Abstract
Human glioblastoma multiforme cells demonstrate varying levels of sensitivity to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis. Endoplasmic reticulum (ER) stress has been shown to trigger cell death through apoptosis. We therefore pursued a strategy of integrating clinically relevant investigational agents that cooperate mechanistically through the regulation of ER stress and apoptosis pathways. Nelfinavir belongs to the protease inhibitor class of drugs currently used to treat patients with HIV and is in clinical trials as an anti-tumor agent. We found that Nelfinavir treatment led to ER stress-induced up-regulation of the DR5 receptor. This transactivation was mediated by the transcription factor CCAAT/enhancer binding protein homologous protein (CHOP). We also determined that ER stress-induced ATF4 up-regulation was responsible for modulation of CHOP. In contrast, DR4 receptor expression was unchanged by Nelfinavir treatment. Combining Nelfinavir with TRAIL led to a significantly enhanced level of apoptosis that was abrogated by siRNA silencing of DR5. We provide evidence that Nelfinavir-induced ER stress modulates DR5 expression in human glioblastoma multiforme cells and can enhance TRAIL efficacy. These studies provide a potential mechanistic rationale for the use of the Food and Drug Administration-approved agent Nelfinavir in combination with DR5 agonists to induce apoptosis in human malignancies.
Keywords: Apoptosis, Cancer Therapy, Cell Death, ER Stress, Signal Transduction, Death Receptor, TRAIL
Introduction
Malignant gliomas account for ∼70% of the 22,500 new cases of malignant primary brain tumors that are diagnosed in adults in the United States each year (1). Glioblastoma multiforme is a high-grade, highly lethal, and frequent brain tumor in adults. Traditional treatments such as DNA damaging agents and radiotherapy have demonstrated modest effects on patient survival, likely because in part of an inherent high-level expression of anti-apoptotic proteins such as Bcl-2, Bcl-XL, and sFLIP (2, 3). Frequent gene amplification and overexpression of the death decoy receptor DcR3 have also been reported in glioblastoma (4). Therefore, new strategies to induce apoptosis may help to overcome therapeutic resistance and improve efficacy.
Nelfinavir is an HIV protease inhibitor and has been used to treat HIV/AIDS patients for over a decade. Recently, several groups reported the anticancer activity of Nelfinavir in a wide range of human cancer cell lines and tumor xenografts. Nelfinavir induces ER2 stress, the unfolded protein response, autophagy, apoptosis, and caspase-independent cell death in various cancer cells (5–9). Multiple mechanisms have been proposed to explain the anticancer activities of the drug, such as inhibition of AKT/protein kinase B (PKB) activation (10), the proteasome (9), or signal transducer and activator of transcription (STAT) 3 (11), and down-regulation of hypoxia-inducible factor 1α (HIF-1α)/VEGF expression (8). However, the exact molecular mechanisms of Nelfinavir efficacy in human cancer cells remains unclear and may be cell type-specific. We currently have an ongoing Phase I trial of Nelfinavir in high-grade brain tumor patients receiving concomitant temozolomide and radiation therapy (clinicaltrials.gov identifier NCT01020292) and sought to study whether Nelfinavir could enhance the anticancer effects of TRAIL.
TRAIL is a member of the tumor necrosis factor superfamily. TRAIL binds to its receptors DR4/TRAIL-R1 and DR5/TRAIL-R2 and activates the extrinsic pathway of apoptosis (12, 13). Cell killing from these receptors occurs because of recruitment to the receptor of the adaptor protein Fas-associated protein with death domain (FADD), which then recruits the proform of caspase 8 with subsequent aggregation and autoactivation of procaspase 8, ultimately leading to activation of effector caspases such as caspase 3 (14). TRAIL selectively and potently induces apoptosis in a wide spectrum of cancer cells but not in normal cells. Importantly, TRAIL also shows selective toxicity to human tumor xenografts in vivo compared with normal tissue (15, 16). These findings have spurred the incorporation of TRAIL into anticancer regimens. Currently, recombinant human TRAIL (rhTRAIL), and several agonistic monoclonal antibodies are in Phase II clinical trials, including mapatumumab, which targets DR4, as well as lexatumumab, Apomab, AMG655, CS-1008, and LBY-135, all of which target DR5 (12). However, increasing evidence indicates that death receptor agonists alone may not be sufficient to effectively activate apoptosis in many types of cancers, including gliomas (17, 18). One therapeutic approach being tested is to induce expression of death receptors, including DR4 and especially DR5, by small molecules resulting in TRAIL-induced tumor cell death or sensitization of TRAIL-resistant cells. Furthermore, DR5 expression levels have been highly correlated with sensitivity to TRAIL in some cell lines (19). Structurally diverse sets of molecules can act through various molecular mechanisms to perturb normal cellular function and induce death receptor expression (17, 18). As DR5 is a target for p53 transcriptional activation, considerable interest has focused on inducing DR5 expression by increasing chemotherapy-induced p53 signaling. One major drawback to this approach is that the majority of tumors do not express wild-type p53. Alternative approaches and pathways to enhance DR5 signaling are therefore needed.
In this study, we show that Nelfinavir sensitizes human glioblastoma cells to TRAIL treatment by up-regulation of the DR5 receptor and thus enhances the extrinsic pathway of apoptosis in U251 cells that harbor mutations in p53. Our data suggest that up-regulation of DR5 is mediated by Nelfinavir-induced ER stress. We also establish CHOP as a critical mediator of the protein kinase RNA-like endoplasmic reticulum kinase (PERK)/eIF2α/ATF4 pathway after Nelfinavir treatment, which ultimately results in up-regulation of DR5 in glioblastoma multiforme cells and sensitization to TRAIL.
EXPERIMENTAL PROCEDURES
Cell Lines and Reagents
U251, A172, and U373 glioma cell lines were cultured in DMEM (4.5g/L, glucose, Mediatech, Manassas, VA) supplemented with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin in a humidified incubator at 37 °C and 5% CO2. DLD1shNT and DLD1shATF4 human colorectal adenocarcinoma cells were cultured in DMEM as above with addition of 1% non-essential amino acids (Invitrogen) and 1% ES cell qualified 2-mercaptoethanol (Millipore, Billerica, MA). Nelfinavir (Toronto Research Chemicals, Inc., Canada), tunicamycin (Sigma-Aldrich), JNK inhibitor SP600125 (EMD Chemicals, Inc., Gibbstown, NJ), and pan-caspase inhibitor z-VAD-fmk (Biomol Research Laboratories, Inc., Plymouth Meeting, PA) were dissolved in dimethyl sulfoxide and stored at −20 °C. Human recombinant TRAIL (PeproTech, Inc., Rocky Hill, NJ) and TRAIL-R2/Fc chimera (R&D Systems, Minneapolis, MN).
Immunoblots and Antibodies
Cells were lysed in 2× Laemmli sample buffer, and an equal amount of cell lysate was processed by immunoblot analysis according to established protocol. Primary antibodies to caspase 3, caspase 8, caspase 9, death receptor 5 (DR5), phosphorylated-eIF2α (Ser-51), poly (ADP-ribose) polymerase were from Cell Signaling Technology, Inc. at 1:1000 dilution. CHOP/GADD153, ATF4, glucose-regulated protein/BiP (GRP78), eukaryotic initiation factor 2α (eIF2α), and Ras-related nuclear protein (Ran) were from Santa Cruz Biotechnology, Inc. and used at a 1:1000 dilution. The horseradish peroxidase-conjugated secondary antibodies were from Sigma Aldrich. Antibody binding was detected by chemiluminescence using ECL Western blotting detection reagents (GE Healthcare).
Cell Viability Assay
A cell viability assay (ATP level assay) was performed using a CellTiter-Glo® luminescent assay kit (Promega). Briefly, cells were seeded at 5 × 103-1 × 104cells/well in black 96-well plates and incubated at 37 °C overnight. Cells were treated with 20 μm Nelfinavir for 24 and 48 h. The viability assays were performed according to manufacturer's protocol, and luminescence of the plates was recorded on a microplate luminometer (Luminoskan Ascent) from Thermo Scientific (Waltham, MA).
Caspase-3/7 Activity Assay
Caspase 3/7 activity was measured by using the Caspase-Glo® 3/7 Assay kit (Promega) according to the manufacturer's protocol. In brief, cells were seeded at 1 × 104cells/well in black 96-well plates and incubated at 37 °C overnight. Cells were treated with 20 μm Nelfinavir for 24 h. Caspase activity was measured on a microplate luminometer (Luminoskan Ascent) from Thermo Scientific (Waltham, MA).
Cell Cycle Analysis
Propidium iodide (PI) staining and flow cytometry were used to determine the degree of cellular apoptosis. Cells were seeded at 3 × 105cells/well in six-well plates and incubated overnight before the experiment. Cells were treated with 20 μm Nelfinavir for 24 h. Floating and adherent cells were collected and resuspended in PBS with 1% FBS. Cold ethanol was added in a dropwise manner while vortexing. Cells were fixed over 20 min at 4 °C, washed with 1 ml PBS, resuspended in 300 ml PBS with 1% FBS, 5 mg RNase A, and 15 mg PI. Then, cells were stored for 30 min at room temperature in the dark. Flow cytometry was done using a Beckman Coulter Elite Epics sorter. The percentage of hypodiploid cells (sub-G1) was used to quantify dead cells in apoptosis assays.
siRNA
Cells were seeded at 3 × 105cells/well in six-well plates and incubated at 37 °C to reach ∼60% confluence on the day of transfection. The following annealed double-stranded siRNAs were obtained from Ambion, Inc. (Applied Biosystems, Inc./Ambion, Inc., Austin, TX): CHOP Silencer® Selected IDs 225792, DR5 Silencer® Selected IDs16757. Negative control siRNA Silencer® Selected, catalog no. 4390844, from Ambion, Inc. was used as a negative control. Cells were transfected with 10–20 nm siRNA diluted in Opti-MEM medium (Invitrogen) using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's transfection protocol. Efficiency of siRNA was measured by Western blot analysis 24 h after transfection.
RT-PCR
Total RNA was isolated with an RNeasy Mini Kit (Qiagen). Reverse transcription PCR was carried out using the Titan one-tube PCR system (Roche). Primer sequences were as follows: DR5, 5′-CAG AGG GAT TGT GTC CAC CT-3′ (sense) and 5′-TAC GGC TGC AAC TGT GAC TC-3′ (antisense); GAPDH, 5′-CCT GAC CTG CCG TCT AGA AA-3′ (sense) and 5′-TTA CTC CTT GGA GGC CAT GT-3′ (antisense) (Integrated DNA Technologies, Coralville, IA). The samples were placed in a thermocycler, 100 ng RNA for GADPH (25 cycles), 500 ng RNA for DR5 (25 cycles), equilibrated at 50 °C, and incubated for 30 min for reverse transcription. The templates were denatured at 94 °C for 2 min. The PCR cycle was as follows: Denaturation at 94 °C for 30 s, annealing at 52 °C for 30 s, and elongation at 68 °C (1 min).
Luciferase Promoter Assay
Plasmid pDR5 contains human DR5 promoter that has a CHOP binding site. The construct pDR5-mCHOP has a mutation in the CHOP binding site at −272/-269. pGL3-Base is an empty vector. The plasmids were constructed as described previously (20). Cells were cultured in 24-well plates and co-transfected with 20 ng of the Renilla luciferase plasmid pRL-SV40 as a normalization control and 200 ng of firefly luciferase constructs containing the DR5 promoter region using Lipofectamine 2000. After 12 h, the cells were transferred into 96-well plates and further cultured for 12 h. Transfected cells were then treated with 20 μm NFV or DMSO for an additional 24 h. Luciferase activity was measured by using the Dual-GloTM luciferase assay system (Promega) according to the manufacturer's instructions. Firefly luciferase activity was normalized by Renilla luciferase activity.
RESULTS
Nelfinavir Treatment Induces DR5 Expression but Not Significant Apoptosis in Glioblastoma Multiforme Cells
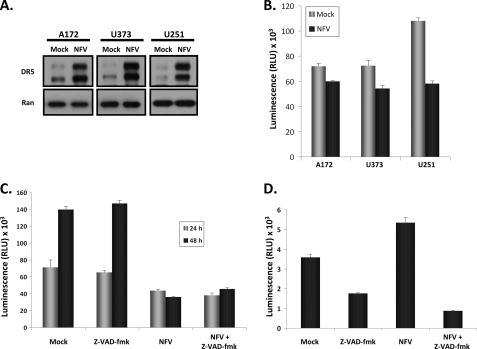
To assess the sensitivity of glioblastoma multiforme to Nelfinavir alone, three glioma cell lines, A172 (p53 wild-type), U373 (p53 mutant), and U251 (p53 mutant), were treated with Nelfinavir. We observed a marked induction of DR5 receptor expression independent of p53 status (Fig. 1A). Flow cytometry and immunofluorescence studies of DR5 demonstrated this expression to be localized to the cell surface (data not shown). Conversely, we did not detect any appreciable increase in the DR4 receptor after Nelfinavir treatment in all cell lines tested (data not shown). We next assessed cellular proliferation and viability in response to Nelfinavir treatment by measuring ATP levels (Fig. 1B). Nelfinavir treatment reduced the viable cell numbers in all cell lines tested, with U251 being the most sensitive (Fig. 1B). To determine whether the Nelfinavir-induced reduction in viable cell number was due to DR5-mediated apoptosis, we assessed U251 viable cell numbers after Nelfinavir treatment in the presence or absence of the pan-caspase inhibitor z-VAD-fmk. As shown in Fig. 1C, inhibition of caspase activation (by z-VAD-fmk pretreatment) did not significantly affect the number of viable cells after exposure to Nelfinavir compared with mock pretreatment, suggesting that the treatment effect on U251 was not directly linked to apoptosis. Furthermore, we also assessed caspase 3/7 activity and found that Nelfinavir led to only a modest increase in caspase activity in U251 cells, which was abrogated by z-VAD-fmk (Fig. 1D). Taken together, these results indicate that Nelfinavir treatment alone inhibits the proliferation of human glioblastoma cells and up-regulates expression of the DR5 receptor but that this increased DR5 expression alone is insufficient for the initiation of apoptosis.
FIGURE 1.
Nelfinavir treatment induces DR5 up-regulation and reduces proliferation in human glioblastoma multiforme cells. A, human glioblastoma multiforme cells A172 (left panel), U373 (center panel), and U251 (right panel) were treated with Nelfinavir (20 μm) for 24 h, and then whole cell lysates were immunoblotted for expression of DR5 (upper panels) or Ran (lower panels) to confirm equal loading of protein. B, A172, U373, and U251 cells were either mock-treated or treated with Nelfinavir (NFV) (20 μm) for 24 h, and then viable cell numbers were determined by the CellTiter-Glo luminescent cell viability assay. C, U251 cells were mock-treated or treated with Nelfinavir (20 μm) in the presence or absence of z-VAD-fmk for 24 or 48 h, and then viable cell numbers were determined by the CellTiter-Glo luminescent cell viability assay. D, U251 cells were mock-treated or treated with Nelfinavir (20 μm) in the presence or absence of z-VAD-fmk for 24 h, and then caspase 3/7 activity was determined by the Caspase-Glo 3/7 assay. All bar graphs and error bars represent means and S.D., respectively. Luminescence assays were conducted in triplicate in two independent experiments.
Nelfinavir Enhances TRAIL-mediated Apoptosis
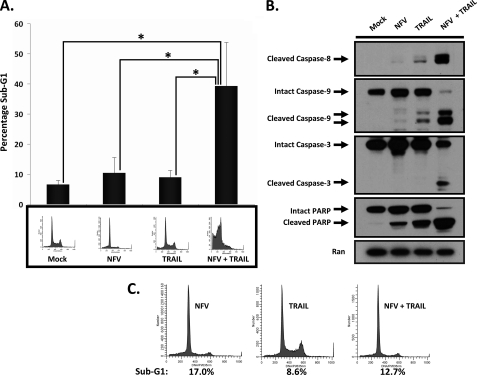
Because Nelfinavir treatment alone was unable to initiate apoptosis, we sought to determine whether Nelfinavir-induced DR5 expression would result in enhanced sensitivity of U251 cells to TRAIL treatment. To assess this, we pretreated U251 cells with Nelfinavir before addition of TRAIL. Analysis of sub-G1 populations demonstrated that Nelfinavir alone did not significantly increase the number of cells with hypodiploid DNA, indicating a lack of apoptotic cell death (Fig. 2A) in agreement with our previous results (Fig. 1). A longer period of drug treatment (48 h) also failed to show a significant increase in cell death when compared with 24 h of treatment (data not shown). As shown in Fig. 2A, Nelfinavir (10.4% sub-G1) or TRAIL (8.9% sub-G1) alone did not induce significant cell death compared with the mock-treated control (6.5% sub-G1), whereas pretreatment with Nelfinavir potently and significantly enhanced TRAIL-induced apoptosis (39.2% sub-G1). Consistent with these findings, pretreatment with Nelfinavir followed by TRAIL treatment markedly increased activation of caspase 8, caspase 9, caspase 3, and cleavage of poly (ADP-ribose) polymerase, as demonstrated in the immunoblot analysis (Fig. 2B). Combined treatment with Nelfinavir and TRAIL augmented the activation of caspase 9, suggesting that Nelfinavir also modulates the intrinsic pathway of apoptosis. In contrast to our findings in U251, normal human astrocytes did not demonstrate significant apoptosis after the combination treatment of Nelfinavir and TRAIL under similar conditions, suggesting a therapeutic window (data not shown). To further establish that TRAIL-mediated apoptosis was primarily due to an increase in DR5 expression, we utilized a human TRAIL-R2/Fc chimera to neutralize TRAIL-induced cell death. The TRAIL-R2/Fc chimera protein has an extracellular domain of DR5 that can bind to TRAIL but does not have a death domain. As shown in Fig. 2C, pretreatment with the TRAIL-R2/Fc chimera effectively abrogated the combined effect of Nelfinavir and TRAIL. These results indicate that Nelfinavir efficiently sensitizes U251 cells to TRAIL-induced apoptosis.
FIGURE 2.
Nelfinavir and TRAIL cotreatment lead to a significant increase in caspase activation and apoptotic cell death in human glioblastoma multiforme cells. A, U251 cells were either mock-treated or treated with Nelfinavir (NFV) (20 μm) alone for 20 h, TRAIL (25 ng/ml) alone for 3 h, or Nelfinavir (20 μm) for 17 h followed by addition of TRAIL (25 ng/ml) for 3 h, and then the percentage of sub-G1 cells was determined by propidium iodide staining and flow cytometry. Sub-G1 analysis was conducted in triplicate in two independent experiments. The bottom panel shows representative flow cytometry results. The bar graph and error bars represent means and S.D., respectively. *, p < 0.05. B, U251 cells were either-mock treated or treated with Nelfinavir (20 μm) alone for 20 h, TRAIL (25 ng/ml) alone for 3 h, or Nelfinavir (20 μm) for 17 h followed by addition of TRAIL (25 ng/ml) for 3 h, and then whole cell lysates were immunoblotted for caspase 8, caspase 9, caspase 3, poly (ADP-ribose) polymerase (PARP), and Ran, as indicated. C, U251 cells in the presence of the TRAIL-R2/Fc chimera (2 ng/ml) were either treated with Nelfinavir (20 μm) alone for 20 h, TRAIL (25 ng/ml) alone for 3 h, or Nelfinavir (20 μm) for 17 h followed by addition of TRAIL (25 ng/ml) for 3 h, and then the percentage of sub-G1 cells was determined by flow cytometry.
Nelfinavir Up-regulates DR5 mRNA Levels and DR5 Expression Is Required for Nelfinavir-mediated Enhancement of TRAIL-induced Apoptosis
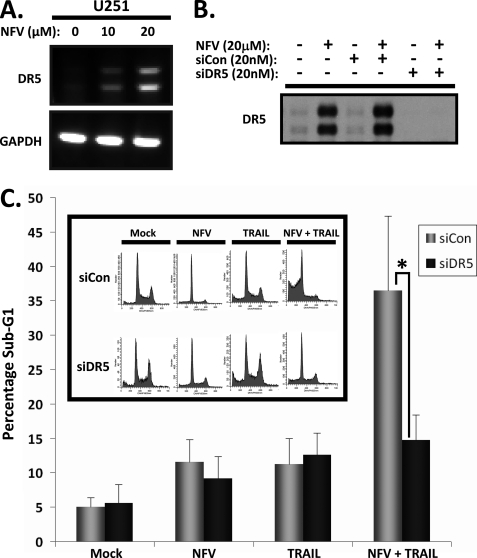
We hypothesized that DR5 up-regulation was required for Nelfinavir-induced enhancement of TRAIL-induced apoptosis. To determine whether Nelfinavir was inducing DR5 on the transcriptional level in glioblastoma cells, we first performed RT-PCR of DR5 mRNA. As shown in Fig. 3A, treatment of U251 cells with Nelfinavir resulted in a dose-dependent increase in the level of DR5 mRNA. To further confirm transcriptional regulation we knocked down DR5 expression utilizing siRNA against DR5. In U251 cells, DR5 siRNA successfully achieved complete knockdown of the minimal baseline DR5 expression as well as that induced by Nelfinavir treatment (Fig. 3B). As expected, cells with silenced DR5 expression (siDR5) showed much lower sub-G1 DNA fragmentation (15.6% sub-G1) compared with those transfected with control siRNA (siCon) (36.6% sub-G1) (Fig. 3C) when treated with both Nelfinavir and TRAIL. This indicates that DR5 knockdown directly results in resistance to Nelfinavir-mediated TRAIL-induced apoptosis. No difference was seen in percentage of sub-G1 cells after treatment with Nelfinavir or TRAIL alone after DR5 silencing (Fig. 3C). We therefore conclude that transcriptionally up-regulated DR5 expression is required for the enhancement of TRAIL-induced apoptosis by Nelfinavir.
FIGURE 3.
Nelfinavir induces transactivation of DR5, and silencing of DR5 receptor expression rescues U251 cells from apoptosis induced by Nelfinavir and TRAIL cotreatment. A, U251 cells were mock-treated (0) or treated with 10 or 20 μm of Nelfinavir for 24 h, and then cells were harvested for RT-PCR evaluation of DR5 mRNA (top panel). RT-PCR of GAPDH mRNA was also performed as indicated to confirm equal loading (bottom panel). B, U251 cells were either transfected with control siRNA (siCon) or DR5-specific siRNA (siDR5), and complete knockdown of Nelfinavir-induced DR5 protein expression was confirmed by immunoblotting of whole cell lysates for DR5. C, following transfection with control siRNA or DR5-specific siRNA, as indicated, U251 cells were either mock-treated or treated with Nelfinavir (20 μm) alone for 20 h, TRAIL (25 ng/ml) alone for 3 h, or Nelfinavir (20 μm) for 17 h followed by addition of TRAIL (25 ng/ml) for 3 h, and then the percentage of sub-G1 cells was determined by flow cytometry. Sub-G1 analysis was conducted in triplicate in two independent experiments. The bar graph and error bars represent means and S.D., respectively. *, p < 0.05. The inset shows representative flow cytometry results after indicated siRNA transfection and treatment.
CHOP Mediates Up-regulation of DR5 in Response to Nelfinavir
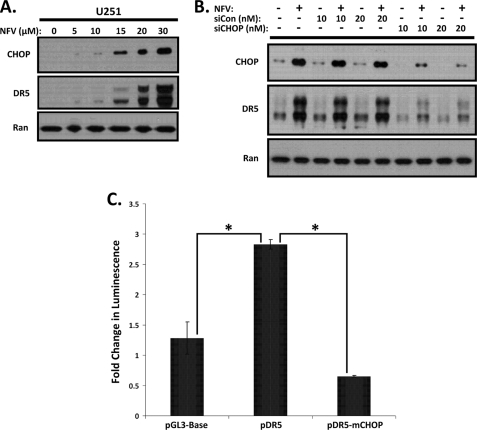
Previous studies have identified CHOP as an upstream regulator of DR5 (20), so we speculated that this member of the CCAAT/enhancer-binding protein family might be responsible for the p53-independent up-regulation of DR5 expression in response to Nelfinavir treatment. We first conducted a dose-response experiment to assess induction of CHOP and DR5 with increasing concentrations of Nelfinavir. As shown in Fig. 4A, expression levels of DR5 correlated closely with increased protein levels of CHOP, suggesting that CHOP was likely responsible for transactivation of DR5. To verify the role of CHOP in the up-regulation of DR5 by Nelfinavir, we silenced CHOP expression with siRNA in U251 cells. As shown in Fig. 4B, immunoblot analysis indicated that siRNA knockdown of CHOP mRNAs significantly inhibited CHOP expression and its subsequent up-regulation of DR5. To confirm that CHOP was responsible for DR5 transcriptional activation, we utilized luciferase reporters driven by a portion of the DR5 promoter containing the CHOP binding site (5′ flanking region of DR5 between −552 and −7). The pDR5 reporter contains the CHOP binding site, whereas the pDR5-mCHOP reporter contains a mutation in the CHOP binding site (20). As shown in Fig. 4C, a significant increase in DR5 promoter activity over the empty luciferase reporter plasmid in transfected U251 cells was seen after Nelfinavir treatment. Mutation of the CHOP binding site led to a pronounced and significant reduction in luciferase reporter activity after Nelfinavir treatment. These results demonstrate that CHOP transactivation is largely responsible for the induction of DR5 after exposure to Nelfinavir.
FIGURE 4.
Nelfinavir treatment leads to CHOP-mediated up-regulation of DR5 in U251 cells. A, U251 cells were mock-treated (0) or treated with increasing concentrations (5, 10, 15, 20, 30 μm) of Nelfinavir as indicated for 24 h, and then whole cell lysates were immunoblotted for dose-dependent expression of CHOP (upper panel). Immunoblotting for the DR5 receptor (center panel) and Ran (lower panel) protein expression was also performed. B, following transfection with 0 (−), 10, or 20 nm of control siRNA (siCon) or CHOP-specific siRNA (siCHOP) as indicated, U251 cells were either mock-treated (−) or treated (+) with Nelfinavir (NFV) (20 μm) for 24 h, and then whole cell lysates were immunoblotted for CHOP (upper panel), DR5 (center panel), or Ran (lower panel). C, U251 cells were transfected with a Renilla luciferase internal control plasmid along with pGL3-Base, pDR5, or pDR5-mCHOP luciferase reporters, as indicated, and then either mock-treated or treated with Nelfinavir (20 μm) for 24 h. The fold change in luminescence of Nelfinavir treatment over mock treatment was calculated for each reporter plasmid after normalizing for transfection efficiency using a Renilla luciferase signal. The bar graph and error bars represent means and S.D., respectively. *, p < 0.05. Luminescence assays were conducted in triplicate in two independent experiments.
Nelfinavir Induces ER Stress and Activates ATF4
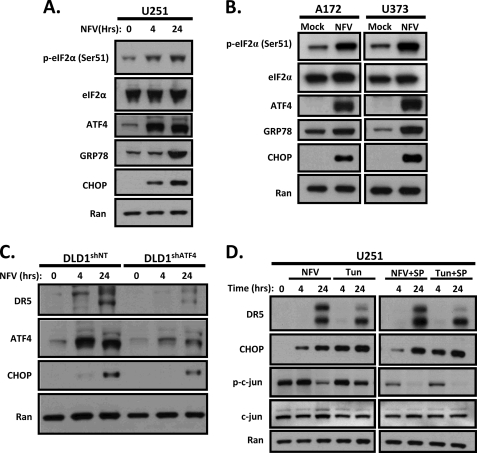
CHOP is a major transcription factor induced by ER stress that has been linked to expression of DR5 in a p53-independent manner. Therefore, we hypothesized that Nelfinavir induces ER stress in glioblastoma cells. To confirm activation of ER stress by Nelfinavir, we probed for three additional markers of ER stress: Ser-51 phosphorylation of eIF2α (P-eIF2α), induction of ATF4 transcription factor and ER chaperone protein GRP78/Bip. As shown in Fig. 5, exposure to Nelfinavir resulted in increased phosphorylation of eIF2α (Ser51) and induction of ATF4, CHOP and GRP78 in U251 (Fig. 5A), A172 and U373 cells (Fig. 5B). The increase in P-eIF2α, ATF4, and CHOP was evident at 4 h post-treatment and sustained at 24 h in U251 cells (Fig. 5A). Expression of DR5 lagged behind phosphorylation of eIF2α and induction of ATF4 and CHOP but was evident at 24 h (Fig. 5, A and D). Because ATF4 is the primary regulator of CHOP transcription during ER stress, we hypothesized that knockdown of ATF4 would result in diminished induction of CHOP and DR5 after Nelfinavir treatment. To test this, we used DLD1 human colorectal cancer cells stably transfected with shRNA against ATF4 (DLD1shATF4) (21). As expected, CHOP induction was attenuated in DLD1shATF4 cells compared with stable DLD1 cells expressing nonspecific shRNA (Fig. 5C). Furthermore, DLD1shATF4 cells demonstrated decreased caspase 8 activation after Nelfinavir + TRAIL treatment compared with their scrambled shRNA-expressing counterparts, suggesting an attenuation of apoptosis (data not shown). Together, these observations support the hypothesis that Nelfinavir treatment leads to an induction of ER stress and up-regulation of DR5, which is mediated by ATF4 and CHOP.
FIGURE 5.
ATF4 mediates Nelfinavir-induced transactivation of CHOP and DR5 that is independent of JNK activation. A, U251 cells were mock-treated (0) or treated with Nelfinavir (NFV) (20 μm) for 4 or 24 h then whole cell lysates were immunoblotted for P-eIF2α, eIF2α, ATF4, GRP78, CHOP, and Ran, as indicated. B, A172 and U373 cells were mock-treated or treated with Nelfinavir (20 μm) for 24 h, and then whole cell lysates were immunoblotted for P-eIF2α, eIF2 α, ATF4, GRP78, CHOP, and Ran, as indicated. C, stable DLD1 cells (shNT or shATF4) were mock-treated (0) or treated with Nelfinavir (20 μm) for 4 or 24 h, and then whole cell lysates were immunoblotted for DR5, ATF4, CHOP, and Ran, as indicated. D, U251 cells were mock-treated (0) or treated with Nelfinavir (20 μm) or tunicamycin (1 μg/ml) for 4 or 24 h in the presence (+ SP) or absence of JNK inhibitor SP600125 (20 μm), and then whole cell lysates were immunoblotted for DR5, CHOP, P-c-jun, c-jun, or Ran, as indicated.
DR5 Up-regulation by Nelfinavir Is Not Dependent on JNK in Human Glioblastoma Cells
ER stress can also trigger the IRE1/ASK1/JNK signaling pathway, and JNK-dependent up-regulation of DR5 has been reported in human non-small cell lung cancer cell lines (22). JNK activates the AP-1 transcription factor, which has binding sites in both the DR5 and CHOP promoters. Additionally, the proteasome inhibitor MG132 can induce ER stress-dependent up-regulation of DR5 through activating the JNK/AP-1 signaling pathway (23). To test if Nelfinavir may also be up-regulating DR5 through activation of the JNK/AP-1 pathway, we studied the effects of Nelfinavir on DR5 and CHOP up-regulation in the presence of JNK inhibitor SP600125. The JNK-specific inhibitor SP600125 abrogates JNK activation and up-regulation of DR5 after C-28 methyl ester of 2-cyano-3,12-dioxoolen-1,9-dien-28-oic acid (CDDO-Me) treatment (22, 24). We also exposed U251 cells to the known ER stress inducer tunicamycin (a glycosylation inhibitor) as a positive control. As shown in Fig. 5D, inhibition of JNK signaling had no apparent effect on Nelfinavir- or tunicamycin-triggered induction of CHOP or DR5. JNK was effectively inhibited by SP600125, as evidenced by the reduced levels of phosphorylated c-Jun (Fig. 5D). Interestingly, Nelfinavir also appeared to inhibit phosphorylation of c-Jun. Together, these findings support the notion that Nelfinavir-induced, CHOP-directed DR5 up-regulation occurs in a JNK-independent manner in human glioblastoma cells.
DISCUSSION
Accumulating evidence supports the notion that ER stress-inducing agents induce up-regulation of DR5 in a wide spectrum of human cancers through various mechanisms (20, 23, 25–28). DR5 modulation may have potential therapeutic value if combined with TRAIL in a clinically relevant manner. Tunicamycin is an antibiotic that blocks the reaction in the first step of N-glycoprotein synthesis, resulting in accumulation of immature proteins that trigger ER stress (29). Thapsigargin induces ER stress by depletion of Ca2+ within the ER lumen through inhibition of the ER Ca2+ pump (30), and MG132 induces ER stress by inhibition of the proteasome, which results in accumulation of unwanted proteins in the cytosol. One major drawback to the utilization of these drugs is that none are Food and Drug Administration-approved or as yet demonstrated to be safe in humans. Recently, it was reported that the Food and Drug Administration-approved drug Nelfinavir could induce up-regulation of DR5 in ovarian cancer cells. However, the mechanism and whether this resulted in TRAIL-sensitization was not addressed (31). We currently have an ongoing clinical trial of Nelfinavir in patients with glioblastoma multiforme (clinicaltrials.gov identifier: NCT01020292) and therefore sought to characterize the mechanism of Nelfinavir-induced DR5 up-regulation.
Our data suggest that the clinically relevant drug Nelfinavir potently induces the expression of DR5 in glioblastoma cells in a CHOP-dependent manner. It has been reported that DR5 is also a target gene of tumor suppressor p53 (32). However, the mutant p53-expressing cell lines U251 and U375 showed similar increases in DR5 up-regulation as the wild-type p53-expressing cell line A172 after exposure to Nelfinavir (Fig. 1A). On the other hand, the p53 wild-type SF767 cell line, after treatment with Nelfinavir, did not show significant induction of CHOP or DR5 (data not shown). Taken together, these data indicated that the observed DR5 induction by Nelfinavir is p53-independent. We also show that the clinically relevant compound Nelfinavir induces ER stress in an ATF4-CHOP-dependent, JNK-independent manner. ER stress can serve dual functions as either cytoprotective or cytotoxic, depending on the length and amplitude of the response (33). It is unclear at this time if Nelfinavir-induced ER stress serves one or both of these functions in glioblastoma multiforme cells. CHOP has traditionally been viewed as a proapoptotic player (34) in response to ER stress but in certain contexts may play an adaptive role (35). In our studies, CHOP induction was primarily responsible for DR5 up-regulation but did not contribute to increased cell death in response to Nelfinavir in the absence of TRAIL. We and others have shown that ATF4 is overexpressed in many human tumors (including gliomas) compared with normal tissues (36), and recently, several groups (37, 38) reported that as part of the unfolded protein response during hypoxia, ATF4 and CHOP up-regulation induce cytoprotective autophagy. It is tempting to speculate that TRAIL treatment may represent a potential therapy to overcome this adaptive survival mechanism in cancer cells.
It is common that human glioma cell lines express low levels of DR5 and are resistant to TRAIL (23, 39–41). In our studies, DR5 up-regulation alone was insufficient for induction of apoptosis in human glioblastoma cells. However, the increased expression of DR5 did lead to enhanced sensitivity to TRAIL, a therapeutic agent currently in late-stage clinical trials. We demonstrated that siRNA directed against DR5 abrogated this increased sensitivity. The fact that Nelfinavir-induced DR5 up-regulation alone was insufficient to induce apoptosis but profoundly sensitized cells to TRAIL strongly suggests that functional DR5 is expressed in response to ER stress. Moreover, flow cytometry studies confirmed localization of DR5 to the cell surface after Nelfinavir treatment. We also found that Nelfinavir treatment led to a decrease in the levels of Bcl-2 expression and increases in both phospho-Bcl-2 and Bim (data not shown). We speculate that Bcl-2 down-regulation could also be a CHOP-dependent process, as reported previously (42). This modulation of Bcl-2 family members likely also contributes to the strong apoptotic response to TRAIL in the setting of ER stress, with DR5 up-regulation being the primary driver.
As mentioned previously, it was reported that the proteasome inhibitor MG132-induced DR5 up-regulation was JNK-dependent and CHOP-independent in U251 and U373 cells (23). Our findings show that in contrast, Nelfinavir-induced DR5 up-regulation is CHOP-dependent and JNK-independent in U251 cells. In addition, a recent publication demonstrated that Nelfinavir promotes proteasome-dependent degradation of Cdc25A phosphatase, which is incompatible with the proteasome inhibition hypothesis (7). Our data and other investigators' work suggest that the mechanism of induction of ER stress by Nelfinavir is likely not generated through inhibition of the proteasome in our experiments in glioblastoma multiforme cells.
In search of coordinated therapies that sensitize glioma cells to TRAIL treatment, Nelfinavir becomes an intriguing candidate. Nelfinavir not only induces ER stress but also has multiple effects of apoptosis, autophagy, caspase-independent cell death in cancer cells (6), and increased oxygenation in the tumor microenvironment (8). Importantly, in contrast to other ER stress inducers mentioned above, Nelfinavir is a Food and Drug Administration-approved drug and has been used to treat AIDS patients safely for more than a decade. Our data clearly show that treatment with Nelfinavir potently increases DR5 expression at both mRNA and protein levels through an ER stress-ATF4-CHOP-DR5 pathway. Furthermore, this death receptor up-regulation potentiated TRAIL-induced apoptosis in human glioblastoma multiforme cells at Nelfinavir concentrations that are likely within the achievable plasma concentration range in humans (6). Resistance to TRAIL is currently one of the hurdles in the development of this agent for cancer therapy. Thus, Nelfinavir or similar Food and Drug Administration-approved agents may potentially represent drugs that are capable of overcoming this resistance by driving ER stress-induced DR5 expression and TRAIL sensitization. Further studies are needed to test this hypothesis and are ongoing.
Acknowledgments
We thank Dr. Hong-Gang Wang (Penn State Milton S. Hershey Medical Center) for graciously providing the pGL3-Base, pDR5-mCHOP, and pDR5 plasmids. We also thank David Dicker for technical assistance with the flow cytometry studies and Drs. Gary Kao and Amit Maity for many helpful discussions.
This work was supported by the Burroughs Wellcome Career Award for Medical Scientists 1006792 (to J. F. D.).
- ER
- endoplasmic reticulum
- TRAIL
- tumor necrosis factor-related apoptosis-inducing ligand
- DR5
- death receptor 5
- eIF2α
- eukaryotic initiation factor 2α
- PI
- propidium iodide.
REFERENCES
- 1. Wen P. Y., Kesari S. (2008) N. Engl. J. Med. 359, 492–507 [DOI] [PubMed] [Google Scholar]
- 2. Rich J. N., Bigner D. D. (2004) Nat. Rev. Drug. Discov. 3, 430–446 [DOI] [PubMed] [Google Scholar]
- 3. Steinbach J. P., Weller M. (2004) J. Neurooncol. 70, 245–254 [DOI] [PubMed] [Google Scholar]
- 4. Arakawa Y., Tachibana O., Hasegawa M., Miyamori T., Yamashita J., Hayashi Y. (2005) Acta Neuropathol. 109, 294–298 [DOI] [PubMed] [Google Scholar]
- 5. Bruning A., Burger P., Vogel M., Rahmeh M., Gingelmaiers A., Friese K., Lenhard M., Burges A. (2009) Cancer Biol. Ther. 8, 226–232 [DOI] [PubMed] [Google Scholar]
- 6. Gills J. J., Lopiccolo J., Tsurutani J., Shoemaker R. H., Best C. J., Abu-Asab M. S., Borojerdi J., Warfel N. A., Gardner E. R., Danish M., Hollander M. C., Kawabata S., Tsokos M., Figg W. D., Steeg P. S., Dennis P. A. (2007) Clin. Cancer Res. 13, 5183–5194 [DOI] [PubMed] [Google Scholar]
- 7. Jiang W., Mikochik P. J., Ra J. H., Lei H., Flaherty K. T., Winkler J. D., Spitz F. R. (2007) Cancer Res. 67, 1221–1227 [DOI] [PubMed] [Google Scholar]
- 8. Pore N., Gupta A. K., Cerniglia G. J., Jiang Z., Bernhard E. J., Evans S. M., Koch C. J., Hahn S. M., Maity A. (2006) Cancer Res. 66, 9252–9259 [DOI] [PubMed] [Google Scholar]
- 9. Pyrko P., Kardosh A., Wang W., Xiong W., Schönthal A. H., Chen T. C. (2007) Cancer Res. 67, 10920–10928 [DOI] [PubMed] [Google Scholar]
- 10. Gupta A. K., Li B., Cerniglia G. J., Ahmed M. S., Hahn S. M., Maity A. (2007) Neoplasia 9, 271–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ikezoe T., Saito T., Bandobashi K., Yang Y., Koeffler H. P., Taguchi H. (2004) Mol. Cancer Ther. 3, 473–479 [PubMed] [Google Scholar]
- 12. Ashkenazi A. (2008) Nat. Rev. Drug Discov. 7, 1001–1012 [DOI] [PubMed] [Google Scholar]
- 13. Ashkenazi A., Dixit V. M. (1998) Science 281, 1305–1308 [DOI] [PubMed] [Google Scholar]
- 14. Thorburn A. (2004) Cell. Signal. 16, 139–144 [DOI] [PubMed] [Google Scholar]
- 15. Ashkenazi A., Pai R. C., Fong S., Leung S., Lawrence D. A., Marsters S. A., Blackie C., Chang L., McMurtrey A. E., Hebert A., DeForge L., Koumenis I. L., Lewis D., Harris L., Bussiere J., Koeppen H., Shahrokh Z., Schwall R. H. (1999) J. Clin. Invest. 104, 155–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Walczak H., Miller R. E., Ariail K., Gliniak B., Griffith T. S., Kubin M., Chin W., Jones J., Woodward A., Le T., Smith C., Smolak P., Goodwin R. G., Rauch C. T., Schuh J. C., Lynch D. H. (1999) Nat. Med. 5, 157–163 [DOI] [PubMed] [Google Scholar]
- 17. Elrod H. A., Sun S. Y. (2008) Cancer Biol. Ther. 7, 163–173 [DOI] [PubMed] [Google Scholar]
- 18. Johnstone R. W., Frew A. J., Smyth M. J. (2008) Nat. Rev. Cancer 8, 782–798 [DOI] [PubMed] [Google Scholar]
- 19. Jang Y. J., Park K. S., Chung H. Y., Kim H. I. (2003) Cancer Lett. 194, 107–117 [DOI] [PubMed] [Google Scholar]
- 20. Yamaguchi H., Wang H. G. (2004) J. Biol. Chem. 279, 45495–45502 [DOI] [PubMed] [Google Scholar]
- 21. Ye J., Kumanova M., Hart L. S., Sloane K., Zhang H., De Panis D. N., Bobrovnikova-Marjon E., Diehl J. A., Ron D., Koumenis C. (2010) EMBO J. 29, 2082–2096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zou W., Yue P., Khuri F. R., Sun S. Y. (2008) Cancer Res. 68, 7484–7492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hetschko H., Voss V., Seifert V., Prehn J. H., Kögel D. (2008) FEBS J. 275, 1925–1936 [DOI] [PubMed] [Google Scholar]
- 24. Zou W., Liu X., Yue P., Zhou Z., Sporn M. B., Lotan R., Khuri F. R., Sun S. Y. (2004) Cancer Res. 64, 7570–7578 [DOI] [PubMed] [Google Scholar]
- 25. Jiang C. C., Chen L. H., Gillespie S., Kiejda K. A., Mhaidat N., Wang Y. F., Thorne R., Zhang X. D., Hersey P. (2007) Cancer Res. 67, 5880–5888 [DOI] [PubMed] [Google Scholar]
- 26. Shiraishi T., Yoshida T., Nakata S., Horinaka M., Wakada M., Mizutani Y., Miki T., Sakai T. (2005) Cancer Res. 65, 6364–6370 [DOI] [PubMed] [Google Scholar]
- 27. Chen L. H., Jiang C. C., Kiejda K. A., Wang Y. F., Thorne R. F., Zhang X. D., Hersey P. (2007) Carcinogenesis 28, 2328–2336 [DOI] [PubMed] [Google Scholar]
- 28. Yoshida T., Shiraishi T., Nakata S., Horinaka M., Wakada M., Mizutani Y., Miki T., Sakai T. (2005) Cancer Res. 65, 5662–5667 [DOI] [PubMed] [Google Scholar]
- 29. Elbein A. D. (1987) Annu. Rev. Biochem. 56, 497–534 [DOI] [PubMed] [Google Scholar]
- 30. Sagara Y., Inesi G. (1991) J. Biol. Chem. 266, 13503–13506 [PubMed] [Google Scholar]
- 31. Brüning A., Vogel M., Burger P., Rahmeh M., Gingelmaier A., Friese K., Lenhard M., Burges A. (2008) Biochem. Biophys. Res. Commun. 377, 1309–1314 [DOI] [PubMed] [Google Scholar]
- 32. Takimoto R., El-Deiry W. S. (2000) Oncogene 19, 1735–1743 [DOI] [PubMed] [Google Scholar]
- 33. Ye J., Koumenis C. (2009) Curr. Mol. Med. 9, 411–416 [DOI] [PubMed] [Google Scholar]
- 34. Zinszner H., Kuroda M., Wang X., Batchvarova N., Lightfoot R. T., Remotti H., Stevens J. L., Ron D. (1998) Genes Dev. 12, 982–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Halterman M. W., Gill M., DeJesus C., Ogihara M., Schor N. F., Federoff H. J. (2010) J. Biol. Chem. 285, 21329–21340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bi M., Naczki C., Koritzinsky M., Fels D., Blais J., Hu N., Harding H., Novoa I., Varia M., Raleigh J., Scheuner D., Kaufman R. J., Bell J., Ron D., Wouters B. G., Koumenis C. (2005) EMBO J. 24, 3470–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rouschop K. M., van den Beucken T., Dubois L., Niessen H., Bussink J., Savelkouls K., Keulers T., Mujcic H., Landuyt W., Voncken J. W., Lambin P., van der Kogel A. J., Koritzinsky M., Wouters B. G. (2010) J. Clin. Invest. 120, 127–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rzymski T., Milani M., Pike L., Buffa F., Mellor H. R., Winchester L., Pires I., Hammond E., Ragoussis I., Harris A. L. (2010) Oncogene 29, 4424–4435 [DOI] [PubMed] [Google Scholar]
- 39. Hao C., Beguinot F., Condorelli G., Trencia A., Van Meir E. G., Yong V. W., Parney I. F., Roa W. H., Petruk K. C. (2001) Cancer Res. 61, 1162–1170 [PubMed] [Google Scholar]
- 40. Li Y. C., Tzeng C. C., Song J. H., Tsia F. J., Hsieh L. J., Liao S. J., Tsai C. H., Van Meir E. G., Hao C., Lin C. C. (2006) Clin. Cancer Res. 12, 2716–2729 [DOI] [PubMed] [Google Scholar]
- 41. Rieger J., Naumann U., Glaser T., Ashkenazi A., Weller M. (1998) FEBS Lett. 427, 124–128 [DOI] [PubMed] [Google Scholar]
- 42. McCullough K. D., Martindale J. L., Klotz L. O., Aw T. Y., Holbrook N. J. (2001) Mol. Cell. Biol. 21, 1249–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]