases, have identified the component residues of the type I active site and have provided a basis from which to probe product specificity. In particular, it has been noted that all the type I PRMTs contain an active site methionine (position 155 in rat PRMT1), which has been hypothesized to dictate the synthesis of ADMA over SDMA by preventing the binding of MMA in a configuration conducive for SDMA formation (12, 13). The idea that a single residue can alter product specificity is reminiscent of the Phe/Tyr “switch” model that is used to explain product specificity in lysine methyltransferases (13–15). Interestingly, we noted a second active site methionine (position 48 in rat) that is also strictly conserved in type I methyltransferases (Fig. 1B). Together, these two methionines are observed to sandwich the incoming substrate arginyl group in the type I methyltransferase active site (Fig. 1C) and thus be positioned to affect the type and degree of methylation. Here, we report that these two active site methionine residues are essential for catalytic activity and product specificity of PRMT1. Our studies show that substitution of Met-48 does not promote the formation of SDMA, ruling out a methionine switch model. Our data are consistent with Met-48 functioning differentially in the methylation of arginine to MMA versus the methylation of MMA to ADMA. Surprisingly, our data also show that Met-48 plays a crucial role in specifying which peptidyl arginine is targeted by PRMT1.
EXPERIMENTAL PROCEDURES
Materials
AdoMet was purchased from Sigma as a chloride salt (≥80%, from yeast). [methyl-3H]AdoMet was purchased from PerkinElmer Life Sciences. All the peptides were synthesized by the Keck Institute and purified to ≥95%. ZipTip®C4/C18 pipette tips were purchased from Millipore. His-hnRNP K was expressed and purified according to Ref. 16.
Expression and Purification of Mutant PRMT1 Proteins
PRMT1 mutant proteins were generated using the QuikChange® site-directed mutagenesis kit (Stratagene) with sets of complementary oligonucleotide primers spanning the desired site of mutation. For each PCR, the pET28b vector (Novagen) containing the gene that codes for N-terminal histidine-tagged rat WT-PRMT1 plasmid (pET28b-PRMT1) (17) was used as a template. Desired mutations (M155A, M48A, M48L, M48F, M48Y, and M48W) were confirmed through DNA sequencing. Mutant proteins were purified using the same methods used to express and purify wild-type His-PRMT1 (described in Ref. 17). Purified proteins were ≥95% pure by SDS-PAGE. Mutant protein sequences were verified using mass spectrometry.
Reverse Phase-HPLC Analysis of Methylated Amino Acids
Assays containing 4 μm WT or mutant PRMT1 proteins, 800 μm AdoMet, 10 nm AdoHcy nucleosidase (MTAN, purified as in Ref. 18), and 50 mm sodium phosphate buffer (pH 7.5) were equilibrated at 37 °C for 3 min. Reactions were initiated with 200 μm R3 peptide and were terminated after 3 h with 10% (v/v, final concentration) trichloroacetic acid (TCA). TCA-precipitated protein was removed through centrifugation, and the supernatant (containing the peptide) was added to a glass vial. An equivalent volume of 12 m HCl was added to each vial. Vials were sealed and heated to 110 °C for ∼24 h for a complete acid hydrolysis. Hydrolyzed amino acids from WT-PRMT1 and the mutant-catalyzed reactions were separated using o-phthaldialdehyde derivatization (19) with a Gemini® 3-μm C18 110-Å LC column 75 × 4.6 mm (Phenomenex). Mobile phase A consisted of 40 mm sodium phosphate buffer, pH 7.8, and mobile phase B was acetonitrile/methanol/H2O (45:45:10, v/v). The HPLC gradient conditions are shown in supplemental Table S1. To verify the presence and peak times of the methylated arginine products, sample reactions were spiked with 2.6 μm [3H]AdoMet (specific activity of 2.02 mCi/μmol). Fractions (83.3 μl) were collected, and radioactivity was counted in 5 ml of scintillation mixture (Fisher). MMA, ADMA, or SDMA standard amino acids were used to verify the identity of the methylated products generated. The detection limit for this method is ∼10 pmol of methylated arginine in a 20-μl sample.
Continuous Spectrophotometric Kinetic Assays of PRMT1 Mutants
A continuous spectrophotometric assay for AdoMet-dependent methyltransferases (17) was used to assay PRMT1 mutants with arginine-containing peptides. Briefly, two coupling enzymes, MTAN and adenine deaminase, were used to hydrolyze and deaminate the AdoHcy generated from methyl group transfer, respectively. This assay avoids any product inhibition that could occur from AdoHcy. Initial rate data representing no more than 10% of product formation were fit to the Michaelis-Menten equation (20) to obtain Km, app and kcat, app values. Each reaction was performed at least in duplicate. The limit of detection for this assay was 0.1 μm CH3/min (which corresponds to a kcat of 0.025 min−1).
Discontinuous Kinetic Assays of PRMT1 Mutants
Another discontinuous but more sensitive assay with ZipTip®C4/C18 was used in testing the enzymatic activity under steady-state conditions (21). Unless noted otherwise in the text, enzyme catalytic activity was tested with 100 nm WT-PRMT1 or mutants, 1 μm AdoMet, and 1 μm [3H]AdoMet, initiated by 200 μm peptide substrates or 1.7 μm hnRNP K protein substrate at 37 °C. At different time points, samples were removed from reactions and processed with ZipTip®C4/C18 pipette tips (for protein or peptide substrates, respectively) to separate the unreacted [3H]AdoMet and the radiolabeled product.
Dissociation Constant Measurement by Intrinsic Fluorescence Quenching
An RF-5301PC spectrofluorophotometer (Shimadzu) was used for fluorescence measurements. For R3 peptide and AdoMet affinity determinations, an excitation wavelength of 290 nm was used, and emission spectra from 300 to 420 nm were collected. The change in fluorescence intensity at the maximum emission (333 nm) was monitored. The excitation and emission slit was 5 nm, and the scan speed was 100 nm/min using 1325 μl containing 1.4 μm PRMT1 in 150 mm sodium phosphate buffer, pH 7.1. Increasing concentrations from 1 to 50 μm peptide ligand (or AdoMet) were added at 2–3-min intervals. Data from at least two titrations were averaged and analyzed using the modified Stern Volmer (22) plots. Data were evaluated by nonlinear regression analysis using SigmaPlot to obtain the dissociation constant (KD) using the following equation: Fc = F (10ϵcd/2), where Fc is the corrected fluorescence; ϵ is the extinction coefficient of AdoMet; c is the concentration of AdoMet, and d is the path length. Finitial/(Finitial − Fc) was then plotted against 1/[AdoMet], and the data were fit to a line where the yintercept = 1/fa, the slope = 1/fa·KQ, and the KQ = 1/KD.
Protein Crystallization
A truncated construct of M48L-PRMT1 was made that lacked the first 13 amino acids of the native sequence. Previous crystallographic studies reported better diffracting crystals using the truncated proteins (11). Histidine-tagged, truncated M48L PRMT1 was expressed and initially purified as described previously (17). In addition to immobilized metal chromatography, M48L was further purified by anion exchange (Mono Q) and gel filtration chromatography as described previously (11). Purified fractions were concentrated to 10–20 mg/ml and incubated with 600 μm AdoHcy and 1 mm peptide (acGGRmeGGFGGKGGFGGKW) for 10–30 min before placing in crystallization trays. Crystallization was performed using sitting drop vapor diffusion. Crystals were grown at room temperature (∼22 °C) in 0.1 m Tris, pH 9.0, and 1.62 m ammonium phosphate monobasic at a 1:1 protein/well drop ratio. Crystals were flash-frozen in a cryo solution containing 0.1 m Tris, pH 9.0, 1.62 m ammonium phosphate monobasic, 0.5 mm peptide, 300 μm AdoHcy, and 20% glycerol.
Data Collection, Structure Determination, and Refinement
Diffraction data were collected using a home source generator and detector (Rigaku RU-200/Raxis IV++). Data were indexed and processed using d*TREK in the program Crystal Clear (24). Molecular replacement was performed using Phaser (25) from the CCP4 suite (26, 27). The search model was wild-type rat PRMT1-AdoHcy-peptide ternary complex (Protein Data Bank entry 1OR8). PHENIX (28) was used to perform positional, b-factor, and TLS refinement. The TLS groups were generated by the TLS Motion Determination server (29). Coot (30) and MolProbity (31) were used for model building and structure validation. The model was refined to an R/Rfree of 20.1:24.5% with no Ramachandran outliers (supplemental Table S2). AdoHcy was observed bound to the active site (supplemental Fig. S1). No compelling electron density was observed for the peptide substrate. Density that could accommodate ∼3 covalently bonded atoms was observed in the active site near Glu-144. The density is inconsistent with solvent molecules included in the crystallization buffer and may correspond to a portion of the peptide substrate. However, because the density could not be confidently assigned, it was not included in the final model. Structure factors and coordinates have been deposited in the Protein Data Bank (Protein Data Bank code 3Q7E). Figures were generated with PyMOL (32).
Mass Spectrometry Peptide Methylation Analysis and de Novo Sequencing Study
A reaction with 4 μm PRMT1, 800 μm AdoMet, and 100 nm MTAN in 50 mm sodium phosphate buffer, pH 7.5, was initiated with 200 μm R3 peptide. At various time points, 10-μl aliquots were quenched with TFA (10% v/v final) and analyzed by LC/MS followed by MS/MS of the desired peaks. 4 μl of reaction solution was injected onto a reverse phase column (75 μm × 150 mm, BEH C18-nanoACQUITY column, Waters, Manchester, UK). Peptides with and without methylation were separated at 40 °C with a nonlinear gradient (supplemental Table S2). Solvent A was 0.1% formic acid in water, and solvent B was 0.1% formic acid in acetonitrile. Monomethylated peptides ([M + H]3+ = 575.9) were then analyzed using a tandem mass spectrometer (Synapt Q-Tof, Waters, Manchester, UK) to determine the position of monomethylation.
Single Turnover Experiments
Single turnover experiments were carried out by mixing a solution containing the preincubated complex of 20 μm wild-type or M48L-PRMT1, 20 μm AdoMet (along with 1 μm of [3H]AdoMet), and 10 nm MTAN in 50 mm sodium phosphate buffer, pH 7.5, at 23 °C for 3 min. Commercially available AdoMet was purified as described previously (33, 34) before use. Reactions were initiated with 200 μm peptide substrates. Radiolabel incorporation over time was measured using the discontinuous ZipTip® assay described previously. Briefly, 10-μl reaction samples of different time points were taken out and quenched by 6 m guanidine HCl solution and processed with ZipTip®C18 assay to quantitate the amount of methylated product (21). The resulting time course of [3H]AdoMet incorporation was fit into single exponential curve, y = a·(1 −exp(−b·x)), to determine the parameters a (maximum product concentration) and b (kchem).
RESULTS
Analysis of the Products Formed by M48A- and M48L-PRMT1
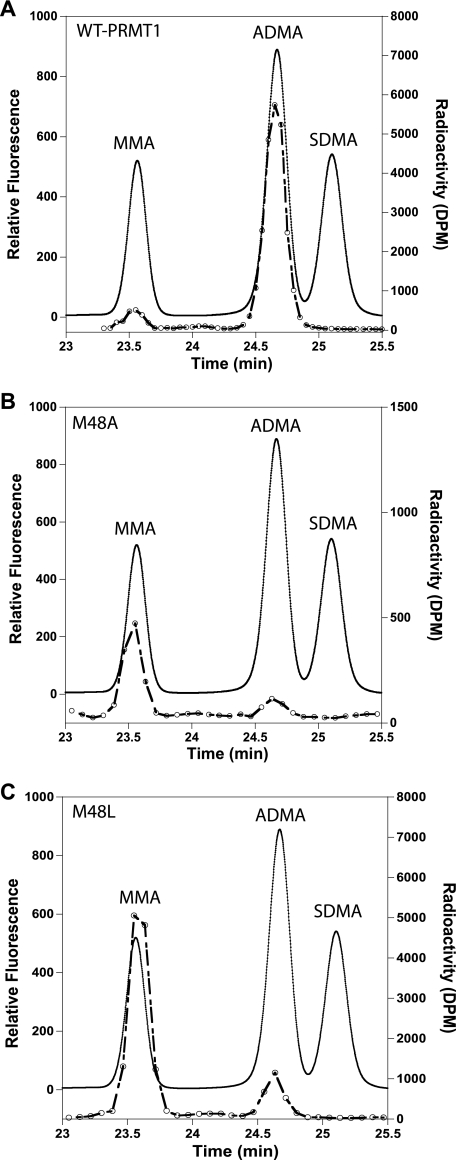
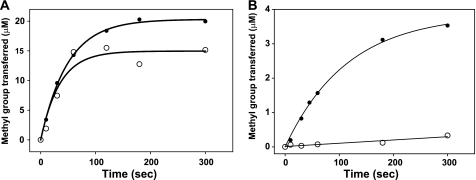
Previous analysis of the PRMT1 crystal structure suggested that the conserved Met at position 155 may be important for governing ADMA formation over SDMA formation by providing steric bulk in the active site and preventing free rotation of the MMA intermediate (12, 13). A closer examination of the PRMT1 structure identified Met-48, a similarly conserved active site residue located opposite Met-155. Given the potential for either methionine to influence substrate geometry in the active site, we investigated whether these residues are involved in specifying ADMA formation versus SDMA. Met-48 and Met-155 were individually mutated to alanines. We analyzed whether ADMA or SDMA was formed in the mutant-catalyzed reaction by amino acid analysis of the acid-hydrolyzed peptide products, using [3H]AdoMet as a tracer. M48A produced only MMA and ADMA, albeit with a higher MMA/ADMA ratio compared with WT-PRMT1 (Fig. 2, A and B). Because of the low activity of M48A observed in the 3-h reaction, Met-48 was further mutated to leucine to determine whether activity could be rescued by reintroducing more steric bulk. Like M48A, M48L also generated only MMA and ADMA, but in a much larger amount, with an even higher ratio of MMA/ADMA than that of M48A (Fig. 2C). No SDMA was detected. Similar results were observed with the M155A mutant, consistent with the recent report that Met-155 is not involved in SDMA formation (35). Standards of MMA, ADMA, SDMA, as well as other amino acids in the R3 peptide (Gly, Arg, and Phe) were analyzed by the same method to verify the methylated product (supplemental Fig. S2). Our results show that removing the steric bulk afforded by Met-155 or Met-48 in the PRMT1 active site was not enough to transform the type I PRMT into a type II PRMT. Thus, Met-48 or Met-155 do not, by themselves, dictate ADMA formation over SDMA formation.
FIGURE 2.
Analysis of Met-48 mutant-catalyzed methylation products by amino acid analysis. Products from reactions catalyzed by wild-type (A), M48A (B), or M48L-PRMT1 (C) were analyzed using reverse phase HPLC. [3H]AdoMet was used as a tracer to verify the presence of the methylated species in each reaction (open circles). MMA, ADMA, and SDMA standard amino acids were used to identify the methylated species in each sample (solid line).
Steady-state Methylation Activities of PRMT1 Mutants
The altered product ratios obtained for M48L and M48A in the previous experiments suggested that the activity of the mutants may be impaired. To determine whether Met-48 has an effect on the ability of PRMT1 to catalyze peptide methylation, the rate of methylation was measured with WT-PRMT1, M155A, and M48A. The activity was followed using an enzyme-coupled continuous spectrophotometric assay and the R3 peptide, a common peptide substrate used in arginine methylation (ac-GGRGGFGGRGGFGGRGGFGG) (Table 1) (17). The R3 peptide is a good substrate for WT-PRMT1 with a catalytic efficiency (kcat/Km) = 4460 m−1 s−1. When the two methionines in question were mutated to small amino acid residues, i.e. alanine, mutants still utilized R3 as a peptide substrate but at varying efficiencies (Table 1 and supplemental Fig. S3). Substitution of Ala for Met at position 155 resulted in a 90% decrease in kcat/Km. The M155A mutation did not affect turnover velocity, but Km was increased by a factor of 10 compared with WT-PRMT1. Substitution of Ala for Met-48 also decreased catalytic efficiency. However, an increase in Km was coupled with a 10-fold drop in kcat, leaving this mutant with just under 2% of WT activity. These results suggest that Met-48 might be more important for activity than Met-155. Increasing the size of the residue at position 48 from Ala to Leu restored 40% of wild-type activity with the R3 peptide. The activity results indicate that the type I conserved Met residues at positions 48 and 155 play important roles in enzymatic activity. Furthermore, the size of Met-48 is critical for maintaining the catalytic activity of PRMT1.
TABLE 1.
Steady-state kinetic activity of PRMT1 mutants with small sized side chains with R3 peptide via an enzyme-coupled continuous spectrophotometric assay
| PRMT1 | Km | kcat | kcat/Km | Activitya of WT-PRMT1 |
|---|---|---|---|---|
| μm | s−1 | m−1s−1 | % | |
| WT | <10 | 0.045 ± 0.003 | 4500 ± 330 | 100 |
| M155A | 120 ± 30 | 0.054 ± 0.005 | 460 ± 120 | 10 |
| M48A | 300 ± 210 | 0.0040 ± 0.0020 | 13 ± 13 | 1.4 |
| M48L | 18 ± 2.7 | 0.030 ± 0.0009 | 1700 ± 260 | 37 |
a The percentage activity is based on the catalytic efficiency kcat/Km of mutants compared with wild type.
To further explore the effect of the residue size at position 48 on the catalytic activity, we further mutated the methionine to amino acids with larger size side chains, such as phenylalanine, tyrosine, and tryptophan. The same kinetic assay was employed to measure the activity of these three mutants with the R3 peptide; however, the turnover rate was very slow and beyond the limit of detection for this method (data not shown). We turned to a more sensitive method that we recently reported on (21) and examined the mutant activity on a protein substrate, hnRNP K (supplemental Table S4). With all larger sized mutations, the activity was severely impaired. M48F had the highest activity among these three mutants, displaying no more than 3% of WT activity. Mutants with the even bigger amino acid tryptophan lost >95% activity. Overall, the kinetic analyses of the mutants show that residues smaller or larger than methionine at position 48 result in less efficient methyl group transfer.
Substrate Binding Affinity of PRMT1 Mutants
The decrease in activity observed with mutations of either Met-155 or Met-48 could be a result of impaired substrate binding or an inability to catalyze methyl transfer. To discern if Met-155 or Met-48 mutations affect the binding of either AdoMet or peptide substrates, we measured the dissociation constants of PRMT1, both wild-type and mutants, with AdoMet and the R3 peptide substrate. The crystal structure of PRMT1 indicates that there are two tryptophans at positions 145 and 294 that lie in the catalytic region near the substrate arginine residue and opposite the AdoMet binding region (11). The intrinsic fluorescence from these residues was exploited in a fluorescence-quenching assay to determine the dissociation constants for the R3 peptide and AdoMet with PRMT1. Lehrer's modified Stern-Volmer plot (22) was used to analyze the fluorescence-quenching data for WT-PRMT1 (supplemental Fig. S4) and the mutants. Dissociation constants of either AdoMet or R3 peptide with WT-PRMT1 and all the mutants were similar to each other in the low micromolar range (Table 2). The observed fluorescent quenching was consistent with the number of active site tryptophans relative to the total number of tryptophans. These results indicate that the decrease in activity observed with the methionine PRMT1 mutants is not due to impaired substrate binding in the enzyme active site.
TABLE 2.
Binding affinity of R3 peptide substrate and AdoMet with intrinsic fluorescence quenching
| PRMT1 | KD, R3 | Fluorescent quenching | KD, AdoMet | Fluorescent quenching |
|---|---|---|---|---|
| μm | % | μm | % | |
| WT | 5.7 ± 0.52 | 28 ± 1.9 | 4.2 ± 1.5 | 17 ± 0.13 |
| M48A | 3.4 ± 0.32 | 37 ± 1.3 | 7.1 ± 1.8 | 15 ± 0.17 |
| M48L | 2.4 ± 0.29 | 28 ± 1.5 | 7.1 ± 0.94 | 29 ± 4.2 |
| M48F | 4.9 ± 0.38 | 30 ± 1.4 | 4.4 ± 0.93 | 23 ± 3.1 |
| M155A | 3.9 ± 0.53 | 34 ± 1.3 | 3.8 ± 0.87 | 19 ± 2.6 |
Automethylation Activity of PRMT1 Mutants
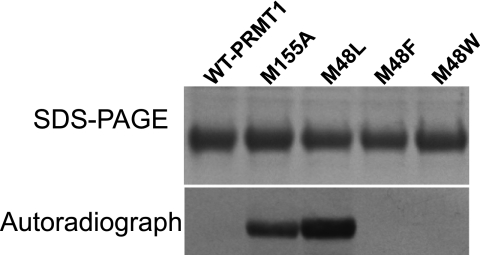
In addition to the methylation of peptide and protein substrates, automethylation of some PRMTs has been documented. For example, automethylation has been observed with PRMT6 and PRMT8 (36, 37). Additionally, Frankel and co-workers (38) recently reported that histidine-tagged human PRMT1 also catalyzes automethylation. Although we have never observed automethylation with His-tagged rat PRMT1, we hypothesized that the methionine mutants of rat PRMT1 may show altered substrate recognition and hence altered product formation. Therefore, the ability of each of the PRMT1 mutants to catalyze automethylation was tested using [3H]AdoMet. Under our reaction conditions, WT-PRMT1 showed no tritium incorporation after 4 h of incubation. Unlike WT-PRMT1, the M48L and M155A mutants clearly showed a strong capacity for automethylation (Fig. 3). We quantified the amount of automethylation using our ZipTip®C4 assay (21). A sample of 15 μm M48L and M155A incorporated 2.14 ± 0.59 μm (14.3%) and 0.68 ± 0.24 μm (4.5%) methyl groups in a 4-h period, respectively. We also confirmed the product of the automethylation by digesting the methylated M48L and resolving the derivatized amino acids by HPLC. The supplemental Fig. S5 shows the presence of radiolabel associated with ADMA. These results show that mutation of either Met-155 or Met-48 gives rise to a PRMT1 enzyme capable of automethylation. We conclude that both Met-155 and Met-48 are important for substrate recognition.
FIGURE 3.
Automethylation of PRMT1 mutants. Reactions were carried out at 37 °C for 4 h with 15 μm PRMT1 (WT or mutants), 8.57 μm AdoMet (0.9 μm [3H]AdoMet added) in 50 mm sodium phosphate buffer, pH 7.5. The top panel is an SDS-polyacrylamide gel stained with Coomassie Blue to detect total protein, and the bottom panel is an autoradiograph to detect radiolabel incorporation. The gel was exposed to film for 5 days.
Analyzing for Changes in Protein Structure
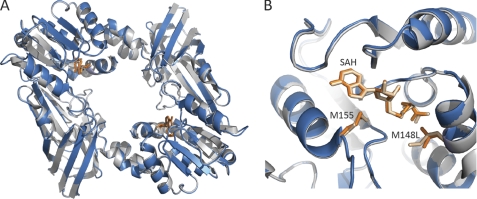
To confirm that mutations of the conserved methionine had no deleterious effect on the PRMT1 structure, we determined the crystal structure of the M48L mutant. Using conditions described for the crystallization of WT-PRMT1, we obtained a 2.2-Å structure of M48L bound to AdoHcy (Fig. 4). No significant differences were observed between the wild-type and mutant structures (root mean square deviation of 0.23 Å over 299 residues), consistent with the kinetic data showing similar binding of AdoMet to WT and M48L.
FIGURE 4.
Superposition of the overall structure (A) and the active site (B) of M48L (blue) and wild-type PRMT1 (gray). Met-48/Leu-48, Met-155, and AdoHcy are shown as sticks with WT-PRMT1 residues colored tan and M48L residues colored orange.
Positional Preference of M48L-PRMT1
Although the crystal structure of M48L indicates that there are no significant changes in the structure of the M48L mutant, the automethylation experiments suggested that substrate recognition is altered in M48L. To further explore the effect that the M48L mutation has on substrate specificity, we asked whether M48L strongly prefers substrate arginines at certain positions and compared the results to WT-PRMT1. We employed the R3 peptide in which all three arginines can be methylated, with one arginine at the N terminus, one in the middle, and one at the C terminus. The short time methylation products of R3 peptide were analyzed by tandem mass spectrometry (MS/MS) to find out which arginine was first methylated by WT-PRMT1 and M48L (supplemental Fig. S6). MS/MS showed that the N-terminal arginine is strongly preferred by both WT-PRMT1 and M48L, with 55 and 65% of the first methylation located on the N-terminal arginine, respectively (Table 3). The middle arginine is the second most preferred; however, the percentage for M48L was reduced by almost 50% compared with the WT-PRMT1. These results suggest that although recognition of the N-terminal arginine of the R3 peptide appears unaltered in M48L, recognition and/or catalysis at the central Arg of R3 is impaired by M48L.
TABLE 3.
First methylation position in the R3 peptide catalyzed by WT-PRMT1 and M48L from MS/MS analysis
| PRMT1 | % of first methylation position |
||
|---|---|---|---|
| 1st Arg | 2nd Arg | 3rd Arg | |
| WT-PRMT1 | 55 | 39 | 6 |
| M48L | 65 | 21 | 14 |
Peptide Substrate Specificity Change for M48L-PRMT1
As a way of complementing the above studies, we also tested the activity of M48L with a series of peptides that differ in the location of the targeted arginine within the peptide and methylation status (listed in Table 4) with the M48L-PRMT1 mutant. All the peptides listed could be easily methylated by wild-type PRMT1 (Table 4). M48L was able to methylate R3 at a relatively lower rate compared with wild-type enzyme. We tested peptides containing a single arginine along with its monomethylated counterpart. M48L could methylate the RKK peptide pair (single arginine at N terminus) at a rate close to the R3 peptide. Surprisingly, even though the sequence of RKK and KRK peptides is the same around the targeted arginyl group (GGRGG), the ability of M48L to methylate the centrally located Arg in KRK was severely hampered. We conclude that the inactivity observed with M48L and KRK peptide is most likely due to the central position of the arginine in KRK peptide pair.
TABLE 4.
Peptide substrate specificity of WT-PRMT1 and M48L under steady-state conditions
| Peptide substrate | Sequence | Activity of WT-PRMT1 | Activity of M48L | Activitya of WT-PRMT1 |
|---|---|---|---|---|
| s−1 | s−1 | % | ||
| R3 | acGGRGGFGGRGGFGGRGG | 0.045 | 0.029 | 65.9 |
| RKK | acGGRGGFGGKGGFGGKW | 0.0051 | 0.0026 | 50.5 |
| RKK-CH3 | acGGRmeGGFGGKGGFGGKW | 0.011 | 0.0053 | 47.9 |
| KRK | acKGGFGGRGGFGGKW | 0.0071 | 0.000075 | 1.06 |
| KRK-CH3 | acKGGFGGRmeGGFGGKW | 0.0039 | 0.000023 | 0.577 |
a The methylation velocity is tested under one saturating condition for both AdoMet and peptide substrates, representing the activity of enzymes. The percentage activity is calculated based on the methylation velocity.
The previous experiments predict that the M48L mutant would be unable to stoichiometrically methylate all arginine residues in the R3 peptide. In other words, the amount of methyl groups transferred by M48L is expected to be lower, although WT-PRMT1 can fully methylate the R3 peptide, transferring six methyl groups to one peptide. When allowed to react with 10, 20, and 30 μm R3 and excess AdoMet (60, 120, and 180 μm total methyl group transfer expected, respectively), M48L catalyzed the transfer of 35, 48, and 83 μm methyl groups, respectively (data not shown). This indicated that each peptidyl arginine was not being fully dimethylated by M48L. Given our current data, it is likely that M48L-PRMT1 methylated the R3 peptide on the first terminal arginine but was unable to fully access or got aborted at the central arginine residue. Overall WT-PRMT1 and M48L mutant strongly prefer the N-terminal arginine, and the methylation activity of M48L decreased dramatically with a single arginine in the middle position.
Single Turnover Experiments with M48L-PRMT1
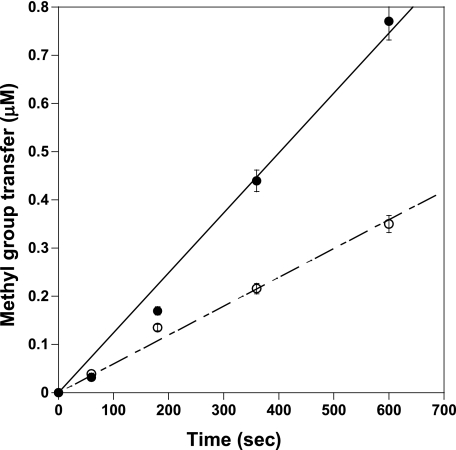
To investigate why the KRK pair cannot be methylated well by M48L-PRMT1, we set up single turnover experiments that would allow us to evaluate the effect of the Met-48 mutation on the conversion of arginine to MMA and the conversion of MMA to ADMA in separate experiments. Reactions were conducted under conditions where the AdoMet concentration was the same as or slightly higher than the enzyme concentration, so as to observe only one turnover of the methylation reaction (Fig. 5). Maximal product concentration and kchem for each reaction were obtained (Table 5).
FIGURE 5.
Single turnover kinetics of WT-PRMT1 (A) and M48L (B) with KRK and KRK-CH3 peptide pair. Filled circles and open circles indicate the KRK peptide and the monomethylated counterpart KRK-CH3 peptide, respectively.
TABLE 5.
Pre-steady-state kinetic parameters of M48L with WT-PRMT1
| Enzyme | Peptide | Maximal product concentration | kchem |
|---|---|---|---|
| μm | s−1 | ||
| 20 μm WT-PRMT1 | KRK | 20.31 ± 0.25 | 0.020 ± 0.0008 |
| KRK-CH3 | 14.96 ± 1.15 | 0.029 ± 0.0082 | |
| RKK | 14.53 ± 1.82 | 0.073 ± 0.037 | |
| RKK-CH3 | 13.08 ± 0.85 | 0.054 ± 0.016 | |
| 20 μm M48L | KRK | 3.85 ± 0.11 | 0.0087 ± 0.0006 |
| KRK-CH3 | 0.16 ± 0.084 | NDa | |
| RKK | 4.71 ± 0.20 | 0.044 ± 0.008 | |
| RKK-CH3 | 9.37 ± 1.43 | 0.018 ± 0.008 |
a ND stands for not detectable.
WT-PRMT1 fully methylated the KRK peptide with the maximum product concentration equal to 20 μm (Table 5). With the KRK-CH3 peptide, the total methyl group transfer decreased slightly, but the turnover rate (0.029 s−1) was very close to the naked peptide (0.020 s−1). With M48L, the reaction only went to 20% completion with the KRK peptide, with the turnover rate reduced by 50%. However, when the M48L reaction was initiated with the monomethylated peptide, hardly any methylation occurred, with the maximum product concentration of around 0.16 μm and the turnover rate kchem close to 0 s−1. Control experiments were set up under the same conditions without the peptide substrates to rule out radiolabel incorporation as a result of M48L automethylation. Our results show impaired monomethylation of the internal arginine by M48L PRMT1 and nearly abolished dimethylation.
The lack of activity that was observed with M48L and KRK-CH3 could be due to the inability of the peptide to bind the mutated active site. To determine whether the KRK monomethylated peptide binds to M48L, we set up a competitive reaction using the RKK-CH3 peptide as a substrate with or without the KRK peptide pair. The concentration of RKK-CH3 peptide was close to its Km value, and KRK-CH3 peptide was one-third the RKK-CH3 concentration. With KRK added into the reaction, the methyl group transfer rate was close to the one without KRK (0.0012 and 0.0011 μm/s, respectively) (Fig. 6). However, with the monomethylated peptide KRK-CH3 added into the preincubation solution, the reaction rate was decreased to half of the original rate (0.0012 and 0.0006 μm/s, respectively). These data suggest that the KRK-CH3 peptide was actually a PRMT1 inhibitor in the methylation reaction. With the KRK peptide in the preincubation mixture, the rate was slightly decreased, possibly because the KRK peptide would bind to M48L but could not be methylated as fast as the RKK-CH3 peptide, as shown in Table 5. The fact that KRK-CH3 could inhibit the methylation of RKK-CH3 suggests that the KRK-CH3 peptide can bind M48L-PRMT1 but is not turned over by the enzyme in appreciable amounts.
FIGURE 6.
Competitive methylation experiments of the KRK-CH3 peptide. The reactions were carried out with 100 nm M48L, 60 μm RKK-CH3 peptide as a substrate, 1 μm AdoMet (1 μm [3H]AdoMet added) in 100 mm HEPES buffer at 37 °C, pH 8.0. 20 μm KRK-CH3 peptide were tested as an inhibitor. The filled circles indicate the control group without the KRK-CH3 peptide, and the open circles indicate the test group with KRK-CH3 added.
We also observed impairment in the rate at which M48L dimethylated argininyl groups located at the N terminus of the peptide, albeit to a lesser extent. Although dimethylation by wild type was 26% slower than monomethylation, dimethylation by M48L was 60% slower than monomethylation of the RKK peptide (Table 5). Clearly, substitution of methionine 48 with leucine in PRMT1 results in differential effects on the conversion of arginine to MMA versus the conversion of MMA to ADMA. Furthermore, the ability of M48L mutants to covalently transfer methyl groups to all targeted arginines in a substrate is impaired. We conclude that the type I-conserved methionine 48 in rat PRMT1 plays a significant role in determining product specificity.
DISCUSSION
In mammals, the PRMT family of enzymes is capable of generating three different methylated arginyl species within protein targets, MMA, ADMA, and SDMA. Coupled with the observation that many PRMT targets have multiple arginines that are methylated (39, 40), the diversity of potential protein products is large and suggests the possibility for a methylarginine code. The biological relevance of such a code has most recently been validated as part of the complex set of post-translational histone modifications affecting gene expression (2, 6, 7). Yet the molecular details of how each of the PRMTs selectively controls the position and degree of methylation have remained elusive. In this work, we provide the first insight into how the type I arginine methyltransferase active site is engineered for product specificity.
Absence of a Methionine Switch to Specify ADMA and SDMA Formation
When the first structures (11, 12) of type I arginine methyltransferases were solved, a strictly conserved methionine (Met-155 in the rat PRMT1 sequence) was located very close to the substrate arginine. Given the fact that this residue is strictly conserved in all type I methyltransferases, but much smaller (e.g. alanine or serine) in type II methyltransferases, a hypothesis was put forth that the methionine provided enough steric bulk to block the binding of monomethylated arginine in a conformation that would allow symmetric dimethylation (12, 13). Likewise, we questioned whether Met-48, which resides on the opposite side of the active site (Fig. 1C), could function in the same manner. The idea of a single residue switch to control product specificity was supported by the elegant studies in the lysine methyltransferase field, which showed that a Phe/Tyr switch in the active site regulates the ability of the SET domain lysine methyl transferases to perform successive methylations (13–15). Surprisingly, M155A and M48A mutants generated only MMA and ADMA after 3-h reactions. Even at the picomole detection limit of our monitoring system, no SDMA was detected (Fig. 2). Therefore, removing the steric bulk afforded by either Met-155 or Met-48 in the active site of PRMT1 is not by itself sufficient to transform PRMT1 into a type II PRMT and rules out the idea of a simple methionine switch. How the type I arginine methyltransferases discriminate between ADMA and SDMA is more complex than previously thought and will likely not be answered until a type II PRMT structure becomes available.
Met-155 and Met-48 Are Important for Enzyme Activity
The kinetic results presented here show that both conserved methionines are important for catalytic activity of PRMT1 but in different ways (Table 1). The similarity between the WT and M48L crystal structures (Fig. 4) indicates that the impaired activity of the mutants is not due to global changes in protein structure. Similar binding affinity of both AdoMet and R3 peptide for WT-PRMT1 and all the mutants (Table 2) illustrates that decreased activity in the mutants is also not due to changes in binding affinity.
As a third possibility to explain the altered activity of the methionine mutants, we considered the mechanism of methyl transfer. Given that methyl transfer occurs by way of an Sn2 reaction (41), the orientation of the arginine substrate and the thioether moiety of AdoMet in the active site become critical. Mutation of the methionines to smaller residues increases the space in the active site, and it potentially allows more orientations to be sampled by the incoming arginyl substrate, not all of which may be productive or as productive as in wild type. Mutation of the methionines may change the ability of the mutant enzyme to undergo compression during catalysis (42) or affect the pKa of the guanidino nitrogen of arginine as it approaches the positively charged sulfonium of AdoMet (43). Consistent with the idea that Met-48 is important for catalysis, our single turnover experiments (Table 5) showed that kchem is decreased in the M48L mutant. With the larger aromatic amino acid mutants, the positioning of the substrate arginine is likely fixed in an orientation that results in slow methyl transfer. We propose that the two type I methionines may work as molecular tweezers to align arginyl substrates at proper orientations. Interestingly, this methionine clamp is not only conserved in type I PRMTs, but it also exists in some other methyltransferases, such as salicylic acid carboxyl methyltransferase, isoflavone O-methyltransferase, and caffeic acid O-methyltransferase (23, 44, 45), suggesting a common mechanism for optimizing methyl transfer among certain AdoMet-dependent methyltransferases.
Met-48 Influences Product Specificity
Accurate product specificity by PRMT1 is a combination of targeting the correct arginine within a protein, as well as catalyzing the correct number of methylations on a particular residue. This study shows that Met-48 plays a role at both levels. Curiously, substitution of Met-48, as well as Met-155, caused the enzyme to recognize a new substrate, observed as automethylation (Fig. 3). Conversely, several experiments (Tables 4 and 5 and Fig. 5) provided results that indicate that the Met-48 mutation differentially impaired the ability of the mutant to dimethylate arginine residues, especially those located near the center of the peptide substrate. Although it is difficult to tease out which step in arginine methylation is hindered in M48L using steady-state data, our single turnover experiments measuring kchem limit the choices to peptide binding or the actual chemical transfer step. When the peptide is already monomethylated, one or both of these steps are hampered. When the monomethylated residue is presented in the center of the peptide substrate, very little if any substrate appears to be bound in the M48L mutant in a catalytically productive manner.
Conclusion
Product specificity as it relates to PRMT1 is multifaceted. Site selection within a protein (sequence and location), degree of methylation (mono- or dimethylation), and number of methylations in each target can produce a dizzying number of different products. How the enzyme is designed to control each one of these aspects is a challenging question that is likely to lead to overlapping/interdependent mechanisms. In this study, we provide the first insight into product specificity in PRMT1. We show that the conserved methionine 48 is important for activity and accurate site selection, which also impacts the degree of methylation in the final product.
Supplementary Material
Acknowledgments
We thank Dr. Alvan Hengge for discussions and critical comments made about the manuscript. We thank Christian Hansen Howell for making some of the mutants in this study.
This work was supported by Herman Frasch Foundation Award 657-HF07 (to J. M. H.), National Science Foundation Award 0920776 (to J. M. H.) funded through the American Recovery and Reinvestment Act, and an American Heart Association undergraduate research fellowship (to P. J. P.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1–S3 and Figs. S1–S6.
The atomic coordinates and structure factors (code 3Q7E) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- PRMT
- protein-arginine methyltransferase
- AdoHcy
- S-adenosylhomocysteine
- AdoMet
- S-adenosyl-l-methionine
- ADMA
- asymmetric dimethyl arginine
- hnRNP K
- heterogeneous nuclear ribonucleoprotein K
- MMA
- monomethyl arginine
- MTAN
- AdoHcy nucleosidase
- SDMA
- symmetric dimethyl arginine.
REFERENCES
- 1. Bedford M. T., Clarke S. G. (2009) Mol. Cell 33, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Huang S., Litt M., Felsenfeld G. (2005) Genes Dev. 19, 1885–1893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Levine T. B., Levine A. B. (2006) Metabolic Syndrome and Cardiovascular Disease, Saunders Elsevier, Philadelphia, PA [Google Scholar]
- 4. Jansson M., Durant S. T., Cho E. C., Sheahan S., Edelmann M., Kessler B., La Thangue N. B. (2008) Nat. Cell Biol. 10, 1431–1439 [DOI] [PubMed] [Google Scholar]
- 5. Bedford M. T. (2007) J. Cell Sci. 120, 4243–4246 [DOI] [PubMed] [Google Scholar]
- 6. Wysocka J., Allis C. D., Coonrod S. (2006) Front. Biosci. 11, 344–355 [DOI] [PubMed] [Google Scholar]
- 7. Kirmizis A., Santos-Rosa H., Penkett C. J., Singer M. A., Green R. D., Kouzarides T. (2009) Nat. Struct. Mol. Biol. 16, 449–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Balakrishnan L., Milavetz B. (2010) Crit. Rev. Biochem. Mol. Biol. 45, 440–452 [DOI] [PubMed] [Google Scholar]
- 9. Cheng X., Blumenthal R. M. (2010) Biochemistry 49, 2999–3008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tang J., Frankel A., Cook R. J., Kim S., Paik W. K., Williams K. R., Clarke S., Herschman H. R. (2000) J. Biol. Chem. 275, 7723–7730 [DOI] [PubMed] [Google Scholar]
- 11. Zhang X., Cheng X. (2003) Structure 11, 509–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang X., Zhou L., Cheng X. (2000) EMBO J. 19, 3509–3519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cheng X., Collins R. E., Zhang X. (2005) Annu. Rev. Biophys. Biomol. Struct. 34, 267–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Takahashi Y. H., Lee J. S., Swanson S. K., Saraf A., Florens L., Washburn M. P., Trievel R. C., Shilatifard A. (2009) Mol. Cell. Biol. 29, 3478–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Collins R. E., Tachibana M., Tamaru H., Smith K. M., Jia D., Zhang X., Selker E. U., Shinkai Y., Cheng X. (2005) J. Biol. Chem. 280, 5563–5570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ostareck D. H., Ostareck-Lederer A., Wilm M., Thiele B. J., Mann M., Hentze M. W. (1997) Cell 89, 597–606 [DOI] [PubMed] [Google Scholar]
- 17. Dorgan K. M., Wooderchak W. L., Wynn D. P., Karschner E. L., Alfaro J. F., Cui Y., Zhou Z. S., Hevel J. M. (2006) Anal. Biochem. 350, 249–255 [DOI] [PubMed] [Google Scholar]
- 18. Cornell K. A., Swarts W. E., Barry R. D., Riscoe M. K. (1996) Biochem. Biophys. Res. Commun. 228, 724–732 [DOI] [PubMed] [Google Scholar]
- 19. Markowski P., Baranowska I., Baranowski J. (2007) Anal. Chim. Acta 605, 205–217 [DOI] [PubMed] [Google Scholar]
- 20. Segel I. H. (ed) (1975) Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady-state Enzyme Systems, John Wiley & Sons, Inc., New York [Google Scholar]
- 21. Suh-Lailam B. B., Hevel J. M. (2010) Anal. Biochem. 398, 218–224 [DOI] [PubMed] [Google Scholar]
- 22. Lehrer S. S. (1971) Biochemistry 10, 3254–3263 [DOI] [PubMed] [Google Scholar]
- 23. Zubieta C., Kota P., Ferrer J. L., Dixon R. A., Noel J. P. (2002) Plant Cell 14, 1265–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rigaku Corporation (2006) CrystalClear: An Integrated Program for the Collection and Processing of Area Detector Data, Version 1.3.6, Rigaku Corp., Tokyo [Google Scholar]
- 25. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bailey S. (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 27. Potterton E., Briggs P., Turkenburg M., Dodson E. (2003) Acta Crystallogr. D Biol. Crystallogr. 59, 1131–1137 [DOI] [PubMed] [Google Scholar]
- 28. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 29. Painter J., Merritt E. A. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 439–450 [DOI] [PubMed] [Google Scholar]
- 30. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 31. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. DeLano W. L. (2007) The PyMOL Molecular Graphics System, Version 1.0r2, DeLano Scientific LLC, Palo Alto, CA [Google Scholar]
- 33. Reich N. O., Mashhoon N. (1991) Biochemistry 30, 2933–2939 [DOI] [PubMed] [Google Scholar]
- 34. Zappia V., Galletti P., Porcelli M., Manna C., Ragione F. D. (1980) J. Chromatogr. 189, 399–405 [DOI] [PubMed] [Google Scholar]
- 35. Rust H. L., Zurita-Lopez C. I., Clarke S., Thompson P. R. (2011) Biochemistry 50, 3332–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Frankel A., Yadav N., Lee J., Branscombe T. L., Clarke S., Bedford M. T. (2002) J. Biol. Chem. 277, 3537–3543 [DOI] [PubMed] [Google Scholar]
- 37. Sayegh J., Webb K., Cheng D., Bedford M. T., Clarke S. G. (2007) J. Biol. Chem. 282, 36444–36453 [DOI] [PubMed] [Google Scholar]
- 38. Lakowski T. M., 't Hart P., Ahern C. A., Martin N. I., Frankel A. (2010) ACS Chem. Biol. 5, 1053–1063 [DOI] [PubMed] [Google Scholar]
- 39. Côté J., Boisvert F. M., Boulanger M. C., Bedford M. T., Richard S. (2003) Mol. Biol. Cell 14, 274–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ostareck-Lederer A., Ostareck D. H., Rucknagel K. P., Schierhorn A., Moritz B., Huttelmaier S., Flach N., Handoko L., Wahle E. (2006) J. Biol. Chem. 281, 11115–11125 [DOI] [PubMed] [Google Scholar]
- 41. Coward J. K. (1977) in The Biochemistry of Adenosylmethionine (Salvatore F., Borek E., Zappia V., Williams-Ashman H. G., Schlenk F. eds) pp. 127–144, Columbia University Press, New York [Google Scholar]
- 42. Olsen J., Wu Y.-S., Borchardt R. T., Schowen R. L. (1979) in Transmethylation (Usdin E., Borchardt R. T., Creveling C. R. eds) Elsevier, New York [Google Scholar]
- 43. Zhang X., Bruice T. C. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 5728–5732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zubieta C., He X. Z., Dixon R. A., Noel J. P. (2001) Nat. Struct. Biol. 8, 271–279 [DOI] [PubMed] [Google Scholar]
- 45. Zubieta C., Ross J. R., Koscheski P., Yang Y., Pichersky E., Noel J. P. (2003) Plant Cell 15, 1704–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.