Abstract
Baeyer-Villiger monooxygenases catalyze the oxidation of carbonylic substrates to ester or lactone products using NADPH as electron donor and molecular oxygen as oxidative reactant. Using protein engineering, kinetics, microspectrophotometry, crystallography, and intermediate analogs, we have captured several snapshots along the catalytic cycle which highlight key features in enzyme catalysis. After acting as electron donor, the enzyme-bound NADP(H) forms an H-bond with the flavin cofactor. This interaction is critical for stabilizing the oxygen-activating flavin-peroxide intermediate that results from the reaction of the reduced cofactor with oxygen. An essential active-site arginine acts as anchoring element for proper binding of the ketone substrate. Its positively charged guanidinium group can enhance the propensity of the substrate to undergo a nucleophilic attack by the flavin-peroxide intermediate. Furthermore, the arginine side chain, together with the NADP+ ribose group, forms the niche that hosts the negatively charged Criegee intermediate that is generated upon reaction of the substrate with the flavin-peroxide. The fascinating ability of Baeyer-Villiger monooxygenases to catalyze a complex multistep catalytic reaction originates from concerted action of this Arg-NADP(H) pair and the flavin subsequently to promote flavin reduction, oxygen activation, tetrahedral intermediate formation, and product synthesis and release. The emerging picture is that these enzymes are mainly oxygen-activating and “Criegee-stabilizing” catalysts that act on any chemically suitable substrate that can diffuse into the active site, emphasizing their potential value as toolboxes for biocatalytic applications.
Keywords: Enzyme Catalysis, Enzyme Mechanisms, Enzyme Structure, Flavin, Oxidation-Reduction, Baeyer-Villiger, Biocatalysis, Monooxygenase
Introduction
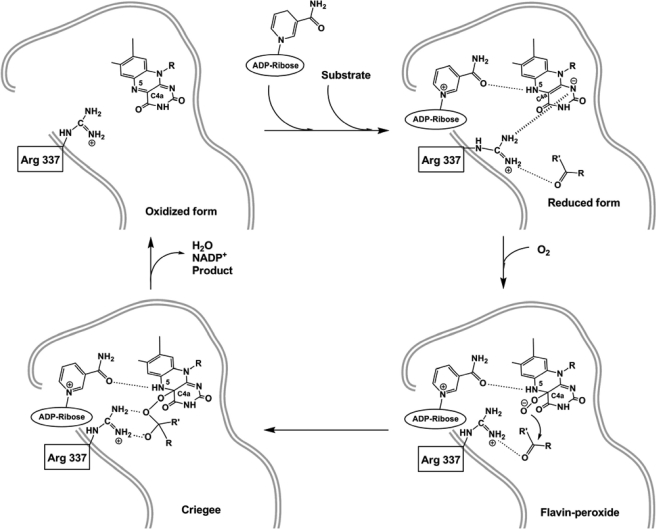
Flavoprotein monooxygenases catalyze the insertion of an oxygen atom into an organic substrate in hydroxylations, epoxidations, Baeyer-Villiger oxidations, and N- and S-oxidations (1–3). These enzymes activate molecular oxygen to overcome its spin-forbidden reaction with an organic molecule through formation of a stable C4a-(hydro)peroxy-flavin adduct (the “flavin-peroxide,” Scheme 1) (4). In the case of Baeyer-Villiger monooxygenases (BVMOs),3 the flavin-peroxide nucleophilically attacks the carbonyl group of ketone substrates with the formation of a covalent tetrahedral Criegee intermediate, analogous to the intermediate that forms in nonenzymatic Baeyer-Villiger oxidations (5). An electronic rearrangement yields an ester or lactone, a water molecule, and the oxidized flavin. A key feature of BVMOs and related monooxygenases is the dual role of NADP(H) in catalysis; it acts as the electron donor to reduce the flavin, and it is required for the stabilization of the essential flavin-peroxide intermediate (6–10).
SCHEME 1.
Schematic representation of the Bayer-Villiger reaction catalyzed by PAMO.
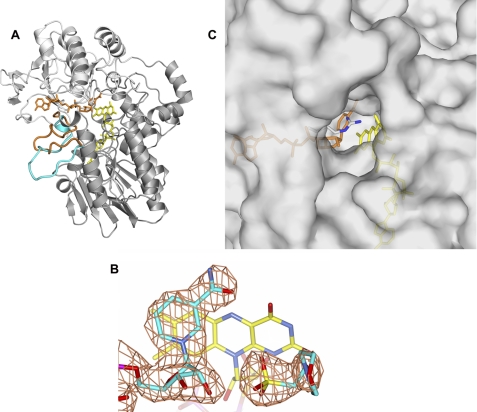
BVMOs are promising targets for biocatalytic applications in synthetic and pharmaceutical chemistry (11, 12). Phenylacetone monooxygenase (PAMO, EC 1.14.13.92) from Thermobifida fusca features several attractive properties: it is thermostable, tolerant to organic solvents (13, 14), and catalyzes enantioselective Baeyer-Villiger oxidations and sulfoxidations on a number of substrates in addition to phenylacetone (15). The three-dimensional structure of PAMO has been solved revealing a two-domain organization (Fig. 1A) (16), and its catalytic cycle has been subject of extensive kinetic studies (17–22). More recently, the structure of cyclohexanone monooxygenase in complex with NADP+ highlighted a proposed mechanism for the dual catalytic role of NADP(H) (9, 23).
FIGURE 1.
Crystallographic studies on PAMO. A, three-dimensional structure of PAMO in complex with NADP+ at 2.2-Å resolution. The NADP- and FAD-binding domains are in light gray and gray, respectively. FAD is in yellow, NADP+ in orange. Upon NADP+ binding, there is rearrangement of the loop 495–515; the loop conformation of the unligated enzyme (Protein Data Bank ID code 1W4X) is in cyan, and that of the NADP+-bound protein is in orange. B, final weighted 2Fo − Fc electron density map for the MES and NADP+ ligands at 1.5 σ level. Oxygens are in red, nitrogens in blue, phosphorus atoms in magenta, sulfurs in yellow, ligand carbons in cyan, and flavin carbons in yellow. C, surface representation of the active-site region. The flavin is yellow, NADP+ is orange, Arg-337 carbons are gray, Arg-337 nitrogens are blue.
Here, we present a biochemical, microspectrophotometric, and structural investigation of PAMO which aims at the identification of the structural and mechanistic features that underlie the complex mechanism of function of BVMOs. The study elucidates key aspects of oxygen activation and substrate oxygenation through the formation of the Criegee intermediate.
EXPERIMENTAL PROCEDURES
Proteins have been purified following published protocols with few modifications (Table 1) (17, 24). Kinetic studies on D66A and R217A mutants were performed as described (17). Wild-type, D66A, and R337K proteins were crystallized by vapor diffusion at 4 °C using 1–3 μl of protein solutions (18 mg/ml in 0.5 μm FAD and 4 mm NADP+, 50 mm Tris/HCl, pH 7.5) mixed with equal volumes of reservoir containing 40% (w/v) PEG4000, 100 mm MES/HCl, pH 6.5, and 100 mm NaCl. The M446G mutant could be crystallized only in the conditions previously used for the ligand-free enzyme (16). All crystal soaking experiments were performed at room temperature in stabilizing solutions containing 4 mm NADP+, 40% (w/v) PEG4000, 100 mm MES/HCl, pH 6.5, and 150 mm NaCl. X-ray diffraction data (Table 1) were collected at 100 K at ESRF (Grenoble, France) and SLS (Villigen, Switzerland) and processed with the CCP4 package (25). The structures were solved and refined using the software PHASER (26), COOT (27), and Refmac5 (28). Three-dimensional structure validation with the program Molprobity (29) indicates that all models fall within the 75th–95th percentiles of the Molprobity score. Supplemental Fig. S1 shows the Ramachadran plot of structure of the R337K mutant bound to MES. Single-crystal microspectrophotometric absorbance spectra were measured on cryo-cooled crystals in both off-line (30) and on-line (31) modes at the Cryobench laboratory and at beam-line ID14-eh1 of ESRF, respectively. Pictures were prepared with PyMOL (40) and CCP4mg (32).
TABLE 1.
Crystallographic data collection and refinement statistics
Protein purification: plasmids were transformed into Escherichia coli TOP10 cells (Invitrogen), and the resulting colonies were preinoculated into LB broth supplemented with 100 μg/ml ampicillin and grown overnight at 37 °C. Cultures were then inoculated (starting A600 0.1) and grown in 2-liter Erlenmeyer flasks at 30 °C in 800 ml of Terrific Broth medium supplemented with 100 μg/ml ampicillin and 0.02% (w/v) l-(+)-arabinose for 20 h. E. coli cells were harvested by centrifugation at room temperature. Cells were resuspended in 50 mm Tris/HCl, pH 7.5, at 25 °C and 10 μm FAD (5 ml of buffer/g of cells). The crude extract obtained by sonication was incubated at 50 °C for 30 min and centrifugated at 70,000 × g. Proteins were loaded onto a HiLoad 16/10 Q-Sepharose column (GE Healthcare) preequilibrated in 50 mm Tris/HCl, pH 7.5, at 25 °C. The enzymes were eluted at 240 mm potassium chloride. The pooled fractions were loaded on a Superdex 200 16/60 column preequilibrated with the same buffer.
| Parameter | WT-NADP+ oxidized | WT-NADP+ reduced | WT-NADP+ bound to MES | R337K-NADP+ oxidized | R337K-NADP+ reduced | R337K-NADP+ bound to MES | D66A-NADP+ bound to MES | M446G |
|---|---|---|---|---|---|---|---|---|
| PDBa code | 2YLR | 2YLS | 2YLT | 2YM1 | 2YM2 | 2YLW | 2YLX | 2YLZ |
| Beam line | ID14eh1 | ID14eh1 | ID14eh1 | ID14eh2 | ID14eh1 | PXI | ID14eh1 | ID23eh2 |
| Unit cell (Å) | a = b = 107.7 c = 107.4 | a = b = 107.9 c = 106.8 | a = b = 107.3 c = 107.7 | a = b = 107.9 c = 107.5 | a = b = 108.4 c = 108.1 | a = b = 107.7 c = 106.0 | a = b = 107.5 c = 107.09 | a = 86.8 b = 115.9 c = 166.6 |
| Space group | P3221 | P3221 | P3221 | P3221 | P3221 | P3221 | P3221 | I222 |
| Resolution (Å) | 2.26 | 2.26 | 2.65 | 2.28 | 2.7 | 2.9 | 2.2 | 2.0 |
| Rsymb,c (%) | 8.4 (26.9) | 9.0 (30.4) | 10.8 (51.6) | 9.6 (49.8) | 11.4 (46.3) | 5.2 (8.4) | 8.5 (44.8) | 8.8 (41.8) |
| Completenessc (%) | 99.9 (99.6) | 99.6 (98.8) | 99.3 (99.8) | 99.8 (100) | 99.8 (99.6) | 98 (97.0) | 100 (100) | 100 (100) |
| Unique reflections | 34,179 | 34,153 | 21,047 | 33,327 | 20,544 | 15,840 | 36,752 | 56,991 |
| Redundancyc | 4.2 (4.0) | 3.0 (2.9) | 3.6 (3.7) | 4.7 (4.7) | 4.2 (4.4) | 4.6 (4.1) | 4.3 (4.3) | 4.0 (4.0) |
| I/σb | 12.6 (4.7) | 8.7 (3.4) | 9.9 (2.6) | 11.6 (3.0) | 9.5 (3.3) | 20.8 (12.9) | 11.8 (3.1) | 11.6 (4.8) |
| No. of atoms | 4,620 | 4,569 | 4,461 | 4,496 | 4,378 | 4,392 | 4,698 | 4,657 |
| Average B value (Å2) | 25.2 | 23.5 | 34.0 | 29.9 | 35.5 | 11.9 | 34.4 | 26.2 |
| Rcrystd (%) | 18.9 | 18.9 | 19.6 | 20.0 | 20.0 | 18.4 | 19.5 | 19.2 |
| Rfreed (%) | 23.8 | 23.5 | 26.1 | 24.5 | 25.9 | 26.6 | 25.2 | 22.6 |
| Rmse bond length (Å) | 0.019 | 0.022 | 0.016 | 0.018 | 0.016 | 0.022 | 0.021 | 0.019 |
| Rms bond angles (°) | 1.8 | 1.9 | 1.7 | 1.7 | 1.7 | 2.0 | 1.9 | 1.6 |
a PDB, Protein Data Bank.
b Rsym = Σ|Ii − 〈I〉|/ΣIi, where Ii is the intensity of ith observation and 〈I〉 is the mean intensity of the reflection.
c Values in parentheses are for reflections in the highest resolution shell.
d Rcryst = Σ|Fobs − Fcalc|/ΣFobs where Fobs and Fcalc are the observed and calculated structure factor amplitudes, respectively. Rcryst and Rfree were calculated using the working and test set, respectively.
e Rms, root mean square.
RESULTS
PAMO in Complex with NADP+
The main goal of our study was to advance our understanding of the enzymology and structural chemistry underlying the function of Baeyer-Villiger flavoenzymes using PAMO as model protein for the enzyme class. Given the essential role of NADP+ in catalysis (Scheme 1), a prerequisite toward this aim was to obtain well diffracting crystals of the enzyme bound to the dinucleotide ligand. The crystallization conditions used to solve the previously reported ligand-free enzyme structure contained large amounts of ammonium sulfate which, as often reported for dinucleotide-binding proteins, prevent NADP+ binding. Optimization of the purification protocol and extensive screenings led us to identify crystallization conditions using PEG as precipitant with protein solutions containing an excess of NADP+. These well diffracting crystals (Table 1) allowed us to solve the structure of the PAMO-NADP+ complex by molecular replacement using the ligand-free structure as search model (Fig. 1) (16). The overall conformations of the individual NADP- and FAD-binding domains are very similar in the ligand-free and NADP+-bound structures as indicated by root mean square deviations of 0.37 and 1.35 Å for the Cα atoms, respectively (Fig. 1A; excluding residues 495–515). The only significant alterations are a 6° rotation of the NADP-binding domain and a drastic conformational change in the loop 495–515 of the FAD-binding domain. This loop forms an elongated β-hairpin in the ligand-free enzyme whereas in the NADP+ complex it undergoes a large (up to 6 Å) shift enabling extensive interactions with the ADP-ribose moiety of NADP+. The same change in loop conformation has been described for cyclohexanone monooxygenase, indicating that these structural alterations are not crystallization artifacts (23). As a consequence of these conformational changes, the cleft at the domain interface becomes narrower, being mostly occupied by the dinucleotide ligand with the nicotinamide ring in direct contact with the dimethylbenzene moiety of the flavin (Fig. 2). In this way, NADP+ together with the surrounding protein residues define the boundaries of the substrate-binding site, which has the shape of a funnel that extends from the protein surface to the flavin (Fig. 1C).
FIGURE 2.
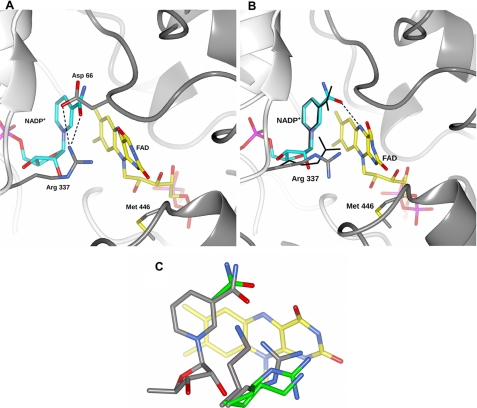
Structural changes upon reduction by dithionite. A, active site of PAMO in complex with NADP+ in the oxidized state. Carbon atoms of NADP+ are cyan, flavin in yellow, protein residues in gray. Nitrogens are in blue, oxygens in red, sulfurs in yellow, and phosphorus atoms in magenta. B, active-site structure of the dithionite-reduced enzyme (2.2 Å). The nicotinamide and Arg-337 of the oxidized enzyme are superimposed and shown as thin lines. We tentatively oriented the carboxamide group with the oxygen atom pointing toward the flavin, but it cannot be ruled out the carboxamide has the opposite orientation with the NH2 group directed toward the flavin. C, superimposition of the oxidized and reduced states of wild-type and R337K structures showing the same type of shift by the side chain at position 337 upon reduction. Carbons of the oxidized proteins are in gray whereas those of the reduced proteins are in green.
The Crystalline Enzyme Is Redox-reactive
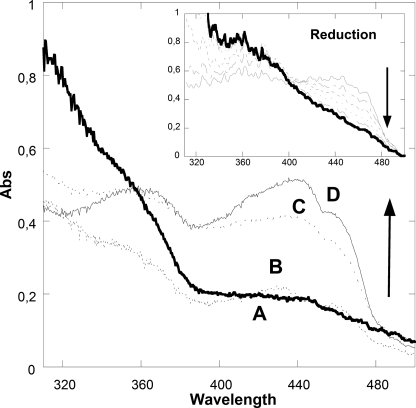
We probed PAMO-NADP+ crystals by microspectrophotometry (30, 31). They exhibited a UV-visible absorbance spectrum very similar to that measured from protein solutions (Fig. 3). Soaking the crystals in NADPH-containing solutions did not cause any bleaching of the flavin, implying that the crystal packing prevents NADP+ displacement by NADPH (Fig. 1A). However, soaking in dithionite-containing solutions led to flavin reduction, which takes place in 1–6 min depending on the crystal size (Fig. 3). The possibility of measuring spectra during data collection allowed us to observe that x-ray irradiation induces rapid crystal reduction, which is completed in relatively short times (5–30 s; Fig. 3 inset) (31). Dithionite-reduced and x-ray- induced crystals exhibit indistinguishable spectra and can be reoxidized in 3–6 min by simple exposure to aerated solutions (Fig. 3). These data demonstrate that the crystalline enzyme is redox-reactive.
FIGURE 3.
Microspectrophotometry of PAMO crystals measured at 100 K. Soaking the crystals at room temperature prior to cryo-cooling in dithionite-containing solutions leads to the reduction of the crystalline enzyme (spectrum A, bold line). Reduced crystals can be reoxidized by soaking at room temperature in aerated solutions as shown by spectra B, C, and D, which were collected on crystals that were cryo-cooled 2, 4, and 6 min after beginning of reoxidation. Spectrum D corresponds to that of the fully reoxidized enzyme, which is indistinguishable from that measured on oxidized crystals directly harvested from the crystallization droplets. The inset shows the flavin reduction during routine x-ray diffraction data collection at 100 K measured at different x-ray exposure times. The flavin is completely reduced, typically after 5–30 s of irradiation.
A point of concern in the crystallographic analysis was whether the structure of the enzyme obtained by collecting x-ray diffraction data on an oxidized crystal would represent the actual conformation of the oxidized enzyme or whether flavin photoreduction triggered by x-ray exposure (Fig. 3 inset) resulted in the structure of the reduced enzyme. Diffraction data are measured at 100 K, which is likely to prevent the conformational changes occurring during reduction (33) (see Fig. 2). In this scenario, the flavin is reduced by x-ray exposure, but the surrounding protein environment does not relax to the actual reduced protein conformation because of the cryo-cooled state of the crystal (33). Several lines of evidence support this interpretation. (i) PAMO crystals are remarkably robust in that they can be subjected to repeated cycles of reduction and reoxidation without any substantial loss of the diffraction power. This property allowed us to perform the following experiment. A crystal was initially used for x-ray data collection at 100 K. The crystal was then thawed and soaked in a dithionite-containing reducing solution for 5–10 min. Next, it was cryo-cooled and used for x-ray data collection. After this second data collection experiment, the crystal was again thawed and soaked in an aerated stabilizing solution to induce crystal reoxidation. This allowed us to measure diffraction data from the same crystal in the oxidized, reduced, and reoxidized state. We could clearly observe that “initially oxidized” (i.e. crystals that were not subject to any treatment before data collection) and reoxidized proteins have the same conformation which differs from that of the reduced enzyme (Fig. 2, B and C). (ii) The same conformational changes observed upon dithionite reduction were observed by photoreducing the crystals. This feature could be observed by performing the following experiment. Crystals were cryo-cooled at 100 K, exposed to x-rays to induce photoreduction, thawed from 100 K to room temperature by a brief (few seconds) soaking in a stabilizing solution, and finally cryo-cooled again. Such a brief “room temperature” annealing of an x-ray reduced crystal is too short to allow reoxidation, but it is long enough to allow the crystalline protein to relax to the reduced-state conformation which, indeed, turns out to be identical to that of exhibited by dithionite-reduced crystals (Fig. 2) (34–36). (iii) We used different data collection strategies to probe the effect of x-ray irradiation. In one experiment, we generated a composite data set by merging diffraction frames measured on several crystals at the beginning of data collection (i.e. when the crystalline proteins are still mostly oxidized). In another experiment, we used the helical data collection strategy (35) that continuously exposes fresh (nonirradiated) crystal regions. For both data collection strategies, the resulting electron densities were identical to those obtained from standard data sets measured on single initially oxidized crystals.
These findings indicate that the atomic model obtained from data measured on initially oxidized crystals visualizes the conformation of the oxidized-state protein. Likewise, the structure obtained from dithionite-reduced crystals is taken as a model of the enzyme in the reduced-state conformation.
Reduced Enzyme Structure
Enzyme reduction by dithionite does not cause overall conformational changes as indicated by the root mean square deviation from the oxidized enzyme Cα positions of 0.3 Å (Fig. 2, A and B). Inspection of the active site shows two localized but functionally highly relevant structural alterations. In the oxidized enzyme, Arg-337 is engaged in H-bonds with the NADP+ carboxamide group and the side chain of Asp-66 (Fig. 2A). Upon enzyme reduction, Arg-337 moves away from NADP+ pointing toward the pyrimidine moiety of the flavin ring. In this way, an electrostatically favored interaction between the positively charged guanidinium group of Arg-337 and the negatively charged reduced flavin can be established (Fig. 2B). At the same time, the carboxamide group of the nicotinamide ring slightly rotates toward the flavin to form an H-bond with the N5 atom of the reduced cofactor. The key point emerging from the reduced enzyme structure is that formation of the negatively charged reduced cofactor is associated with a localized rearrangement of the two central elements of the catalytic site; NADP+ and Arg-337 (Scheme 1).
The Complex with MES Reveals an Oxyanion Hole
In the course of the crystallographic studies, we often observed a strong electron density peak, which was particularly evident in crystals soaked in solutions containing increased MES buffer concentrations.4 Motivated by this observation, we performed soaking experiments in highly concentrated (400 mm) MES solutions. The resulting electron densities clearly indicated that a MES molecule binds in the active site in direct contact with Arg-337 and the ribose group of NADP+ (Fig. 1B). Such a binding mode positions the ligand morpholino ring in the funnel leading to the flavin cofactor (Fig. 4A). We also found that MES is a competitive inhibitor of PAMO although with low affinity (Ki in the millimolar range).4 The electron density obtained from dithionite-reduced crystals soaked in MES solutions did not indicate the presence of any ligand molecule, whose binding is evidently impeded by the negatively charged reduced flavin. The finding that MES specifically binds to the active site of the oxidized protein is particularly insightful in that the ligand sulfonic group is H-bonded to the side chain of Arg-337 and the 2′-hydroxyl group of the NADP+ ribose (Fig. 4A and Scheme 1). This observation suggests that one of the elements of the catalytic armamentarium of BVMOs is the presence of a sort of oxyanion hole in the catalytic center, formed by the Arg-337-NADP+ pair.
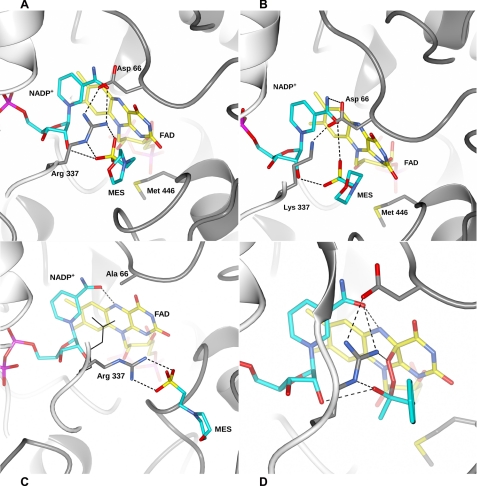
FIGURE 4.
MES binding to PAMO. A, in the wild-type protein, binding involves H-bonding with Arg-337, which further interacts with Asp-66. B, in R337K, MES binds in a similar position but is unable to directly H-bond to Lys-337. C, structure of D66A mutant shows a MES molecule in the funnel that leads to the active site. Arg-337 of the oxidized wild-type enzyme is superimposed and shown in thin lines. D, model of Criegee intermediate (see also Scheme 1) in BVMOs. Colors are as in Fig. 2A.
R337K and Active-site H-bond Network
Mutagenesis studies have shown that Arg-337 is essential for catalysis in a remarkable way (17). The Arg-337 mutants are reduced by NADPH and form an extremely stable flavin-peroxide intermediate (Scheme 1). Their lack of catalytic activity results from impaired ability to oxygenate the organic substrate. Given these functional features, we performed the structural elucidation of the R337K mutant in both oxidized and reduced states. The mutation causes virtually no conformational changes with respect to the wild-type structures (Fig. 2C). In the oxidized state, Lys-337 amino group interacts with the carboxamide of the NADP+ and the side chain of Asp-66. Upon reduction, Lys-337 moves toward the flavin, adopting a conformation similar to that of Arg-337 in the reduced wild-type protein. Furthermore, R337K retains the ability to bind MES which, however, can form an H-bond only with the NADP+ ribose because of the absence of the guanidinium group from the side chain in position 337 (Fig. 4B). Different from Arg, the Lys amino group is unable to simultaneously engage NADP+, Asp-66, and active-site ligand in H-bonding interactions (Fig. 4, A and B). As a result, the fine geometry of the oxyanion hole formed by Arg-337 and NADP+ ribose is significantly affected, which prohibits oxygenations to occur.
D66A Mutant and Substrate Entry
Having found that the essential Arg-337 interacts with Asp-66 (in the oxidized protein; Fig. 2A), we studied the D66A mutant to probe the role of this side chain in catalysis. The mutant protein is enzymatically active with the oxidative step (the reaction with oxygen; Scheme 1) being unaffected by the mutation as indicated by a second-order rate constant of 720 mm−1 s−1 (850 mm−1 s−1 in the wild type) (17). The only step that is perturbed by the mutation is the oxidation of NADPH which occurs with a rate constant (0.5 s−1) that is 24-fold lower than that of the wild-type enzyme, making this step rate-limiting for the overall reaction. Thus, Asp-66 plays a main (but not essential) role for proper NADPH binding and/or oxidation possibly because its negative charge may help in the proper positioning of NADPH within the active site (Scheme 1) and/or because it favors formation of the positively charged oxidized NADP+. The crystal structure of the oxidized D66A shows that the mutation leads to a shift in the Arg-337 position. This side chain cannot bind to the side chain in position 66 and adopts an extended conformation, similar to that observed in the reduced wild-type protein (Fig. 4C). Remarkably, the electron density indicates that a MES molecule is bound in the active site but in a position different from that observed in the wild-type. This ligand is located at the entrance of the funnel, and its sulfonic group is anchored to the guanidinium of the extended Arg-337. It is tempting to speculate that this MES-binding mode represents a position transiently adopted by the substrate during admission into the catalytic site (see “Discussion”).
Reshaping the Substrate Binding Site
Met-446 is located at the bottom of the substrate site (in the orientation of Figs. 2A and 3A) (24). This residue is located in a loop (the so-called “bulge”) that is critical for the enzyme substrate specificity (21). The structure of the enzyme in complex with MES (Fig. 4A) shows that the Met side chain interacts extensively with the ligand on one side of the binding site. The M446G mutant shows profoundly altered properties enabling the enzyme to act on several aromatic compounds that are not substrates of the wild-type protein (e.g. indigo) (24). We were only able to obtain crystal of M446G in the ligand-free form (no NADP+ bound). The structure shows no detectable conformational changes with respect to the wild-type enzyme crystallized under the same conditions. The only effect is that the removal of Met-446 side chain widens the active site, whose shape is substantially altered by the mutation.
DISCUSSION
The binding of NADP+ to PAMO is virtually the same as that observed in the structures of cyclohexanone monooxygenase (23) and flavin-containing monooxygenases (6, 7), which all share the property of utilizing NADP+ to stabilize the crucial flavin-(hydro)peroxide. The data on cyclohexanone monooxygenase suggested a “sliding” process during catalysis. NADPH first binds in the position competent for hydride donation to the flavin N5 atom. Next, NADP+ slides over the flavin to adopt the conformation observed in the NADP+-bound crystal structures. The elucidated PAMO-NADP+ structure fully conforms to this proposal (Scheme 1).
A fundamental question for the functioning of BVMOs and, more generally, for flavin and oxygen enzymology, is to understand the nature of the essential role played by NADP+ in flavin-peroxide stabilization (Scheme 1). This flavin intermediate is characterized by an absorption peak in the 360–390-nm region, often with an extinction coefficient similar to that of the ∼370-nm peak of the fully oxidized flavin (3, 9). As shown in Fig. 3, spectra measured during reoxidation (see spectrum C) are nearly identical to the spectrum of the oxidized enzyme from 360 nm to 390 nm, whereas their absorbance in the 450-nm region is well below that of the oxidized flavin. These findings suggest that a fraction (∼20%) of the crystalline enzyme molecules might be in the flavin-peroxide form during reoxidation. However, crystal spectra never indicated substantial (>50% occupancy) accumulation of the intermediate in the crystals. Apparently, the crystalline environment and highly viscous crystallization solutions prevent intermediate accumulation, possibly because the rate of its formation in the crystals is slower than that of its decay. Furthermore, the putative low occupancy flavin-peroxide intermediate seen in the spectrum is immediately reduced by x-rays. Nevertheless, the structure of reduced PAMO with NADP+ provides considerable insight into flavin-peroxide formation. A key observation is that the carboxamide group of NADP+ establishes an H-bond with the N5 atom of the reduced flavin (Fig. 2B). Such an H-bond protects the reduced flavin from losing its N5 proton to the surrounding medium, and its has been recently highlighted by studies on various enzymes that stabilize the flavin-(hydro)peroxide, including flavin-containing monooxygenases (7), p-hydroxyphenylacetate hydroxylase, and pyranose oxidase (37, 38). Transient proton exchange (with solvent or neighboring groups) is thought to favor the decay of the intermediate, whose stability requires a stably protonated N5 atom (8). In particular, an “unprotected” N5 could donate the proton directly to the distal oxygen of the flavin-peroxide adduct, inevitably leading to intermediate collapse. Thus, the emerging fundamental concept is that NADP+ is required for oxygen-activation primarily by providing essential H-bonding interactions that modulate flavin reactivity.
Another insightful observation from the structure of the reduced enzyme-NADP+ complex concerns Arg-337, which adopts an extended conformation to interact with the negatively charged reduced flavin (Fig. 2B). This conformation obstructs the site for the organic substrate, whose catalytically competent binding is prevented prior to the reaction with dioxygen to maintain the C4a-N5 locus of the reduced flavin readily accessible for dioxygen. Upon formation of the flavin-peroxide and the concomitant loss of the negative charge on the flavin ring, Arg-337 shifts back to the conformation interacting with the NADP+ nicotinamide, making the catalytic center accessible to the substrate (Figs. 2, A and B, and 4, A and C, and Scheme 1). This observation is consistent with multisubstrate kinetic studies. The order of NADPH and substrate binding was studied for the R217A mutant of PAMO, which features a Km for NADPH (320 μm) suited for this type of experiments (the mutation is unlikely to affect the reaction mechanism because Arg-217 is located far away from the flavin) (39). When the observed reaction rates were plotted according to Lineweaver-Burk method, the characteristic pattern of parallel lines was observed, indicating an ordered binding of substrates. In particular, the parallel lines are consistent with a sequential mechanism with an intervening irreversible step (i.e. flavin reoxidation). The binding location of MES in the D66A structure further suggests that the extended conformation of Arg-337 might represent a steering element for the substrate, guiding it into the active site upon flavin-peroxide formation (Scheme 1).
The structure of the enzyme in complex with MES indicates that Arg-337 and the NADP+ ribose cooperate in forming a binding site for a negative charge. We modeled the Criegee intermediate using the complex with MES as reference structure (Fig. 4D). The model was built by positioning the intermediate tetrahedral group on the site occupied by the sulfonic moiety of MES and by positioning the phenylacetone ring in a location overlapping that of MES morpholino group. Such a modeling experiment reveals that the Criegee intermediate can be perfectly accommodated in the active site with Arg-337 and NADP+-ribose acting as the H-bond donors for the negatively charge oxygen of the tetrahedral group. At the same time, the sulfur atom of the MES sulfonic group mimics the carbonyl carbon of the substrate that is covalently attacked by the terminal oxygen of the flavin-peroxide (Fig. 4D). In this scenario, Arg-337 has a dual role in substrate oxygenation: (i) it H-bonds to the carbonyl oxygen of the substrate enhancing the propensity of the carbonyl carbon to undergo the nucleophilic attack by the flavin-peroxide, and (ii) together with the NADP+ ribose, it compensates for the negative charge of the Criegee intermediate.
It is worth noting that the active-site architecture does not identify clear elements for recognition of specific substrate(s). Rather, the active site appears to be primarily designed to activate dioxygen through flavin-peroxide stabilization and to stabilize the Criegee intermediate. In other words, PAMO and similar BVMOs are mainly oxygen-activating and “Criegee-stabilizing” catalysts that act on any chemically suitable substrate that can diffuse into the active site and reach the catalytic center where the flavin-peroxide and oxyanion hole are positioned. Along these lines, it is not surprising that substrate specificity of PAMO and similar enzymes can be manipulated by simple mutations such as those targeting Met-446. These data emphasize the potential of using PAMO and similar BVMOs as toolboxes for biocatalytic applications and support the idea that protein engineering and directed evolution methods can be used effectively to develop finely tuned biocatalysts.
Supplementary Material
Acknowledgments
We thank Dale E. Edmondson (Emory University) and Stefano Franceschini (Pavia) for helpful advice. We acknowledge the European Synchrotron Radiation Facility for provision of beam time.
This work was supported by European Union Project Oxygreen 212281 and Fondazione Cariplo Grant 2008.3148.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
The atomic coordinates and structure factors (codes 2YLR, 2YLS, 2YLT, 2YM1, 2YM2, 2YLW, 2YLX, and 2YLZ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
We repeatedly attempted crystal soaking in phenylacetone or similar substrates without detecting clear binding in the crystals based on the inspection of the electron density maps. Because of the low affinity, it was impossible to obtain an accurate measurement of the Ki for MES.
- BVMO
- Baeyer-Villiger monooxygenase
- PAMO
- phenylacetone monooxygenase.
REFERENCES
- 1. Massey V. (2000) Biochem. Soc. Trans. 28, 283–296 [PubMed] [Google Scholar]
- 2. Ghisla S., Edmondson D. E. (2009) Flavin Coenzymes, John Wiley & Sons, Inc., New York [Google Scholar]
- 3. Mattevi A. (2006) Trends Biochem. Sci. 31, 276–283 [DOI] [PubMed] [Google Scholar]
- 4. van Berkel W. J., Kamerbeek N. M., Fraaije M. W. (2006) J. Biotechnol. 124, 670–689 [DOI] [PubMed] [Google Scholar]
- 5. Criegee R. (1948) Justus Liebigs Ann. Chem. 560, 127–135 [Google Scholar]
- 6. Alfieri A., Malito E., Orru R., Fraaije M. W., Mattevi A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 6572–6577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Orru R., Torres Pazmiño D. E., Fraaije M. W., Mattevi A. (2010) J. Biol. Chem. 285, 35021–35028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jones K. C., Ballou D. P. (1986) J. Biol. Chem. 261, 2553–2559 [PubMed] [Google Scholar]
- 9. Sheng D., Ballou D. P., Massey V. (2001) Biochemistry 40, 11156–11167 [DOI] [PubMed] [Google Scholar]
- 10. van den Heuvel R. H., Tahallah N., Kamerbeek N. M., Fraaije M. W., van Berkel W. J., Janssen D. B., Heck A. J. (2005) J. Biol. Chem. 280, 32115–32121 [DOI] [PubMed] [Google Scholar]
- 11. Mihovilovic M., Müller B., Stanetty P. (2002) Eur. J. Org. Chem. 2002, 3711–3730 [Google Scholar]
- 12. de Gonzalo G., Mihovilovic M. D., Fraaije M. W. (2010) Chembiochem 11, 2208–2231 [DOI] [PubMed] [Google Scholar]
- 13. Fraaije M. W., Wu J., Heuts D. P., van Hellemond E. W., Spelberg J. H., Janssen D. B. (2005) Appl. Microbiol. Biotechnol. 66, 393–400 [DOI] [PubMed] [Google Scholar]
- 14. de Gonzalo G., Ottolina G., Zambianchi F., Fraaije M. W., Carrea G. (2006) J. Mol. Catal. B Enzym. 39, 91–97 [Google Scholar]
- 15. de Gonzalo G. d., Torres Pazmiño D. E., Ottolina G., Fraaije M. W., Carrea G. (2005) Tetrahedron Asymmetry 16, 3077–3083 [Google Scholar]
- 16. Malito E., Alfieri A., Fraaije M. W., Mattevi A. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 13157–13162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Torres Pazmiño D. E., Baas B. J., Janssen D. B., Fraaije M. W. (2008) Biochemistry 47, 4082–4093 [DOI] [PubMed] [Google Scholar]
- 18. Torres Pazmiño D. E., Dudek H. M., Fraaije M. W. (2010) Curr. Opin. Chem. Biol. 14, 138–144 [DOI] [PubMed] [Google Scholar]
- 19. Torres Pazmiño D. E., Snajdrova R., Baas B. J., Ghobrial M., Mihovilovic M. D., Fraaije M. W. (2008) Angew. Chem. Int. Ed. Engl. 47, 2275–2278 [DOI] [PubMed] [Google Scholar]
- 20. Torres Pazmiño D. E., Winkler M., Glieder A., Fraaije M. W. (2010) J. Biotechnol. 146, 9–24 [DOI] [PubMed] [Google Scholar]
- 21. Reetz M. T., Wu S. (2009) J. Am. Chem. Soc. 131, 15424–15432 [DOI] [PubMed] [Google Scholar]
- 22. Wu S., Acevedo J. P., Reetz M. T. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 2775–2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mirza I. A., Yachnin B. J., Wang S., Grosse S., Bergeron H., Imura A., Iwaki H., Hasegawa Y., Lau P. C., Berghuis A. M. (2009) J. Am. Chem. Soc. 131, 8848–8854 [DOI] [PubMed] [Google Scholar]
- 24. Torres Pazmiño D. E., Snajdrova R., Rial D., Mihovilovic M., Fraaije M. W. (2007) Adv. Synth. Catal. 349, 1361–1368 [Google Scholar]
- 25. CCP4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 26. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 29. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Royant A., Carpentier P., Ohana J., McGeehan J., Paetzold B., Noirclerc-Savoye M., Vernede X., Adam V., Bourgeois D. (2007) J. Appl. Crystallogr. 40, 1105–1112 [Google Scholar]
- 31. McGeehan J., Ravelli R. B., Murray J. W., Owen R. L., Cipriani F., McSweeney S., Weik M., Garman E. F. (2009) J. Synchrotron Radiat. 16, 163–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., Murshudov G. N., Cohen S., Perrakis A., Noble M. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2288–2294 [DOI] [PubMed] [Google Scholar]
- 33. Røhr A. K., Hersleth H. P., Andersson K. K. (2010) Angew. Chem. Int. Ed. Engl. 49, 2324–2327 [DOI] [PubMed] [Google Scholar]
- 34. Weik M., Colletier J. P. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Flot D., Mairs T., Giraud T., Guijarro M., Lesourd M., Rey V., van Brussel D., Morawe C., Borel C., Hignette O., Chavanne J., Nurizzo D., McSweeney S., Mitchell E. (2010) J. Synchrotron Radiat. 17, 107–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schlichting I., Berendzen J., Chu K., Stock A. M., Maves S. A., Benson D. E., Sweet R. M., Ringe D., Petsko G. A., Sligar S. G. (2000) Science 287, 1615–1622 [DOI] [PubMed] [Google Scholar]
- 37. Ruangchan N., Tongsook C., Sucharitakul J., Chaiyen P. (2011) J. Biol. Chem. 286, 223–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sucharitakul J., Wongnate T., Chaiyen P. (2011) J. Biol. Chem. 286, 16900–16909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dudek H. M., Torres Pazmiño D. E., Rodríguez C., de Gonzalo G., Gotor V., Fraaije M. W. (2010) Appl. Microbiol. Biotechnol. 88, 1135–1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.