Abstract
Glutamate dehydrogenase (GDH) catalyzes the oxidative deamination of l-glutamate and, in animals, is extensively regulated by a number of metabolites. Gain of function mutations in GDH that abrogate GTP inhibition cause the hyperinsulinism/hyperammonemia syndrome (HHS), resulting in increased pancreatic β-cell responsiveness to leucine and susceptibility to hypoglycemia following high protein meals. We have previously shown that two of the polyphenols from green tea (epigallocatechin gallate (EGCG) and epicatechin gallate (ECG)) inhibit GDH in vitro and that EGCG blocks GDH-mediated insulin secretion in wild type rat islets. Using structural and site-directed mutagenesis studies, we demonstrate that ECG binds to the same site as the allosteric regulator, ADP. Perifusion assays using pancreatic islets from transgenic mice expressing a human HHS form of GDH demonstrate that the hyperresponse to glutamine caused by dysregulated GDH is blocked by the addition of EGCG. As observed in HHS patients, these transgenic mice are hypersensitive to amino acid feeding, and this is abrogated by oral administration of EGCG prior to challenge. Finally, the low basal blood glucose level in the HHS mouse model is improved upon chronic administration of EGCG. These results suggest that this common natural product or some derivative thereof may prove useful in controlling this genetic disorder. Of broader clinical implication is that other groups have shown that restriction of glutamine catabolism via these GDH inhibitors can be useful in treating various tumors. This HHS transgenic mouse model offers a highly useful means to test these agents in vivo.
Keywords: Allosteric Regulation, Glutamate, Glutamate Dehydrogenase, Insulin Secretion, Structural Biology, Tumor Metabolism, EGCG
Introduction
Glutamate dehydrogenase (GDH)3 is a homohexameric enzyme found in all organisms and catalyzes the reversible oxidative deamination of l-glutamate to 2-oxoglutarate using NAD(P)+ as coenzyme (1). Animal GDH is composed of ∼500 residues and, unlike GDH from other kingdoms, is allosterically regulated by a wide array of metabolites. The major allosteric activators are ADP (2, 3) and leucine (4), whereas the major inhibitors include GTP (5), ATP (2), and palmitoyl-CoA (6).
GDH is composed of two trimers of subunits stacked directly on top of each other with at least three domains in each subunit (7–14). The bottom domain makes extensive contacts with a subunit from the other trimer. On top of this domain is the “NAD binding domain” that is composed of the conserved nucleotide-binding motif. Animal GDH has a long protrusion, the “antenna,” rising above the NAD binding domain that is not found in the other kingdoms (11–14). The antenna from each subunit lies immediately behind the adjacent, counter-clockwise neighbor within the trimer. GDH undergoes large, concerted conformational changes during each catalytic turnover event. The NAD binding domain rotates by ∼18° about the “pivot helix” that runs along the back of this domain. The intertwined antennae rotate about each other, and the inner core of the hexamer expands and contracts as the mouth opens and closes.
Allosteric regulators control GDH activity by affecting these large conformational changes (15). GTP binds at the base of the antenna, wedged in between the NAD binding domain and the pivot helix (11, 12), only when the catalytic cleft is closed. In this way, GTP slows down product release by increasing the binding affinity of substrate and coenzyme (5, 16, 17). In contrast, the activator ADP binds behind the NAD binding domain and immediately under the pivot helix (14). ADP may facilitate the rotation of the NAD binding domain, thereby decreasing the energy required to open the catalytic cleft and release the product (14).
Although the complex allosteric regulation of mammalian GDH has been studied for decades, recent studies on hyperinsulinism/hyperammonemia (HHS) have clearly linked GDH regulation to insulin and ammonia homeostasis (18). This disorder is caused by mutations that desensitize GDH to the effects of GTP inhibition. This gain of function increases the rate of mitochondrial oxidation of glutamate in the β-cells, thereby increasing the ATP4−/MgADP2− ratio. This closes the plasma membrane KATP channels, opens voltage-gated Ca2+ channels, and leads to insulin granule exocytosis. HHS patients have increased β-cell responsiveness to leucine and susceptibility to hypoglycemia following high protein meals (19). The elevation of serum ammonia levels is probably due to increased ammonia production from glutamate oxidation and possibly also impaired urea synthesis by carbamoylphosphate synthetase due to reduced formation of its activator, N-acetylglutamate, from glutamate. Studies have also suggested a high correlation between HHS and childhood onset epilepsy, learning disabilities, and seizures (20). The current treatment for HHS is to control insulin secretion with compounds such as diazoxide (21), but this does not address the serum ammonium and CNS pathologies.
We previously demonstrated that two of the four major catechins from green tea were effective inhibitors of GDH (22). Epigallocatechin gallate (EGCG) and epicatechin gallate (ECG) both inhibited both wild type and the HHS form of GDH with nanomolar ED50 values in vitro. Further, we demonstrated that EGCG inhibits BCH-stimulated insulin secretion, a process that is mediated by GDH, under conditions where GDH is no longer inhibited by high energy metabolites. EGCG does not affect glucose-stimulated insulin secretion under high energy conditions where GDH is probably fully inhibited. These compounds act in an allosteric manner independent of their antioxidant activity, and the β-cell stimulatory effects are directly correlated with glutamine oxidation.
Presented here is the atomic structure of ECG bound to GDH and a demonstration that these compounds ameliorate the effects of HHS in transgenic mice expressing the dysregulated human enzyme. The atomic structure of the GDH-ECG complex and site-directed mutagenesis of the binding site demonstrate that these compounds target the ADP allosteric site. We also demonstrate that EGCG blocks the hyperresponse to glutamine by isolated islets from a transgenic mouse expressing the human HHS form of GDH. In addition, hexachlorophene, which binds to a different and internal allosteric site in the GDH hexamer, also blocks GDH-mediated insulin secretion. Consistent with EGCG inhibition of GDH activity, the addition of EGCG increases the pool of glutamine and glutamate in the transgenic β-islets. Finally, we demonstrate that oral administration of EGCG to HHS mice prevents hypersinsulinemic (or hypoglycemic) response to amino acids and improves the abnormally low basal blood glucose levels in the transgenic mice. Therefore, this safe compound holds promise that the dysregulated HHS form of GDH can be controlled allosterically in vivo.
EXPERIMENTAL PROCEDURES
Site-directed Mutagenesis
Site-directed mutagenesis on three residues that interact with ECG was performed using the previously described pET20a-hGDH expression vector (23), and primers are shown in Table 1. A series of mutants, R90S, D123A, and S397I, were constructed using the overlapping PCR method and confirmed by DNA sequencing.
TABLE 1.
Primers used to construct the R90S, D123A, and S397I mutations in the human GDH heterologous expression vector (23)
| Primer name | Primer sequence (5′ to 3′) |
|---|---|
| c427a (R90S) | 5′-cacagccagcacagcacgccctgca-3′ |
| c427a_antisense | 5′-tgcagggcgtgctgtgctggctgtg-3′ |
| a527c (D123A) | 5′-gtgtgcagtggttgctgtgccgtttgggg-3′ |
| a527c_antisense | 5′-ccccaaacggcacagcaaccactgcacac-3′ |
| g1349t (S397I) | 5′-gaagaatctaaatcatgtcatctatggccgtttgaccttc-3′ |
| g1349t_antisense | 5′-gaaggtcaaacggccatagatgacatgatttagattcttc-3′ |
Protein Expression and Purification
Escherichia coli BL21 (DE3) star cells (Invitrogen) were sequentially transformed with chaperon plasmids pGro7 harboring GroES and GroEL (Takara Bio Inc., Otsu, Japan) and pET20a-hGDH or its mutants, respectively. Transformed cells were cultured at 37 °C in LB medium containing 100 μg/ml ampicillin, 20 μg/ml chloramphenicol, and 1 mg/ml arabinose for inducing expression of GroES and GroEL. After the A600 of the culture reached 0.6–0.8, medium was cooled to room temperature, and 0.2 mm isopropyl 1-thio-β-d-galactopyranoside, final concentration, was added to induce expression. After 20 h, cells were harvested by centrifugation and resuspended in buffer A (0.1 m sodium phosphate, pH 7.5, 0.6 m ammonium sulfate, and 1 mm EDTA). The cells were ultrasonicated, and cellular debris was removed by centrifugation. GDH was precipitated with a 60% saturated ammonium sulfate, final concentration. The pellets were resuspended into buffer A, and loaded onto a butyl-S-Sepharose (GE Healthcare) column equilibrated with the same buffer. After extensive washing, a gradient elution with buffer B containing 50% ethylene glycol and 20 mm sodium phosphate (pH 7.5) was applied. Fractions containing GDH activity (from 35% B to 85% B) were combined and concentrated with a pressurized Amicon (Millipore Corp., Beverly, MA) stir cell. Concentrated samples were rapidly diluted over 20-fold with 20 mm sodium phosphate buffer (pH 7.8) and loaded onto an equilibrated Source Q column, and GDH was eluted with a gradient of 0–0.3 m NaCl over 20 column volumes in 20 mm sodium phosphate buffer (pH 7.8). The mutant, S397I, always lost its activity during the low salt buffer exchange. Therefore, only hydrophobic interaction column-purified S397I GDH was used in the steady state assays.
Assays were conducted using a Beckman Coulter DU800 spectrophotometer. The reaction mixture solution contained 10 mm 2-oxoglutarate, 0.2 mm NADH, 50 mm NH4Cl, and 0.1 m sodium phosphate (pH 7.5) buffer. The reactions were initiated by adding the appropriate amount of enzyme solution, and the steady state rate was measured by monitoring NADH oxidation at 340 nm.
Crystallization of the GDH-ECG Complex
Co-crystallization of GDH with ECG was performed using the hanging drop vapor diffusion method at room temperature. Drops were formed using a 2:1 mix of protein and reservoir solutions. The reservoir solution contained 0.9 m sodium chloride, 50 mm triethylammonium-HCl (pH 7.0), 5 mm reduced glutathione, 8–9% polyethylene glycol, 1 m 1,6-hexanediol. The protein stock solution contained 5 mg/ml GDH, 0.4 mm ECG, 3 mm NADPH, and 20 mm sodium glutamate.
Data Collection and Structure Determination
Crystals were flash-frozen in liquid nitrogen after stepwise transfer with a series of cryoprotectant solutions with increasing glycerol concentrations of 4, 9, 15, and 20% (v/v). The artificial reservoir solutions were composed of 18% PEG 8000, 0.4 m 1,6-hexanediol, 0.4 m NaCl, saturated ECG, 5 mm reduced glutathione, 2 mm NADPH, 20 mm glutamate, and 50 mm buffer triethylammonium-HCl (pH 7.0). Data sets were collected at the Advanced Photon Source beam line 19ID and processed with the HKL2000 program suite (24). The abortive complex (GDH + GTP + NADPH + glutamate) structure of 1HYZ was used as initial model for molecular replacement with the program CNS (25), the program COOT (26) was used for model building, and all inhibitor topologies were produced using the program PRODRG (27). The initial locations and positions of ECG were identified as peaks in difference maps (Fo − Fc) with maximum values of at least ∼5σ. For refinement, 12-fold non-crystallographic restraints were applied to four sections of the protein: residues 10–208, 209–392, 393–444, and 445–489. These segments correspond to the glutamate binding domain, the NAD binding domain, the antenna, and the pivot helix, respectively. These restraints greatly improved the geometry of the model and yielded superior results compared with using the entire subunit as a single segment for non-crystallographic restraints. Final refinement statistics are shown in Table 2.
TABLE 2.
Refinement statistics of the ECG-GDH complex
The numbers in parentheses denote the values in the highest resolution shells.
| GDH-ECG complex | |
|---|---|
| Protein Data Bank code | 3QMU |
| Wavelength (Å) | 0.97915 |
| Space group | P21 |
| Unit cell a, b, c (Å) | 95.1, 433.2, 94.2 |
| β (degrees) | 118.74 |
| Resolution range (Å) | 50–3.6 (3.73–3.6) |
| Unique reflections | |
| R(I)sym (%) | 9.3 (50.7) |
| Completeness (%) | 99.7 (98.9) |
| Redundancy | |
| I/σ(I) | 14.8 (2.2) |
| Refinement statistics | |
| Rwork (%) | 25.7 (31.8) |
| Rfree (%) | 31.2 (37.1) |
| No. of reflections | 72,322 (6332) |
| Completeness (%) | 95.0 (84.0) |
| No. of protein atoms | 46,560 |
| No. of NADPH atoms | 576 |
| No. of ECG atoms | 384 |
| Average B values (Å2) | |
| Protein atoms | 146 |
| NADPH atoms | 185 |
| ECG atoms | 130 |
| Root mean square deviations | |
| Bond length (Å) | 0.01 |
| Bond angles (degrees) | 1.3 |
| Ramachandran results (%) | |
| Most favored | 65.9 |
| Additionally allowed | 28.7 |
| Generously allowed | 4.3 |
| Disallowed | 1.1 |
For density improvement, non-crystallographic real space averaging was applied using the refined coordinates and combined with the observed structure factors using DM in CCP4i (28). The mask including one of the 12 drug molecules and phase recombination was disabled in DM to help remove model bias. The averaged map was then back-transformed, and the phases were applied to the observed structure factors for subsequent averaging. Each map was averaged 10 times, and the final correlation coefficients were ∼99% for each subunit.
GDH Transgenic Mice
Generation of GDH transgenic (TG) mice that express the H454Y human GDH mutation driven by the rat insulin promoter was described previously (29). Both GDH TG mice and their control wild type (WT) littermates were fed a standard rodent chow diet, maintained on a 12-h light/dark cycle.
Amino Acid Tolerance Test and EGCG Treatment in Vivo
The procedure of amino acid tolerance test was described previously (30). In brief, overnight fasted mice were administered with EGCG or water control with the same volume via oral gavage at a dose of 100 mg/kg of body weight; 30 min later, all mice were administered a physiological amino acid mixture via oral gavage with a dose of 1.5 g/kg. Blood samples were taken from the tail at the indicated times. Plasma glucose concentrations were measured using a Freestyle flash glucose meter.
Effects of EGCG on Basal Glucose Levels under Fasting Conditions
GDH TG and WT mice were fasted for 18 h and then administered with two doses of EGCG (100 mg/kg) or water (same volume/kg of body weight as EGCG) at fasting time of 18 and 24 h. Blood glucose was measured at 18 and 24 h of fasting.
GDH Enzyme Assay, Insulin Secretion, and Cytosolic Calcium Measurements in Isolated Islets
Islets were isolated by collagenase digestion and cultured for 3 days in RPMI 1640 medium containing 10 mm glucose (31). GDH enzyme assays were performed in fresh isolated islets; the assay procedures were described previously (30). Insulin secretion was tested by islet perifusion and batch incubations (29, 30, 32). Cytosolic calcium ([Ca2+]i) levels were measured by dual wavelength fluorescence microscopy using fura-2 acetoxymethylester in perifused islets (30). Insulin was measured by radioimmunoassay at the University of Pennsylvania radioimmunoassay core.
Intracellular Amino Acid Measurements
50 islets/batch of isolated cultured islets were preincubated with glucose-free Krebs-Ringer bicarbonate buffer for 60 min at 37 °C in a shaking water bath and then incubated with various treatments for another 60 min. The supernatants were collected for insulin assays. Islets were washed twice with cold PBS and then homogenized in PBS buffer, and 2 μl of homogenate was taken for protein determination. To these samples, 10% sulfosalicylic acid was added to precipitate the protein, and 1 μm norvaline was used as an internal standard. Intracellular amino acids were then measured with an UltraPerformance LC system (Waters) (33).
RESULTS
Structure of ECG Bound to GDH
Although it has been clearly demonstrated that ECG/EGCG inhibits GDH in tissue culture (22, 34, 35), these polyphenols are reactive molecules (36–39) that have been shown to affect a number of cellular systems (40, 41). Therefore, the localization of the ECG/EGCG binding site is important to validate the specificity of polyphenol binding, identifies mutation sites for probing the mechanism of inhibition, and is invaluable for drug design.
Mammalian GDH crystallizes in an open or closed conformation depending upon the ligands that are added during crystallization. When coenzyme from one side of the reaction is combined with substrate from the other (e.g. NADPH + Glu), the active site is usually closed down upon this tightly bound abortive complex. These crystals typically contain one hexamer per crystallographic asymmetric unit and diffract to better than 2.5 Å resolution. In contrast, without such an abortive complex, there are usually two entire hexamers in the asymmetric unit, the 12 different subunits are often opened to differing degrees, and the crystals do not diffract to resolutions better than 3.2 Å. It was therefore unexpected that GDH + ECG crystallized in the open conformation although high concentrations of NADPH and Glu were added to the crystallization solutions (Table 2).
The bound ECG was made evident by a strong (6σ) electron density peak in the difference map (Fig. 1). Although the resolution of these crystals was limited, the electron density was significantly improved by 12-fold non-crystallographic electron density averaging applied to the individual domains. The electron density envelope matches that of the ECG molecule extremely well and allowed for unique positioning of the molecule into its binding pocket. Notably, the active site clefts are partially open, electron density for Glu in the active site is not observed, and the electron density for the bound NADPH is extremely weak and not contiguous. Because the concentrations of NADPH and Glu should have been sufficient to produce crystals of GDH in the closed conformation, this suggests that the addition of ECG decreases the affinity for substrate and coenzyme. This is also consistent with our finding that ECG and EGCG could not be soaked into crystals of GDH in the closed conformation.
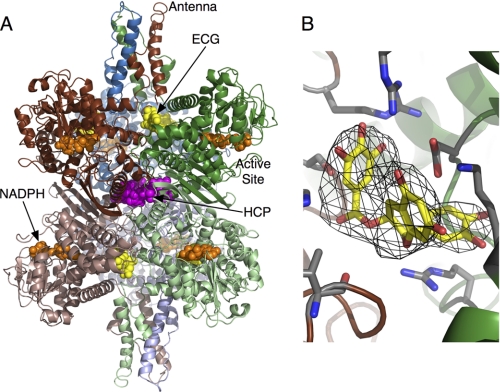
FIGURE 1.
Structure of the GDH-ECG complex. A, the entire GDH hexamer shown as a ribbon diagram. Yellow and orange spheres represent the bound ECG and NADPH molecules, respectively. For comparison, the HCP binding site from previous structural studies (42) is highlighted by mauve spheres. B, an averaged omit map showing the electron density for the bound ECG molecule.
The location of this peak is beneath the pivot helix and essentially in the same location as the major allosteric activator, ADP (Fig. 2). This location is quite distal to where another GDH inhibitor, hexachlorophene (HCP), binds to the core of the enzyme and does not favor the open conformation like ECG and ADP (42). As shown in Fig. 2, ECG binds at the interface between adjacent subunits within the hexameric trimers and, via a number of hydrogen bonds, effectively cross-links the subunits together. It is notable that our recent studies have shown that this interface expands and contracts as the catalytic mouths open and close, respectively (42). For comparison, ADP and GTP molecules from previous structures (12, 14) were overlaid onto this ECG complex. Interestingly, the side chain of His-209 in the GTP complex is rotated toward and interacts with GTP.
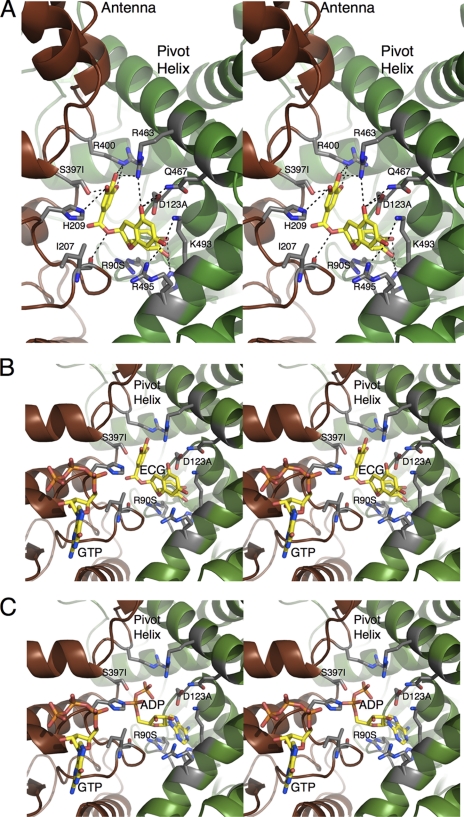
FIGURE 2.
GDH-ECG interactions. A, details of the interactions between the bound ECG molecule and GDH. Dashed lines represent the possible hydrogen bonds. Also noted are the locations of the mutations analyzed in this study; S397I, R90S, and D123A. B, stereo image showing the relationships between the ECG and GTP binding sites. C, stereo image showing the relationship between the ADP and GTP sites. The orientation is identical to B to show the overlap between the ADP and ECG binding contacts.
Mutagenesis Analysis of the ECG Binding Site
From the GDH-ECG interactions shown in Fig. 2, three mutations were selected to further dissect the similarities and differences between ECG, ADP, and GTP allosteric regulation: S397I, D123A, and R90S (Fig. 3). As shown in this figure, none of these residues make direct contact with bound GTP. The guanidinium moiety of Arg-90 stacks against the purine ring of ADP as well as one of the phenolic groups of ECG. Asp-123 forms hydrogen bonds with the flavonoid ring of ECG and the ribose ring of ADP. Finally, Ser-397 was chosen because it represents one of the hydrogen bonds provided by the adjacent subunit. The D123A and the R90S mutations produced stable enzyme that could be fully purified as described under “Experimental Procedures.” However, the S397I mutation that lies at the subunit interface yielded unstable GDH that denatured at low ionic strength. Therefore, crude extract from the E. coli heterologous expression system could only be purified using hydrophobic interaction chromatography because the enzyme could always be kept in high salt buffers. Because of this, only GTP and ADP dose-response assays could be performed on this mutant because the contaminants in this partially purified sample interfered with the ECG and EGCG.
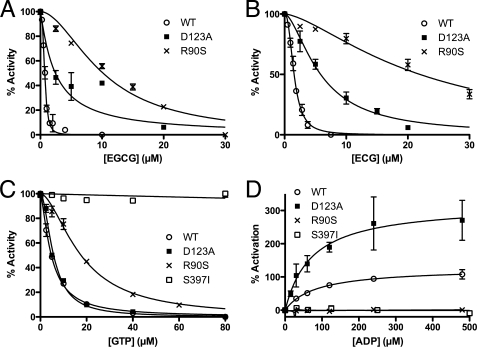
FIGURE 3.
Effects of ECG binding site mutations on ECG/EGCG, ADP, and GTP allosteric regulation. A and B compare sensitivity of purified WT, D123A, and R90S forms of GDH to ECG and EGCG inhibition. Both mutants partially abrogate ECG/EGCG inhibition. C and D include the partially purified S397I mutant that was not included in the polyphenol assays because contaminants reacted with EGCG/ECG. The mutation at the subunit interface, S397I, blocks GTP inhibition, whereas the other two mutations in the ECG/ADP binding site have little or no effect on GTP inhibition. The S397I and R90S mutations block ADP activation, whereas the D123A mutation appears to make ADP a more effective activator. Error bars, S.E.
The D123A mutation increased the ED50 for EGCG and ECG by ∼6-fold but had no effect on GTP inhibition. However, this mutation did not appear to affect the ED50 for ADP activation but apparently doubled the extent of ADP activation. This shows that although ADP and ECG/EGCG share some contact residues, there are differences in what role those residues play in the allosteric processes. The R90S mutation had the most profound effect on ECG/EGCG inhibition by increasing the ED50 by 10–20-fold. Interestingly, this mutation had a modest effect (∼3-fold increase) on the ED50 for GTP but completely eliminated ADP activation. This clearly shows the overlap between the ADP and ECG/EGCG sites and the importance of the interactions between the aromatics of ECG/EGCG and ADP with the guanidinium moiety of Arg-90. Finally, the unstable S397I mutation is apparently insensitive to both GTP and ADP regulation. This suggests that this region is involved in subunit-subunit communication that may be essential for allosteric regulation. This is akin to our previous studies showing that removal of the antenna domain also abrogated ADP and GTP regulation (43). These results provide strong affirmation of the location of ECG and ligand-protein interactions observed in the GDH-ECG complex structure.
EGCG Inhibits GDH Enzyme Activity in Isolated Islets
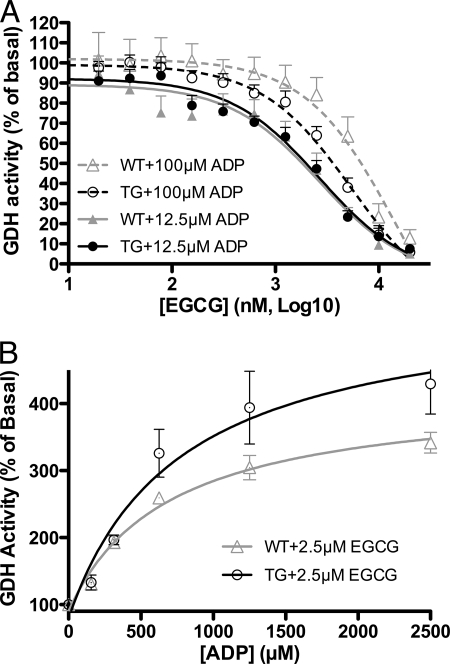
Previously, we demonstrated that EGCG blocked GDH-mediated stimulated insulin secretion from rat islets (22). To validate this finding and to ascertain whether there is any difference between how wild type and HHS islets respond to EGCG, GDH activity in isolated islets was measured. In the presence of 12.5 μm ADP, the ED50 for ADP stimulation in islets (30), EGCG inhibited GDH activity similarly in TG and WT (Fig. 4A). However, the ED50 for EGCG is significantly higher than when measured with purified GDH (22). When ADP concentration increased to 100 μm, the EGCG inhibition curve shifted to high EGCG concentration, but TG and WT islets have undistinguishable responses to EGCG inhibition. The HHS GDH H454Y mutation has impaired sensitivity to GTP inhibition (18, 23) but responds to EGCG in a manner nearly identical to that of wild type tissue. This is consistent with the structural results showing that ECG/EGCG does not interact with the GTP binding site. Also consistent with the structural result is that ADP reverses EGCG inhibition (Fig. 4B). However, in the presence of 2.5 μm EGCG, the ED50 for ADP rises from 12 to 400 μm. Therefore, these results also demonstrate marked antagonism in vivo between EGCG and ADP.
FIGURE 4.
Effects of EGCG on whole cell GDH activity. A, EGCG inhibits both TG and WT GDH with nearly identical efficacy. The higher concentration of ADP appears to interfere with EGCG inhibition. B, ADP can overcome EGCG inhibition. As a sign of ADP-EGCG antagonism, the ED50 for ADP under these conditions is more than 20 times higher than in the absence of EGCG (n = 4). Error bars, S.E.
EGCG Inhibits Glutamine-stimulated Insulin Secretion in GDH TG Islets
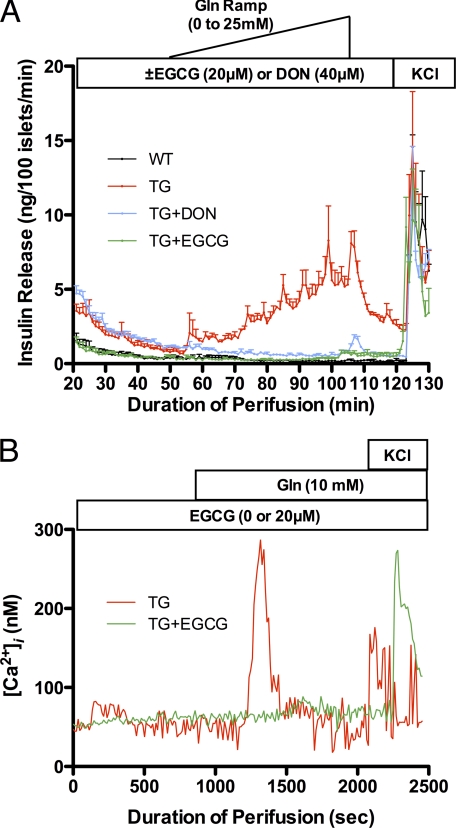
As we reported previously, GDH TG islets are sensitive to glutamine-stimulated insulin secretion due to increased glutamine oxidation (29, 31). From the results above, it follows that EGCG should effectively control the aberrant glutamine stimulation of the H454Y TG islets. To this end, islets from TG and WT were perifused with a glutamine ramp (Fig. 5A). As observed previously (29), GDH TG but not WT islets were sensitive to glutamine stimulation. 6-diazo-5-oxo-norleucine (DON), a glutaminase inhibitor, blocked the response of TG islets to glutamine stimulation but failed to lower the basal insulin secretion to the same level as WT controls (the higher level of insulin release at T = 20 min). In contrast, 20 μm EGCG not only blocked TG islet response to glutamine but also lowered basal insulin secretion to the same levels as WT. This is probably due to the fact that inhibition of glutaminase by DON only eliminates glutamine flux to glutamate and does not block conversion of the other amino acid conversion to glutamate through transamination. In contrast, EGCG inhibition of GDH blocks oxidation of amino acids immediately before they enter the TCA cycle and therefore lowered even basal insulin secretion in TG islets. As an additional control, Fig. 5B shows, as expected, that EGCG also blocked glutamine-induced calcium influx that is the prelude to insulin exocytosis.
FIGURE 5.
The effect of EGCG on Gln-stimulated insulin secretion in H454Y transgenic mouse β-cell islets. A, TG tissue secretes insulin in response to a Gln ramp stimulation. This is not observed in WT islets, and glutamine-stimulated insulin secretion in TG islets is blocked by the glutaminase inhibitor, DON, and by the GDH inhibitor, EGCG. Note that EGCG, but not DON, brings the basal insulin secretion levels (T = 20 min) down to that of WT (data are mean ± S.E. (error bars), n = 3 for each group). The black line representing WT tissue may be difficult to see because it lies directly under the TG + EGCG line (green). B, Gln stimulates Ca2+ influx in response to Gln, and this is also blocked by EGCG (representative data are shown; all experiments were repeated three times, and all showed comparable results). In both A and B, the addition of KCl serves as a control to demonstrate that none of the treatments affect insulin secretion per se.
The GDH Inhibitor, Hexachlorophene, Also Blocks Glutamine-stimulated Insulin Secretion in TG Islets
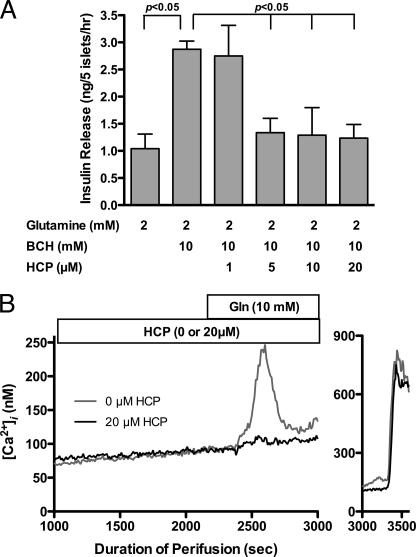
A major drawback in using polyphenols as pharmaceutical agents is that they are relatively unstable in solution (36, 37), and their hydrophilic nature makes for inefficient movement across the cell membrane. Therefore, as an additional control, one of the other recently identified GDH inhibitors (44), HCP, was tested for possible inhibition of GDH-mediated insulin secretion in the HHS β-cell. HCP is more hydrophobic and is chemically inert in solution. In contrast to ECG, HCP binds to the inner core of the hexamer (Fig. 1) and inhibits purified GDH with an ED50 of ∼2 μm (42, 44). As shown in Fig. 6A, HCP inhibits BCH/glutamine-stimulated insulin secretion in a dose-dependent manner with an IC50 value that appears to be close to that measured with purified GDH. Finally, Fig. 6B shows that HCP, as with EGCG/ECG, inhibits glutamine-induced calcium influx in TG islets.
FIGURE 6.
The effects of HCP on GDH-mediated insulin secretion from wild type and HHS tissue. A, in the presence of Gln, BCH stimulates insulin secretion from wild type islets via stimulation of GDH activity. This response is completely blocked by the addition of 5 μm HCP. This concentration is consistent with our previous in vitro studies using purified GDH (42). B, HCP inhibition is also manifested as a block in the calcium influx upon the addition of Gln in HHS islets. Error bars, S.E.
EGCG Blocked Glutamine Consumption in TG Islets
EGCG inhibition of glutamine-stimulated insulin secretion in GDH TG islets is probably due to blockage of glutamine oxidation driven by gain of function mutation of GDH. To confirm this hypothesis, intracellular amino acid levels were measured in isolated islets. As shown in Table 3, compared with WT islets, 10 mm glutamine stimulates insulin secretion in TG islets, but both WT and TG islets have a similar response to 10 mm glucose. TG islets have decreased intracellular aspartate and glutamate and glutamine + glutamate levels, suggesting that elevated glutaminolysis is responsible for glutamine-stimulated insulin secretion. At 20 μm, EGCG blocked glutamine consumption in TG islets. However, because aspartate and glutamate levels are still lower in TG islets compared with controls, this dose of EGCG is apparently sufficient to block glutamine-induced insulin secretion but apparently cannot completely inhibit catabolic flux through GDH. However, glucose response was similar in TG and WT islets, and EGCG had little or no effect on GSIS. Our previous findings that suggested that high ATP/GTP levels due to glucose oxidation probably block GDH-mediated catabolism may explain the limited effect of EGCG on GSIS.
TABLE 3.
Insulin secretion and intracellular amino acid concentrations of islets from transgenic and wild type islets
| n = 4 | Gln, 10 mm |
Gln, 10 mm/EGCG, 20 μm |
Glc (10 mm) |
Glc (10 mm)/EGCG (20 μm) |
||||
|---|---|---|---|---|---|---|---|---|
| TG | WT | TG | WT | TG | WT | TG | WT | |
| Insulin secretion (ng/μg protein/h) | 3.9 ± 0.6a | 1.6 ± 0.2 | 2.3 ± 0.3b | 2.5 ± 0.4 | 3.7 ± 0.5 | 2.8 ± 0.3 | 3.3 ± 0.2 | 4.0 ± 0.3 |
| Amino acids (nmol/mg protein) | ||||||||
| Aspartate | 62 ± 10a | 134 ± 16 | 81 ± 23a | 153 ± 3 | 16 ± 2 | 15 ± 1 | 15 ± 1 | 30 ± 2 |
| Glutamate | 53 ± 8c | 128 ± 15 | 73 ± 16c | 149 ± 8 | 24 ± 5 | 20 ± 1 | 23 ± 3 | 35 ± 2 |
| Glutamine | 278 ± 26 | 640 ± 171 | 412 ± 45b | 479 ± 49 | 36 ± 7 | 10 ± 3 | 47 ± 13 | 18 ± 2 |
| Gln + Glu | 332 ± 19a | 768 ± 179 | 484 ± 56b | 628 ± 57 | 60 ± 11 | 27 ± 3 | 70 ± 16 | 53 ± 1 |
a p < 0.05 compared with WT.
b p < 0.05 compared with group without EGCG.
c p < 0.01 compared with WT.
EGCG Controls Amino Acid-induced Hypoglycemia in TG Mice
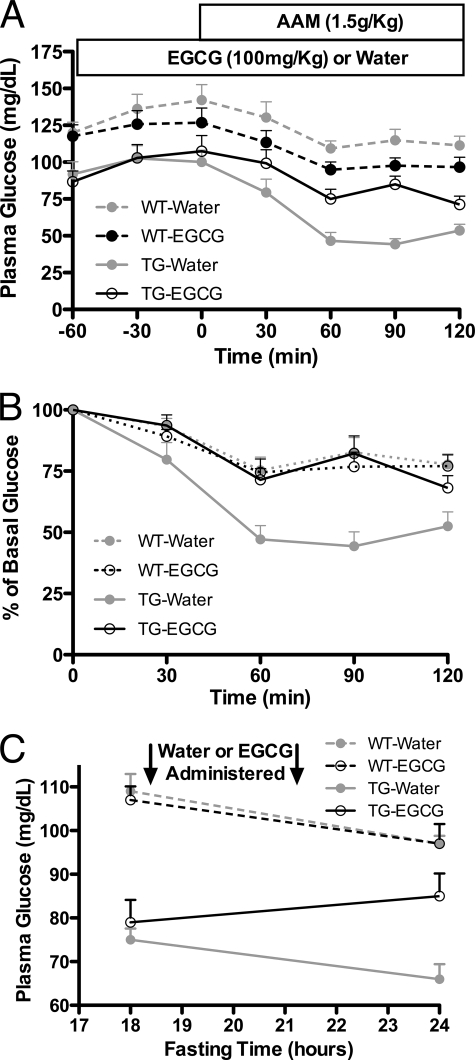
All of the above experiments confirmed the inhibitory effect of EGCG on GDH, including GDH enzyme kinetics, insulin secretion, and glutaminolysis in isolated islets. Because of the gain of function mutations in GDH, HHS patients exhibit protein-induced hypoglycemia due to an exaggerated insulin response. Although both lead compounds, HCP and ECG/EGCG, are effective in blocking the HHS symptoms in isolated tissue, the tea polyphenols have minimal in vivo side effects and were selected for whole animal studies. In a manner identical to HHS patients, TG mice exhibit a rapid decline in blood glucose levels within 30 min of consumption of an amino acid mixture (Fig. 7). This hyperresponse was effectively blocked by oral administration of EGCG prior to challenge. The data on islets suggested that EGCG might be effective at controlling the basal insulin levels in HHS tissue (Fig. 5), and therefore the effects of EGCG on fasting mice were tested. Compared with WT mice, TG mice had significantly lower glucose level after 18 h of fasting (*, p < 0.01) due to higher basal levels of insulin secretion. When water was administered to the TG mice, the glucose level continued to drop over time. However, when EGCG was administered, there was marked improvement in the plasma glucose levels of the HHS mice, approaching that of the WT group. As expected, EGCG did not have any effect on WT mice, probably due to the tonic state of WT GDH under these conditions. Therefore, these results demonstrate that EGCG can control the hyperresponse to a bolus of amino acids in HHS animals as well as improve the basal plasma glucose levels when administered chronically.
FIGURE 7.
The effects of oral administration of EGCG on the hypersecretion of insulin in HHS transgenic mice. A, plasma glucose levels in WT mice (n = 12 for water- or EGCG-treated mice) are essentially unaffected by oral administration of water or EGCG prior to the administration of the amino acid mixture. However, the plasma glucose levels rapidly drop in the HHS TG mice (n = 12) upon the administration of the amino acid mixture, but this is blocked when the animals are fed EGCG (n = 16) prior to the amino acid challenge. B, the data here are the same as in A except that they are presented as percentage of basal levels of glucose. C, the effects of chronic administration of EGCG on the basal glucose levels during fasting rather than amino acid challenge (n = 6 for each treatment). Here, administration of EGCG to the HHS mice significantly improves plasma glucose levels. Error bars, S.E.
DISCUSSION
There is a great deal of interest in developing allosteric regulators as therapeutics for a variety of drug targets. Allosteric regulators can be highly effective while not mimicking common substrates and coenzymes. A number of GDH allosteric inhibitors were found by screening under high substrate and coenzyme conditions, where competitive inhibitors were unlikely to be effective (42, 44). Indeed, the EGCG/ECG site identified here represents the third drug target site identified that is entirely distal to the active site. As shown in Fig. 1, the HCP site is in the core of the GDH hexamer and forms a ring of aromatic interactions that cross-links all of the six subunits together. HCP is hydrophobic and binds into hydrophobic cavities at this subunit interface. Because these interfaces expand and contract as the catalytic cleft opens and closes, respectively, we proposed that these compounds act by inhibiting this “breathing” process that is apparently part of the catalytic turnover process.
EGCG/ECG is quite different from HCP. These polyphenols are extremely hydrophilic, and ECG interactions with GDH are dominated by polar interactions. Although the other inhibitors were able to bind to GDH in the closed conformation, ECG appears to have pushed the structural equilibrium toward the open conformation despite the presence of high concentrations of Glu and NADPH. Indeed, ECG was never observed bound to GDH in the closed conformation even when the crystals were soaked in very high concentrations of ECG. This is the same as what was observed when crystallizing the ADP-GDH complex.
Our previous studies demonstrated that a single mutation (R463A) on the pivot helix abrogated ADP activation without affecting ADP binding (as per TNP-ADP binding). From this, we suggested that ADP might be facilitating enzymatic turnover by decreasing the energy required to open the catalytic cleft (14). From the mutagenesis studies presented here, it may be more complicated that that. The R90S mutation yielded the clearest results in that its guanidinium group stacks up against the aromatic rings in both ECG and ADP. This mutation essentially eliminated polyphenol inhibition as well as ADP activation. This mutation, however, does have some effect on GTP inhibition. This may be due to the fact that Arg-90 hydrogen-bonds to a loop in the adjacent subunit that lies immediately beneath the GTP binding site. Asp-123 lies beneath the pivot helix and hydrogen-bonds with the ribose ring on ADP and with a phenolic group on ECG. From this location, it is not surprising that the D123A mutation had no effect on GTP inhibition but did affect polyphenol inhibition. What is surprising, however, is that this mutation actually accentuated ADP activation without significantly affecting its Kact. This may be due to the interactions between Asp-123 and Arg-463. These two side chains form a salt bridge, and Asp-123 may shield some of the charge on Arg-463. By removing Asp-123, the Arg-463 interaction with the β-phosphate on ADP may be strengthened, thereby improving the ability of ADP to open the catalytic cleft. This is essentially the opposite as the R463A mutation that eliminates ADP activation by eliminating the charge interaction between Arg-463 and ADP (14). Ser-397 lies at the base of the antenna, and the S397I mutation greatly destabilizes the enzyme while abrogating both GTP and ADP regulation. This may simply be due to the marked sensitivity of the antenna region, as exemplified by the fact that removing the antenna also eliminates GTP and ADP activation (43).
These results pose a number of questions with regard to the mechanism of ADP activation. Under conditions where product release is not the rate-limiting step (e.g. low substrate or coenzyme concentrations), ADP inhibits the reaction. In contrast, ECG/EGCG inhibits under all conditions (22). There are at least two possible reasons for this difference. The first model is a more global energy effect on the protein structure. The fact that ECG/EGCG has a much tighter binding (as per the much lower ED50) may suggest that these compounds decrease the Gibbs free energy of the protein and in turn may make conformational changes more difficult. In this case, the lower energy conformation is that of the “open” state. This implies that there may not be a particular interaction with ECG/EGCG responsible for inhibition but rather a series of interactions that increase affinity and decrease the energy state of the protein. Alternatively, it may be that interactions like that found with Asp-123 specifically interfere with the more subtle conformational changes that occur in this area as the catalytic cleft opens and closes. This latter model suggests that improving binding affinity of an ADP analog without these specific interactions would not improve the activation efficacy. From work with drugs against the common cold virus that block uncoating by increasing entropy (45–47), it seems more likely that ECG/EGCG inhibits GDH mainly by changing the overall energy state of the enzyme rather than specific interactions.
The whole cell activity measurements validate the EGCG inhibitory effect on GDH. The ED50 for EGCG is significantly higher than was found when purified GDH was used (22). This is probably due to the fact that the polyphenols have anti-oxidant activity, and their highly soluble nature makes for poor membrane penetration. Nevertheless, these results clearly demonstrate that EGCG inhibits GDH in situ and that, as was found in vitro (22), there is clear allosteric antagonism between the activator ADP and EGCG.
Our previous studies demonstrated that EGCG blocked GDH-dependent insulin secretion in normal tissue (22). As shown in Fig. 5, this also extends to TG mouse tissue. As expected, the glutaminase inhibitor, DON, and EGCG are both able to block the HHS hyperresponse to the addition of Gln. However, only EGCG was able to bring down the basal level of insulin release to that of WT tissue. This, in addition to the fact that DON is significantly more toxic in vivo than EGCG (48), suggests that it is possible to treat HHS in a non-toxic manner by directly targeting GDH. As shown here, EGCG does not decrease insulin levels in WT tissue. This is probably due to GDH being kept mostly in a tonic state in the pancreas and its allosteric inhibition being alleviated only when the energy state of the mitochondria is low. Indeed, the ADP/EGCG antagonism may allow for an allosteric “release valve” whereby even EGCG inhibition is abrogated by ADP when the need for amino acid catabolism is strong enough. This model is further supported by the metabolism studies that measure amino acids levels in the pancreatic tissue (Table 3). Under glucose-rich conditions, there is no significant effect of EGCG on Glu/Gln levels in either TG or WT cells. This is probably due to nearly quiescent GDH activity because of the elevated levels of GTP and ATP. However, when Gln is the major carbon source, EGCG significantly blocks Glu metabolism in TG tissue while not having significant effects on WT tissue. Under such conditions, the GDH activity is expected to increase to respond to the energy needs of the mitochondria. Because the GDH activity is much higher in TG tissue, it follows that it will be more sensitive to EGCG inhibition.
Although EGCG is a natural product with extremely low toxicity issues, it has several problems if it is to be used as a therapeutic agent (49). It is poorly absorbed in the intestinal tract, it is rapidly modified by enzymes such as catechol-O-methyltransferease, and its anti-oxidant activity makes it relatively unstable in solution. To validate our findings with EGCG, a more stable GDH inhibitor, HCP, identified in our previous HTS studies (44), was also examined. Exactly as was found with EGCG, HCP was very effective at blocking the hyperresponse to Gln in TG tissue (Fig. 6). However, probably due to its greater stability and hydrophobicity, the approximate EC50 for HCP in tissue is nearly the same as was found in vitro with purified GDH (44). This demonstrates the balance between stability, toxicity, and bioavailability that will eventually need to be found in developing therapeutics for HHS.
These results point to clear directions for further drug design. This is an interesting starting point, where two very different compounds bind to the same site with quite different affinities. Analogs of ADP can be made that create interactions with Asp-123 like ECG. The D123A mutation abrogated ECG inhibition but actually improved ADP activation. Therefore, Asp-123-ECG interactions may partially account for either the higher affinity or the inhibition. This could have an added benefit in that it would modify the purine ring such that it might not compete with ADP-binding proteins. It may also be possible to modify the β-phosphate of ADP to better mimic ECG. The β-phosphate of ADP interacts with Arg-400 and Arg-463 but is in close proximity to Asp-123. Perhaps an ADP analog with a polar but not a phosphate moiety in that position would improve binding and allow it to inhibit like ECG. Conversely, an EGCG mimic might be made by replacing the reactive polyphenols with polar moieties that interact with the crucial residues as per the mutational studies.
The remaining question was whether either of these lead compounds could control the HHS symptoms in the TG mice when administered orally. Due to its low toxicity, EGCG was selected for in vivo application. An optimal drug for HHS should be able to block the hyperinsulinism response upon the consumption of amino acids as well as elevate basal serum glucose levels. As shown in Fig. 7, when EGCG is orally administered before challenging the TG mice with an amino acid mixture, the GDH-mediated hyperinsulism is blocked. In addition, as was first observed in the islet perifusion assays (Fig. 5), chronic administration of EGCG during fasting improved the basal plasma glucose levels in the TG mice (Fig. 7). Together, these results clearly demonstrate that it is possible to directly target the dysregulated form of GDH in HHS in vivo. It remains to be seen whether such compounds can also elevate serum ammonium levels and prevent the CNS pathology caused by HHS.
It is important to note that GDH inhibitors may have more applications than just treating HHS. Recent studies confirmed our observation that EGCG inhibits GDH in situ and may be useful in treating glioblastoma (34). In this work, EGCG was found to sensitize glioblastoma cells to glucose withdrawal and to inhibitors of Akt signaling and glycolysis. Subsequently, others demonstrated that EGCG inhibition of GDH activity may be useful in treating the tuberous sclerosis complex (TSC) disorder (35). Nearly all of the TSC1/2−/− cells that were deprived of glucose and given rapamycin died upon administration of EGCG. As expected, EGCG effects were reversed if GDH-mediated oxidation of glutamate was circumvented by the addition of 2-oxoglutarate, pyruvate, or aminooxyacetate. Not only do these studies validate our findings, but they also demonstrate that a non-toxic GDH inhibitor could be a synergistic tool in treating tumors.
Acknowledgment
We thank Blue California for the generous gift of EGCG used for the whole animal studies.
This work was supported, in whole or in part, by National Institutes of Health Grants DK072171 (to T. J. S.), DK53012 (to C. A. S.), and DK19525 (for the islet biology and radioimmunoassay cores). This work was also supported by American Diabetes Association Research Award 1-05-RA-128 (to C. A. S.).
The atomic coordinates and structure factors (code 3QMU) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- GDH
- glutamate dehydrogenase
- HCP
- hexachlorophene
- HHS
- hyperinsulinemia/hyperammonemia
- EGCG
- epigallocatechin gallate
- ECG
- epicatechin gallate
- DON
- 6-diazo-5-oxo-norleucine
- TG
- transgenic
- BCH
- 2-aminobicyclo[2,2,1]heptane-2-carboxylic acid
- TSC
- tuberous sclerosis complex.
REFERENCES
- 1. Hudson R. C., Daniel R. M. (1993) Comp. Biochem. Physiol. B 106, 767–792 [DOI] [PubMed] [Google Scholar]
- 2. Frieden C. (1965) J. Biol. Chem. 240, 2028–2037 [PubMed] [Google Scholar]
- 3. George A., Bell J. E. (1980) Biochemistry 19, 6057–6061 [DOI] [PubMed] [Google Scholar]
- 4. Yielding K. L., Tomkins G. M. (1961) Proc. Natl. Acad. Sci. U.S.A. 47, 983–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Iwatsubo M., Pantaloni D. (1967) Bull. Soc. Chem. Biol. 49, 1563–1572 [PubMed] [Google Scholar]
- 6. Fahien L. A., Kmiotek E. (1981) Arch. Biochem. Biophys. 212, 247–253 [DOI] [PubMed] [Google Scholar]
- 7. Baker P. J., Britton K. L., Engel P. C., Farrants G. W., Lilley K. S., Rice D. W., Stillman T. J. (1992) Proteins Struct. Funct. Genet. 12, 75–86 [DOI] [PubMed] [Google Scholar]
- 8. Dean J. L., Wang X. G., Teller J. K., Waugh M. L., Britton K. L., Baker P. J., Stillman T. J., Martin S. R., Rice D. W., Engel P. C. (1994) Biochem. J. 301, 13–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Smith C. A., Norris G. E., Baker E. N. (1996) in Acta Crystallogr. A 52, (suppl.) C226–C227 [Google Scholar]
- 10. Stillman T. J., Baker P. J., Britton K. L., Rice D. W. (1993) J. Mol. Biol. 234, 1131–1139 [DOI] [PubMed] [Google Scholar]
- 11. Peterson P. E., Smith T. J. (1999) Structure 7, 769–782 [DOI] [PubMed] [Google Scholar]
- 12. Smith T. J., Peterson P. E., Schmidt T., Fang J., Stanley C. A. (2001) J. Mol. Biol. 307, 707–720 [DOI] [PubMed] [Google Scholar]
- 13. Smith T. J., Schmidt T., Fang J., Wu J., Siuzdak G., Stanley C. A. (2002) J. Mol. Biol. 318, 765–777 [DOI] [PubMed] [Google Scholar]
- 14. Banerjee S., Schmidt T., Fang J., Stanley C. A., Smith T. J. (2003) Biochemistry 42, 3446–3456 [DOI] [PubMed] [Google Scholar]
- 15. Smith T. J., Stanley C. A. (2008) Trends Biol. Chem. 33, 557–564 [DOI] [PubMed] [Google Scholar]
- 16. Frieden C. (1963) Biochem. Biophys. Res. Commun. 10, 410–415 [Google Scholar]
- 17. Koberstein R., Sund H. (1973) Eur. J. Biochem. 36, 545–552 [DOI] [PubMed] [Google Scholar]
- 18. Stanley C. A., Lieu Y. K., Hsu B. Y., Burlina A. B., Greenberg C. R., Hopwood N. J., Perlman K., Rich B. H., Zammarchi E., Poncz M. (1998) N. Engl. J. Med. 338, 1352–1357 [DOI] [PubMed] [Google Scholar]
- 19. Hsu B. Y., Kelly A., Thornton P. S., Greenberg C. R., Dilling L. A., Stanley C. A. (2001) J. Pediatr. 138, 383–389 [DOI] [PubMed] [Google Scholar]
- 20. Bahi-Buisson N., Roze E., Dionisi C., Escande F., Valayannopoulos V., Feillet F., Heinrichs C., Chadefaux-Vekemans B., Dan B., de Lonlay P. (2008) Dev. Med. Child Neurol. 50, 945–949 [DOI] [PubMed] [Google Scholar]
- 21. Stanley C. A., Baker L. (1976) Adv. Pediatr. 23, 315–355 [PubMed] [Google Scholar]
- 22. Li C., Allen A., Kwagh J., Doliba N. M., Qin W., Najafi H., Collins H. W., Matschinsky F. M., Stanley C. A., Smith T. J. (2006) J. Biol. Chem. 281, 10214–10221 [DOI] [PubMed] [Google Scholar]
- 23. Fang J., Hsu B. Y., MacMullen C. M., Poncz M., Smith T. J., Stanley C. A. (2002) Biochem. J. 363, 81–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 25. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Acta Crystallogr. D 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 26. Emsley P., Cowtan K. (2004) Acta Crystallogr. D 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 27. Schüttelkopf A. W., van Aalten D. M. (2004) Acta Crystallogr. D 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
- 28. Bailey S. (1994) Acta Crystallogr. D 50, 760–763 15299374 [Google Scholar]
- 29. Li C., Matter A., Kelly A., Petty T. J., Najafi H., MacMullen C., Daikhin Y., Nissim I., Lazarow A., Kwagh J., Collins H. W., Hsu B. Y., Nissim I., Yudkoff M., Matschinsky F. M., Stanley C. A. (2006) J. Biol. Chem. 281, 15064–15072 [DOI] [PubMed] [Google Scholar]
- 30. Li C., Chen P., Palladino A., Narayan S., Russell L. K., Sayed S., Xiong G., Chen J., Stokes D., Butt Y. M., Jones P. M., Collins H. W., Cohen N. A., Cohen A. S., Nissim I., Smith T. J., Strauss A. W., Matschinsky F. M., Bennett M. J., Stanley C. A. (2010) J. Biol. Chem. 285, 31806–31818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li C., Najafi H., Daikhin Y., Nissim I. B., Collins H. W., Yudkoff M., Matschinsky F. M., Stanley C. A. (2003) J. Biol. Chem. 278, 2853–2858 [DOI] [PubMed] [Google Scholar]
- 32. Li C., Buettger C., Kwagh J., Matter A., Daikhin Y., Nissim I. B., Collins H. W., Yudkoff M., Stanley C. A., Matschinsky F. M. (2004) J. Biol. Chem. 279, 13393–13401 [DOI] [PubMed] [Google Scholar]
- 33. Di Gangi I. M., Chiandetti L., Gucciardi A., Moret V., Naturale M., Giordano G. (2010) Anal. Chim. Acta 677, 140–148 [DOI] [PubMed] [Google Scholar]
- 34. Yang C., Sudderth J., Dang T., Bachoo R. G., McDonald J. G., DeBerardinis R. J. (2009) Cancer Res. 69, 7986–7993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Choo A. Y., Kim S. G., Vander Heiden M. G., Mahoney S. J., Vu H., Yoon S. O., Cantley L. C., Blenis J. (2010) Mol. Cell 38, 487–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hong J., Lu H., Meng X., Ryu J. H., Hara Y., Yang C. S. (2002) Cancer Res. 62, 7241–7246 [PubMed] [Google Scholar]
- 37. Sang S., Lee M. J., Hou Z., Ho C. T., Yang C. S. (2005) J. Agric. Food Chem. 53, 9478–9484 [DOI] [PubMed] [Google Scholar]
- 38. Hou Z., Sang S., You H., Lee M. J., Hong J., Chin K. V., Yang C. S. (2005) Cancer Res. 65, 8049–8056 [DOI] [PubMed] [Google Scholar]
- 39. Mochizuki M., Yamazaki S., Kano K., Ikeda T. (2002) Biochim. Biophys. Acta 1569, 35–44 [DOI] [PubMed] [Google Scholar]
- 40. Yang C. S., Wang X., Lu G., Picinich S. C. (2009) Nat. Rev. Cancer 9, 429–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Feng W. Y. (2006) Curr. Drug Metab. 7, 755–809 [DOI] [PubMed] [Google Scholar]
- 42. Li M., Smith C. J., Walker M. T., Smith T. J. (2009) J. Biol. Chem. 284, 22988–23000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Allen A., Kwagh J., Fang J., Stanley C. A., Smith T. J. (2004) Biochemistry 43, 14431–14443 [DOI] [PubMed] [Google Scholar]
- 44. Li M., Allen A., Smith T. J. (2007) Biochemistry 46, 15089–15102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Smith T. J., Kremer M. J., Luo M., Vriend G., Arnold E., Kamer G., Rossmann M. G., McKinlay M. A., Diana G. D., Otto M. J. (1986) Science 233, 1286–1293 [DOI] [PubMed] [Google Scholar]
- 46. Smith T. J., Baker T. S. (1999) Adv. Virus Res. 52, 1–23 [DOI] [PubMed] [Google Scholar]
- 47. Katpally U., Smith T. J. (2007) J. Virol. 81, 6307–6315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Earhart R. H., Amato D. J., Chang A. Y., Borden E. C., Shiraki M., Dowd M. E., Comis R. L., Davis T. E., Smith T. J. (1990) Invest. New Drugs 8, 113–119 [DOI] [PubMed] [Google Scholar]
- 49. Smith T. J. (2011) Expert Opin. Drug Discov. 6, 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]