Background: Covalent modification of mRNA translation initiation factors regulates gene expression.
Result: The translational repressor 4E-BP1 is covalently modified by O-GlcNAcylation.
Conclusion: The function of 4E-BP1 is regulated not only by phosphorylation but also by O-GlcNAcylation.
Significance: These findings reveal a novel mechanism through which hyperglycemia-mediated 4E-BP1 O-GlcNAcylation represses cap-dependent mRNA translation and protein synthesis.
Keywords: Diabetes, Glycosylation, Post-translational Modification, Translation Initiation Factors, Translation Regulation, O-linked Glycosylation, eIF4E, eIF4E Binding Protein 1, Hyperglycemia, Protein Truncation
Abstract
4E-BP1 is a protein that, in its hypophosphorylated state, binds the mRNA cap-binding protein eIF4E and represses cap-dependent mRNA translation. By doing so, it plays a major role in the regulation of gene expression by controlling the overall rate of mRNA translation as well as the selection of mRNAs for translation. Phosphorylation of 4E-BP1 causes it to release eIF4E to function in mRNA translation. 4E-BP1 is also subject to covalent addition of N-acetylglucosamine to Ser or Thr residues (O-GlcNAcylation) as well as to truncation. In the truncated form, it is both resistant to phosphorylation and able to bind eIF4E with high affinity. In the present study, Ins2Akita/+ diabetic mice were used to test the hypothesis that hyperglycemia and elevated flux of glucose through the hexosamine biosynthetic pathway lead to increased O-GlcNAcylation and truncation of 4E-BP1 and consequently decreased eIF4E function in the liver. The amounts of both full-length and truncated 4E-BP1 bound to eIF4E were significantly elevated in the liver of diabetic as compared with non-diabetic mice. In addition, O-GlcNAcylation of both the full-length and truncated proteins was elevated by 2.5- and 5-fold, respectively. Phlorizin treatment of diabetic mice lowered blood glucose concentrations and reduced the expression and O-GlcNAcylation of 4E-BP1. Additionally, when livers were perfused in the absence of insulin, 4E-BP1 phosphorylation in the livers of diabetic mice was normalized to the control value, yet O-GlcNAcylation and the association of 4E-BP1 with eIF4E remained elevated in the liver of diabetic mice. These findings provide insight into the pathogenesis of metabolic abnormalities associated with diabetes.
Introduction
The best characterized mechanism for regulating the mRNA binding step in translation initiation involves the reversible binding of the translational repressor 4E-BP1 to the mRNA cap-binding protein eIF4E. Binding of 4E-BP1 to eIF4E blocks its association with eIF4G and disrupts assembly of the cap-binding complex, eIF4F (1). As a result, a shift from cap-dependent to cap-independent translation occurs, leading to an altered pattern of selection of mRNAs to be translated (2). Binding of 4E-BP1 to eIF4E is reversible and is controlled by the sequential phosphorylation of multiple serine and threonine residues, several of which are regulated by the mammalian target of rapamycin in complex 1 (mTORC1)2 signaling pathway (3). Hypophosphorylated 4E-BP1 binds to eIF4E strongly; however, upon phosphorylation the interaction of 4E-BP1 with eIF4E is dramatically reduced.
It has long been established that diabetes is associated with increased binding of 4E-BP1 to eIF4E (4). In part, the increased binding to eIF4E is due to a reduction in phosphorylation of 4E-BP1 resulting from diminished mTORC1 signaling in response to the deficiency of insulin. Another mechanism that could account for the diabetes-induced increase in the association of 4E-BP1 with eIF4E is the addition of N-acetylglucosamine to serine or threonine residues (O-GlcNAcylation) at or near some specific phosphorylation site(s) (5). Protein O-GlcNAcylation is driven by the flux of glucose through the hexosamine biosynthetic pathway, which converts the sugar to UDP N-acetylglucosamine (UDP-GlcNAc), the donor substrate for the addition of O-GlcNAc to serine and threonine residues (6). UDP-GlcNAc synthesis is regulated by the enzyme glutamine-fructose-6-phosphate amidotransferase (GFAT), which catalyzes the first and rate-determining step in the hexosamine biosynthetic pathway. This reaction involves the irreversible transfer of the amino group from glutamine and the isomerization of fructose-6-phosphate to produce glucosamine-6-phosphate. The reversible addition of O-GlcNAc to proteins is catalyzed by O-GlcNAc transferase, whereas O-GlcNAcase catalyzes removal in a dynamic relationship that is reminiscent of protein phosphorylation/dephosphorylation (7–9). In fact, O-GlcNAcylation often is found on or near phosphorylation sites, and there is ample evidence for extensive crosstalk between O-GlcNAcylation and phosphorylation in controlling biological processes (10–14). Although only 2–5% of intracellular glucose enters the hexosamine biosynthetic pathway under normal conditions (15), both hyperglycemia and diabetes have been shown to elevate flux through the pathway and increase the production UDP-GlcNAc (16). Additionally, increased O-GlcNAcylation has been linked to the complications of diabetes (17).
Another mechanism that impairs 4E-BP1 phosphorylation is p53-mediated truncation, which rapidly leads to dephosphorylation of 4E-BP1, sequestration of eIF4E into an inactive 4E-BP1·eIF4E complex, and decreased assembly of the eIF4F complex (18–20). Activation of p53 induces a specific N-terminal cleavage of 4E-BP1, which results in an isoform that is ∼3 kDa smaller than the full-length protein (20). Intriguingly, O-GlcNAcylation of p53 at Ser149 stabilizes p53 and blocks ubiquitin-dependent proteolysis (21). Thus elevated flux through the hexosamine biosynthetic pathway may not only inhibit 4E-BP1 phosphorylation by promoting O-GlcNAcylation but may also enhance p53 activity, resulting in increased levels of the phosphorylation resistant truncated 4E-BP1.
In the present study, Ins2Akita/+ mice were used as a model of type 1 diabetes to evaluate the hypothesis that in the liver of diabetic mice elevated flux of glucose through the hexosamine biosynthetic pathway leads to increased O-GlcNAcylation and concomitant reduced phosphorylation of 4E-BP1 resulting in increased association with eIF4E. Ins2Akita/+, a C57BL/6 mutant mouse, spontaneously develops diabetes with significant early loss of pancreatic β-cell mass as a result of a single missense mutation (C96Y) in the insulin 2 gene (22, 23). As a result, misfolded proinsulin accumulates and promotes endoplasmic reticulum stress in β-cells that leads to apoptosis through induction of the ER stress-associated apoptosis factor Chop (24). Ins2Akita/+ mice develop progressive hyperglycemia secondary to hypoinsulinemia. We report that 4E-BP1 is O-GlcNAcylated in both liver and H4IIE cells in culture. Our results explore the regulation of 4E-BP1 in the liver of diabetic mice and provide evidence for a model wherein hyperglycemia-driven 4E-BP1 O-GlcNAcylation could inhibit cap-dependent mRNA translation and protein synthesis.
EXPERIMENTAL PROCEDURES
Materials
Protease inhibitor mixture was purchased from Sigma and ECL Western blotting detection reagent was purchased from Pierce. Anti-phospho-S6K1(Thr389), goat anti-mouse, and goat anti-rabbit IgG horseradish peroxidase-conjugated antibodies were purchased from Bethyl Laboratories. Anti-O-GlcNAc antibody was purchased from Covance, anti-GAPDH antibody was purchased from Santa Cruz Biotechnology, and all other antibodies were purchased from Cell Signaling Technology. Preparation of the 4E-BP1 and eIF4E antibodies has been described previously (25, 26). 4E-BP1-phosphospecific antibodies were purchased from Cell Signaling.
Animals
Male Ins2Akita/+ mice and Ins2+/+ littermates (C57BL/6 background) were bred in the Diabetic Complications Animal Models Core using procedures approved by The Pennsylvania State University College of Medicine Institutional Animal Care and Use Committee. Diabetic phenotype and genotype were confirmed 4.5 weeks after birth by blood glucose concentrations >250 mg/dl (One-Touch Lifescan meter; Lifescan, Inc., Milpitas, CA) via tail puncture. To determine the effects of normalizing glucose levels in Ins2Akita/+ mice, phlorizin (0.4 g/kg in 10% EtOH, 15% dimethyl sulfoxide, and 75% saline) was administered subcutaneously twice daily for 7 days.
In Situ Liver Perfusion
Ins2Akita/+ mice and Ins2+/+ littermates were maintained on a 12:12 h light:dark cycle, with food and water provided ad libitum. Livers were perfused in situ with a nonrecirculating medium for 30 min at a flow rate of 4 ml/min, as described previously (27), with the following modifications. In an attempt to match physiological glucose concentrations, the perfusate contained either 33 or 11 mm glucose for Ins2Akita/+ or Ins2+/+ mice, respectively. Where indicated, O-(2-acetamido-2-deoxy-d-glucopyranosylidenamino)N-phenylcarbamate (PUGNAc) (1.4 mm in PBS with 1% dimethyl sulfoxide) was infused directly into the inflow line at a rate of 0.2 ml/min to produce a final calculated concentration of 40 μm. The perfusate also contained amino acids at the concentrations present in the arterial plasma of a fasted rat (28).
Processing of Liver Samples
For the analysis of protein phosphorylation state, a portion (∼0.3 g) of liver was homogenized in 7 volumes of homogenization buffer (20 mm HEPES, 2 mm EGTA, 50 mm NaF, 100 mm KCl, 0.2 mm EDTA, 50 mm β-glycerophosphate, 1 mm benzamidine, 200 mm sodium vanadate, and 10 μl/ml Sigma protease inhibitor mixture, pH 7.4) using a Polytron homogenizer. The homogenate was centrifuged at 1000 × g for 3 min at 4 °C, and the resulting supernatant was subjected to SDS-PAGE and Western blot analysis (29). Phosphorylation of S6K1, 4E-BP1, and mTOR was measured in the supernatants using phospho-specific antibodies as described previously (29).
Quantitation of Total 4E-BP1 Protein Expression
Livers were homogenized in 7 volumes of λ-phosphatase buffer (30) containing 10 μl/ml Sigma protease inhibitor mixture, pH 7.4, and the homogenates were centrifuged at 1000 × g for 3 min. Fifty μl of supernatant were combined with 10 μl λ-phosphatase (New England Biolabs) for 1 h at 37 °C, and the sample was subjected to SDS-PAGE and Western blot analysis.
Immunoprecipitations
Immunoprecipitations were performed by incubating 1000 × g supernatants of liver homogenates with monoclonal anti-eIF4E or anti-4E-BP1 antibody. Homogenate containing ∼1 mg of protein was incubated with 4 μg antibody for 2 h at 4 °C. Then, 500 μg BioMag goat anti-mouse IgG beads (Qiagen), previously blocked with low salt buffer (20 mm Tris-HCl, 150 mm NaCl, 5 mm EDTA, 0.5% Triton X-100, 0.1% β-mercaptoethanol, pH 7.4) containing 1% BSA, was added to each sample with 175 μl of PBS and 12.5 μl of Triton X-100, and the suspension was rocked at 4 °C for 1 h. Beads were washed twice with 1 ml of cold low salt buffer, once with high salt buffer (50 mm Tris HCl, 500 mm NaCl, 5 mm EDTA, 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS, 0.04% β-mercaptoethanol, pH 7.4), resuspended in 1× SDS sample buffer, and then boiled for 5 min. Supernatants were subjected to Western blot analysis using antibodies to 4E-BP1, eIF4E, and O-GlcNAc, and the results were normalized for the amount of eIF4E in the immunoprecipitate.
Quantitative Real-time PCR
RNA was isolated from livers, following a standard TRIzol protocol (Invitrogen) according to the manufacturer's instructions. Total RNA was reverse transcribed using an ABI High Capacity cDNA reverse transcription kit (Applied Biosystems). For quantitation of mRNA, quantitative RT-PCR was performed using a QuantiTech SYBR Green PCR kit (Qiagen) and the following primers for GFAT: 5′-TGGCGTGGCCAGTACAAA-3′ and 5′-GGAGATCCTGTCATCACACATCA-3′.
Statistical Analysis
The data are expressed as mean ± S.E. One-way analysis of variance and Student's t test were used to compare differences among groups. p < 0.05 was considered statistically significant.
RESULTS
4E-BP1 Is O-GlcNAcylated in Mouse Liver
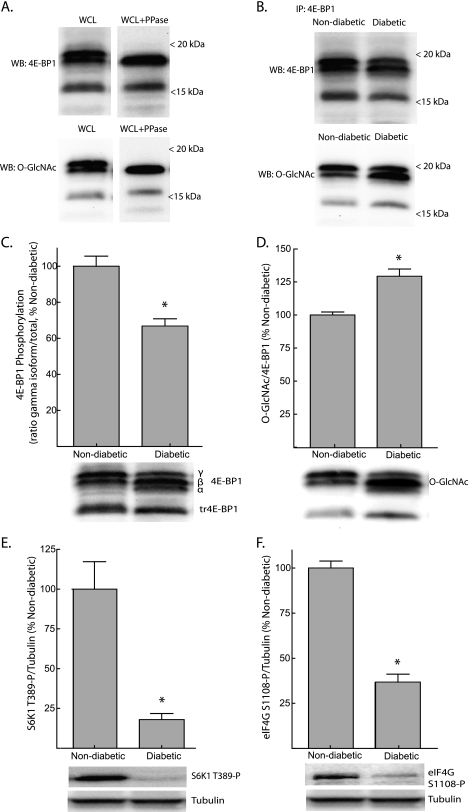
SDS-PAGE of liver supernatants resolved full-length 4E-BP1 into multiple isoforms based upon phosphorylation state in addition to a protein that was ∼3 kDa smaller (Fig. 1A, top). Western blot analysis for O-GlcNAcylated proteins revealed a signal at 15–20 kDa that was reminiscent of the banding pattern of 4E-BP1 (Fig. 1A, bottom). When liver supernatants were treated with λ-phosphatase, the phospho-isoforms of full-length 4E-BP1 collapsed into a single band at ∼18 kDa. In the presence of λ-phosphatase, a similar shift was seen with the O-GlcNAcylation bands, suggesting that the protein recognized by the antibody was 4E-BP1. Further evidence supporting such a conclusion is provided by the results shown in Fig. 1B in which 4E-BP1 was immunoprecipitated from liver supernatants using a monoclonal anti-4E-BP1 antibody. As seen in the figure, O-GlcNAc-modified 4E-BP1 was specifically detected in immunoprecipitates of liver supernatants. Detection of O-GlcNAc on both the upper and lower isoforms of 4E-BP1 suggests that the protein may be phosphorylated and O-GlcNAcylated simultaneously. In addition, O-GlcNAcylation of the truncated protein (tr4E-BP1) suggests that the modification is not restricted to the N-terminal portion of the protein.
FIGURE 1.
Reduced phosphorylation and increased O-GlcNAcylation of 4E-BP1 in the liver of Ins2Akita/+ mice. A, Western blot analysis of supernatants from liver whole cell lysates (WCL) using anti-4E-BP1 (top panels) or anti-O-GlcNAc (bottom panels) antibodies, before (left panels) or after (right panels) treatment with λ-phosphatase (PPase). B, 4E-BP1 was immunoprecipitated (IP) from liver supernatants of non-diabetic or diabetic mice and subsequently subjected to Western blot (WB) analysis for 4E-BP1 (top panel) or O-GlcNAc (bottom panel) content. 4E-BP1 phosphorylation (C) and O-GlcNAcylation (D) in the liver of Ins2Akita/+ diabetic mice was assessed by Western blot analysis. Phosphorylation was assessed as the proportion of the protein present in the γ-form relative to the total amount of 4E-BP1 in all forms (α+β+γ). Activity of mTORC1 was assessed by Western blot analysis of S6K1 phosphorylation on Thr389 (E) and eIF4G on Ser1108 (F). Representative blots are show. Values are means + S.E. (n = 5). *, p < 0.05 versus non-diabetic.
Diabetes Enhances O-GlcNAcylation of 4E-BP1 and Promotes Binding to eIF4E
In the present study, the Ins2Akita/+ mouse was selected as a model of type 1 diabetes. Compared with non-diabetic Ins2+/+ littermates, the relative phosphorylation of 4E-BP1 in the liver of diabetic mice was reduced to 66% of the non-diabetic value (Fig. 1C). In addition to reduced phosphorylation, O-GlycNAcylation of 4E-BP1 was increased by 29% (Fig. 1D). It is well established that 4E-BP1 is phosphorylated on multiple residues by mTORC1 (3). To more clearly evaluate mTORC1 signaling, the phosphorylation of two additional targets of the kinase was measured. In diabetic mice, phosphorylation of S6K1 on Thr389 and eIF4G on Ser1108 were both significantly reduced (Fig. 1, E and F), suggesting that mTORC1 signaling was repressed in the liver of diabetic compared with non-diabetic mice.
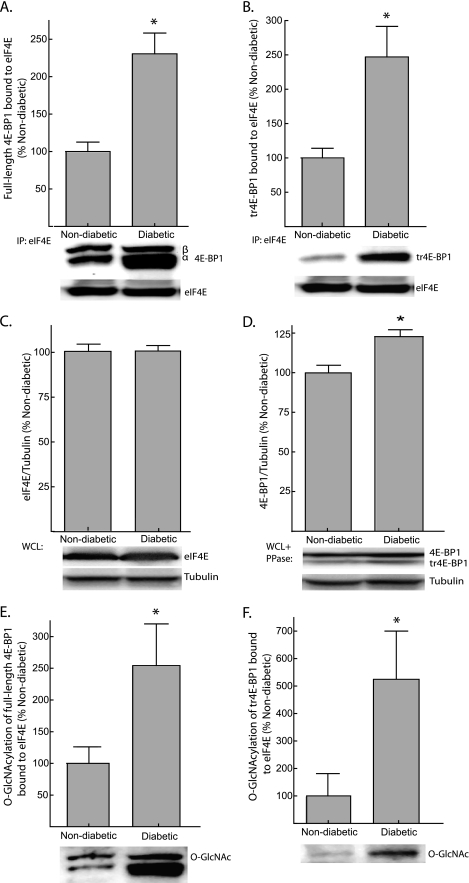
The association of 4E-BP1 with eIF4E was evaluated by immunoprecipitating eIF4E from liver supernatants and measuring the amount of 4E-BP1 present in the immunoprecipitate by Western blot analysis. The amounts of both full-length 4E-BP1 (Fig. 2A) and tr4E-BP1 (Fig. 2B) bound to eIF4E were elevated by >2-fold in the liver of diabetic Ins2Akita/+ mice when compared with non-diabetic mice. The expression of eIF4E in the liver of diabetic and non-diabetic mice was not different (Fig. 2C); however, the enhanced association of eIF4E with 4E-BP1 is potentially influenced by a 24% elevation in the expression of 4E-BP1 in the liver of diabetic mice (Fig. 2D). Because the expression of 4E-BP1 in the liver of diabetic mice is only modestly elevated compared with non-diabetic mice, yet the amount of 4E-BP1 bound to eIF4E is increased by >2-fold, it is likely that other factors, such as changes in phosphorylation or O-GlcNAcylation, play an important role in influencing the interaction of 4E-BP1 with eIF4E. Indeed, O-GlcNAcylation of full-length and truncated 4E-BP1 in the eIF4E immunoprecipitate was elevated 2.5- and 5-fold, respectively (Fig. 2, E and F).
FIGURE 2.
Increased association of full-length and truncated 4E-BP1 (tr4E-BP1) with eIF4E in the liver of Ins2Akita/+ mice. The association of full-length 4E-BP1 (A) and tr4E-BP1 (B) with eIF4E was examined by immunoprecipitating eIF4E from liver supernatants and measuring the amount of 4E-BP1 in the immunoprecipitate (IP) by Western blot analysis. C, total eIF4E was measured in WCL by Western blot analysis. D, 4E-BP1 content was assessed by treating supernatants from liver WCL with λ-phosphatase (PPase) followed by Western blot analysis as described under “Experimental Procedures.” O-GlcNAcylation of full-length 4E-BP1 (E) and tr4E-BP1 (F) bound to eIF4E was also measured by Western blot analysis. Representative blots are shown. Values are means + S.E. (n = 5). *, p < 0.05 versus non-diabetic.
A similar effect on the modification of 4E-BP1 was observed when streptozotocin was used to chemically induce diabetes in Dilute Brown Non-Agouti mice (supplemental Fig. S1). The liver of streptozotocin treated mice exhibited increased O-GlcNAcylation of 4E-BP1 and increased association of 4E-BP1 with eIF4E (supplemental Fig. S1, C and D). Notably, O-GlcNAcylation of 4E-BP1 bound to eIF4E was increased by >3-fold compared with control mice (supplemental Fig. S1, E and F). These findings confirm that increased O-GlcNAcylation of 4E-BP1 occurs in other models of diabetes. Together, these findings suggest that the changes in 4E-BP1 phosphorylation and O-GlcNAcylation that occur with diabetes may differentially affect the function of the protein.
Role of Hyperglycemia in Regulation of 4E-BP1
The role of hyperglycemia in mediating the diabetes-induced alterations in 4E-BP1 was assessed using phlorizin to reduce plasma glucose concentrations. Previous studies have shown that phlorizin treatment rapidly lowers plasma glucose concentrations in diabetic animals by producing renal glycosuria and blocking intestinal glucose absorption through inhibition of sodium-glucose symporters (e.g. Ref. 31). Ins2Akita/+ mice have elevated concentrations of blood glucose compared with non-diabetic mice (32). In the present study, 11-week-old Ins2Akita/+ mice and Ins2+/+ littermates were injected twice daily for 7 days with phlorizin or vehicle alone. Phlorizin reduced the plasma glucose of diabetic mice from a concentration in excess of 600 mg/dl (upper level of detection) to 310 ± 30 mg/dl over the 7-day treatment period (supplemental Fig. S2).
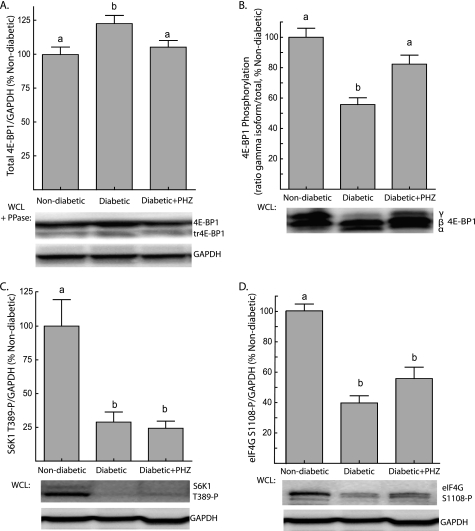
Total hepatic 4E-BP1 expression was modestly increased in diabetic animals, and upon treatment with phlorizin, the amount of 4E-BP1 in livers of diabetic animals returned to near control values (Fig. 3A). Hyperphosphorylation of 4E-BP1 favors its polyubiquitination and degradation (33); thus, these changes likely reflect alterations in phosphorylation of the protein. Notably, phlorizin treatment restored phosphorylation of 4E-BP1 in the liver of diabetic mice to a value not significantly different than non-diabetic mice (Fig. 3B). To determine whether the increase in 4E-BP1 phosphorylation was simply due to restoration of mTORC1 signaling, the effect of phlorizin treatment on phosphorylation of S6K1 and eIF4G was evaluated. Unlike 4E-BP1 phosphorylation, phlorizin treatment of diabetic mice did not result in a significant increase the phosphorylation of either S6K1 on Thr389 (Fig. 3C) or eIF4G on Ser1108 (Fig. 3D). This finding suggests that the restoration of 4E-BP1 phosphorylation by phlorizin was not due to increased mTORC1 signaling, but instead may be the result of removing a competing modification such as O-GlcNAc that inhibits 4E-BP1 phosphorylation at one or more specific sites. In fact, O-GlcNAcylation of 4E-BP1 was reduced when diabetic mice were treated with phlorizin (Fig. 4A), such that the extent of 4E-BP1 O-GlcNAcylation was positively correlated with blood glucose concentration following 7 days of treatment (supplemental Fig. S3).
FIGURE 3.
Phlorizin treatment of diabetic mice lowers blood glucose concentrations and alters hepatic 4E-BP1 phosphorylation without changing mTORC1 signaling. Ins2Akita/+ diabetic mice and non-diabetic littermates were subcutaneously injected with solvent or phlorizin (PHZ) twice daily for 7 days to lower blood glucose concentrations (see supplemental Fig. S1). A, 4E-BP1 content was assessed by treating supernatants from liver WCL with λ-phosphatase (PPase) followed by Western blot analysis as described under “Experimental Procedures.” B, 4E-BP1 phosphorylation was assessed by Western blot analysis as described in the legend to Fig. 1. Western blot analysis of S6K1 phosphorylation on Thr389 (C) and eIF4G phosphorylation on Ser1108 (D). Samples were blotted for GAPDH to serve as a loading control. Representative blots are shown. Values are means + S.E. (n = 10). Statistical significance is denoted by the presence of different letters above the bars on the graphs. Bars with different letters are statistically different, p < 0.05.
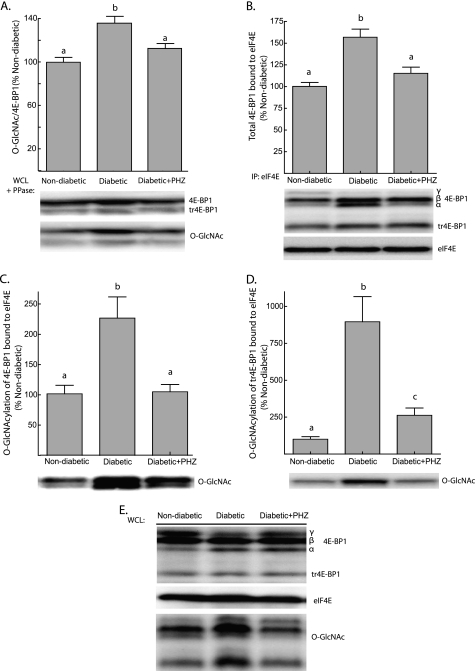
FIGURE 4.
Phlorizin treatment of diabetic mice inhibits hepatic O-GlcNAcylation of 4E-BP1 and reduces the amount of 4E-BP1 bound to eIF4E. Ins2Akita/+ diabetic mice and non-diabetic littermates were treated with phlorizin (PHZ) or solvent only twice daily for 7 days. A, 4E-BP1 content was assessed by treating supernatants from liver WCL with λ-phosphatase (PPase) followed by Western blot analysis as described under “Experimental Procedures.” B, the association of 4E-BP1 with eIF4E was examined by immunoprecipitating (IP) eIF4E from liver supernatants and measuring the amount of 4E-BP1 in the immunoprecipitate by Western blot analysis and normalizing the results to the amount of eIF4E in the immunoprecipitate. O-GlcNAcylation of full-length 4E-BP1 (C) and truncated 4E-BP1 (tr4E-BP1; D) bound to eIF4E was also measured by Western blot analysis. E, total eIF4E, 4E-BP1, and 4E-BP1 O-GlcNAcylation was assessed in WCL by Western blot analysis. Representative blots are shown. Values are means + S.E. (n = 10). Statistical significance is denoted by the presence of different letters above the bars on the graphs. Bars with different letters are statistically different, p < 0.05.
To further evaluate the possibility that reducing O-GlcNAcylation may improve phosphorylation of a specific site(s), 4E-BP1 was analyzed with phosphospecific antibodies. Interestingly, phosphorylation of 4E-BP1 at Thr70 was increased when diabetic mice were treated with phlorizin, whereas there was no change in phosphorylation at either Thr36/47 or Ser65 (supplemental Fig. S4, A–C). Additionally, when O-GlcNAcylated 4E-BP1 was immunoprecipitated from liver supernatants, it was observed to have reduced phosphorylation at Thr70 compared with total liver supernatant (supplemental Fig. S4D). Together, these findings suggest that O-GlcNAcylation of 4EBP1 inhibits phosphorylation at Thr70. One potential residue that could be O-GlcNAcylated to inhibit Thr70 phosphorylation is Thr81, which in silico O-GlcNAcylation site prediction scores as the most likely target of modification (supplemental Fig. S4E). When Thr81 was mutated to alanine, O-GlcNAcylation of the variant protein in HEK293T cells was reduced by 62% compared with wild-type 4E-BP1 (supplemental Fig. S4E). Interestingly, 4E-BP1 phosphorylation at Thr81 has been previously identified by large scale phosphoproteomic analysis (34).
As a consequence of the increase in 4E-BP1 phosphorylation, it was anticipated that phlorizin treatment of diabetic mice would also reduce the interaction of 4E-BP1 with eIF4E. Immunoprecipitation of eIF4E from liver supernatants revealed this to be the case, as phlorizin treatment reduced the amount of 4E-BP1 in immunoprecipitates from the liver of Ins2Akita/+ diabetic compared with non-diabetic littermates (Fig. 4B). Analysis of 4E-BP1 in the eIF4E immunoprecipitate also revealed a reduction in the O-GlcNAcylation of 4E-BP1 bound to eIF4E following phlorizin treatment of diabetic mice. Indeed, the O-GlcNAcylation of full-length 4E-BP1 bound to eIF4E in the liver of diabetic mice following treatment with phlorizin was not significantly different than non-diabetic values (Fig. 4C), whereas O-GlcNAcylation on tr4E-BP1 was reduced from 9-fold to just 2.5-fold above non-diabetic values (Fig. 4D). These data suggest that 4E-BP1 O-GlcNAcylation may limit the amount of eIF4E available for eIF4F assembly.
Hyperglycemia Increases 4E-BP1 O-GlcNAcylation by Increasing Flux through Hexosamine Biosynthetic Pathway
A number of studies have shown that flux through the hexosamine biosynthetic pathway increases during diabetes and in response to hyperglycemic conditions, leading to enhanced O-GlcNAcylation of proteins (15, 35, 36). High plasma glucose concentrations provide additional substrate for both glycolysis and the hexosamine biosynthetic pathway; however, because hyperglycemia inhibits glycolysis through GAPDH (37) and up-regulates the expression and activity of GFAT, the rate-limiting enzyme of the hexosamine biosynthetic pathway (38), the net result is a substantial increase in UDP-GlcNAc production. In support of this possibility, in H4IIE rat liver heptoma cells incubated in medium containing 25 mm glucose, both the amount of 4E-BP1 associated with eIF4E and its O-GlcNAcylation were elevated compared with cells maintained in medium containing 5 mm glucose (supplemental Fig. S5A).
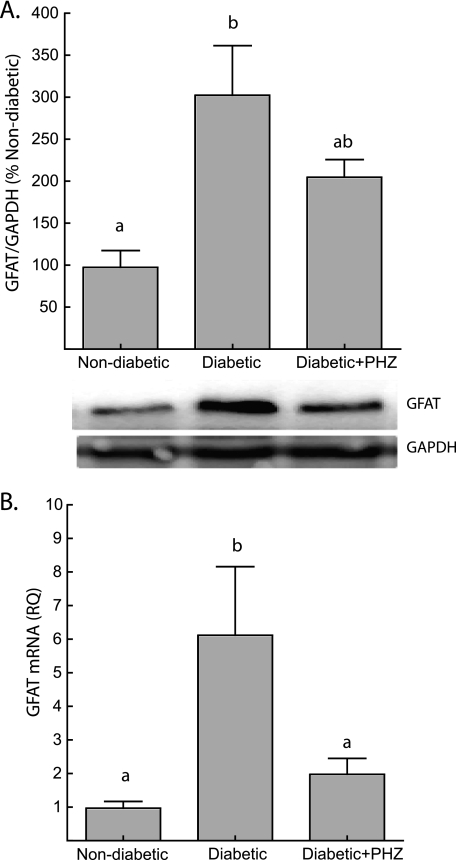
To determine whether increased flux through the hexosamine biosynthetic pathway could be responsible for alterations in 4E-BP1 O-GlcNAcylation, we measured the expression of GFAT in the liver of diabetic and non-diabetic mice and determined the effect of phlorizin treatment on its expression. The expression of both GFAT protein (Fig. 5A) and mRNA (Fig. 5B) in the liver of Ins2Akita/+ mice was increased significantly compared with the non-diabetic value. In addition, phlorizin treatment and consequent reduction of blood glucose concentrations resulted in down-regulation of GFAT protein and mRNA expression in the liver of diabetic mice to values not significantly different than non-diabetic controls after 7 days of treatment. These changes in GFAT expression suggest that flux through the hexosamine biosynthetic pathway could play an important role in mediating the effects of hyperglycemia on 4E-BP1. This conclusion is also supported by the finding that treating H4IIE cells with azaserine, an inhibitor of GFAT, blocked the high glucose-mediated increase in the association of 4E-BP1 with eIF4E (supplemental Fig. S5B).
FIGURE 5.
Increased expression of GFAT in the liver of Ins2Akita/+ diabetic mice is mediated by hyperglycemia. A, the expression of GFAT in liver supernatants was assessed by Western blot analysis and normalized to GAPDH content. Representative blots are shown. B, RNA was extracted from liver homogenates following a standard TRIzol protocol. GFAT mRNA expression was normalized to an internal GAPDH control. Values are means + S.E. (n = 10). Statistical significance is denoted by the presence of different letters above the bars on the graphs. Bars with different letters are statistically different, p < 0.05.
O-GlcNAcylation of 4E-BP1 Increases Its Association with eIF4E
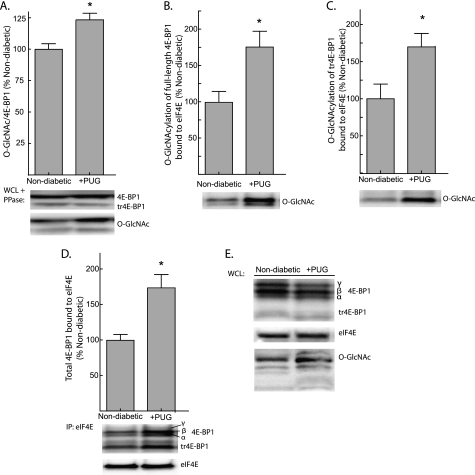
To evaluate the influence of O-GlcNAcylation on the interaction of 4E-BP1 with eIF4E, livers from Ins2+/+ non-diabetic mice were perfused in situ with perfusate containing PUGNAc, a non-metabolizable analog of glucosamine that forms a stable complex with and inhibits O-GlcNAcase, the enzyme that catalyzes the removal of O-GlcNAc residues from protein. By disrupting O-GlcNAc cycling, PUGNAc increases O-GlcNAcylation of nuclear and cytoplasmic proteins (39). Livers from non-diabetic mice were perfused in situ with a non-recirculating medium containing 11 mm glucose and either 40 μm PUGNAc or vehicle alone. Perfusing livers with PUGNAc increased total 4E-BP1 O-GlcNAcylation by 24% (Fig. 6A). The O-GlcNAcylation of both full-length (Fig. 6B) and truncated 4E-BP1 (Fig. 6C) associated with eIF4E was increased by 75 and 70%, respectively, when non-diabetic livers were perfused with PUGNAc. In addition, PUGNAc also produced a 74% increase in the association of 4E-BP1 with eIF4E (Fig. 6D) but did not alter the expression of eIF4E or 4E-BP1 in the liver of non-diabetic mice (Fig. 6E). Similar results were obtained by treating H4IIE cells with PUGNAc (supplemental Fig. S5C). Together, these findings suggest an association between O-GlcNAcylation of 4E-BP1 and its interaction with eIF4E.
FIGURE 6.
PUGNAc, an inhibitor of O-GlcNAcase, increases the binding of 4E-BP1 to eIF4E in perfused liver. Livers from non-diabetic Ins2+/+ mice were perfused in situ with a nonrecirculating medium containing either 40 μm PUGNAc (PUG) or vehicle alone. A, O-GlcNAcylation of total 4E-BP1 was assessed by treating supernatants from liver WCL with λ-phosphatase (PPase) followed by Western blot analysis. O-GlcNAcylation of full-length 4E-BP1 (B) and truncated 4E-BP1 (tr4E-BP1; C) bound to eIF4E was measured by Western blot analysis. D, the association of 4E-BP1 with eIF4E was examined by immunoprecipitating eIF4E from liver supernatants and measuring the amount of 4E-BP1 in the immunoprecipitate by Western blot analysis. E, total eIF4E, 4E-BP1, and 4E-BP1 O-GlcNAcylation was assessed in WCL by Western blot analysis. Representative blots are shown. Values are means + S.E. (n = 8). Statistical significance is denoted by *, p < 0.05.
In a second set of experiments, in situ perfusion was performed on livers from both Ins2Akita/+ diabetic and non-diabetic mice. Due to the absence of insulin from the perfusion medium, mTORC1 signaling in the liver of non-diabetic mice was reduced by in situ perfusion, such that there was no significant difference in the phosphorylation of 4E-BP1 (Fig. 7A), S6K1 on Thr389 (Fig. 7B) nor eIF4G on Ser1108 (Fig. 7C) between the non-diabetic and diabetic conditions following a 30-min perfusion. However, O-GlcNAcylation of 4E-BP1 remained elevated in the liver of diabetic mice (Fig. 7D). Despite similar 4E-BP1 phosphorylation, the amount of 4E-BP1 bound to eIF4E remained elevated by 65% in livers from diabetic mice in comparison to non-diabetic mice (Fig. 7E). Similarly, the association of tr4E-BP1 with eIF4E remained elevated by 110% in diabetic as compared with non-diabetic mice (Fig. 7F). Whereas reduced phosphorylation of 4E-BP1 did not appear to be responsible for alterations in the association of 4E-BP1 with eIF4E, the O-GlcNAcylation of both full-length 4E-BP1 (Fig. 7G) and tr4E-BP1 (Fig. 7H) bound to eIF4E remained elevated in livers from diabetic mice. These results suggest that O-GlcNAcylation of 4E-BP1 may enhance its interaction with eIF4E independent of 4E-BP1 phosphorylation. Thus, O-GlcNAcylation and phosphorylation of 4E-BP1 may independently control binding of the protein to eIF4E in response to a variety of changes in the cellular environment.
FIGURE 7.
O-GlcNAcylation of hepatic 4E-BP1 increases its association with eIF4E. Livers from Ins2Akita/+ diabetic mice and non-diabetic littermates were perfused in situ with a nonrecirculating medium in the absence of insulin. A, phosphorylation of 4E-BP1 was evaluated by Western blot analysis as described in the legend to Fig. 1. Phosphorylation of S6K1 on Thr389 (B) and eIF4G on Ser1108 (C) was measured by Western blot using phosphospecific antibodies. D, O-GlcNAcylation of total 4E-BP1 was measured by Western blot analysis. The association of 4E-BP1 (E) and truncated 4E-BP1 (tr4E-BP1; F) with eIF4E was examined by immunoprecipitating (IP) eIF4E from liver supernatants and measuring the amount of 4E-BP1 in the immunoprecipitate by Western blot analysis. O-GlcNAcylation of full-length 4E-BP1 (G) and tr4E-BP1 (H) bound to eIF4E was measured by Western blot analysis. I, total eIF4E, 4E-BP1, and 4E-BP1 O-GlcNAcylation was assessed in WCL by Western blot analysis. Representative blots are shown. Values are means + S.E. (n = 8). Statistical significance is denoted by *, p < 0.05.
DISCUSSION
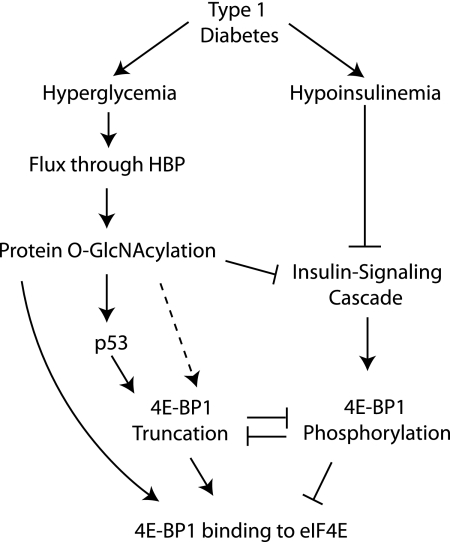
The findings of the present study demonstrate that in addition to the well characterized regulation of 4E-BP1 by mTORC1-mediated phosphorylation, O-GlcNAcylation also plays an important role in controlling its association with the cap-binding protein eIF4E. In the liver of diabetic mice that are hypoinsulinemic and hyperglycemic, 4E-BP1 exhibited both reduced phosphorylation and increased O-GlcNAcylation compared with the non-diabetic condition. In addition, higher levels of O-GlcNAc were particularly evident on hypophosphorylated forms of 4E-BP1 found to be associated with eIF4E, as was the case with tr4E-BP1. When hyperglycemic conditions were attenuated with phlorizin, mTORC1 signaling remained repressed in the liver of diabetic mice, yet the association of 4E-BP1 with eIF4E returned to non-diabetic values, as did the O-GlcNAcylation of 4E-BP1. In addition, perfusing livers of non-diabetic mice with PUGNAc increased both O-GlcNAcylation of 4E-BP1 and its association with eIF4E. Furthermore, when 4E-BP1 phosphorylation was normalized in the liver of non-diabetic and diabetic mice, O-GlcNAcylation and the association of 4E-BP1 with eIF4E remained elevated in the livers of diabetic mice. Together, these data provide evidence for a model wherein hyperglycemia-induced O-GlcNAcylation of 4E-BP1 increases its association with eIF4E in a manner that is not entirely dependent on a reduction in 4E-BP1 phosphorylation (Fig. 8).
FIGURE 8.
Model for hyperglycemia-induced 4E-BP1 O-GlcNAcylation and truncation. Under diabetic conditions, a combination of hyperglycemia and hypoinsulinemia produce increased 4E-BP1 binding to eIF4E to inhibit the translation of mRNAs through a cap-dependent process. Hyperglycemia-driven increased flux through the hexosamine biosynthetic pathway (HBP) leads to O-GlcNAcylation of numerous proteins. Repressed insulin-signaling occurs in response to both hypoinsulinemia and O-GlcNAcylation of IRS-1 and Akt, leading to reduced phosphorylation of 4E-BP1 by the mTORC1 signaling pathway. Hypophosphorylated 4E-BP1 binds strongly to eIF4E, whereas phosphorylation leads to dissociation. Reductions in 4E-BP1 phosphorylation are associated with increased 4E-BP1 O-GlcNAcylation, which enhances the interaction of 4E-BP1 with eIF4E independent of the phosphorylation state. In addition, O-GlcNAcylation of p53 blocks its proteasomal degradation, potentially increasing p53-mediated truncation of 4E-BP1. Truncated 4E-BP1 is almost completely unphosphorylated and interacts strongly with eIF4E. Although phosphorylation of 4E-BP1 inhibits its truncation, it is possible that O-GlcNAcylation may serve a role in directly promoting truncation, thereby further enhancing its interaction with eIF4E (dashed line).
The O-GlcNAc cycling enzymes O-GlcNAc transferase and O-GlcNAcase strongly associate with cytosolic ribosomes (40). In addition, a number of key components of the translational machinery, including numerous ribosomal proteins and translation factors, have been identified as being O-GlcNAc-modified in global glycoproteomic studies (40). In total, O-GlcNAcylation has been detected on 34 of the ∼80 proteins that compose the mammalian ribosome (40). Among translation initiation factors, evidence exists that eIF4G (41), eIF3g (42), eIF4A (43), and poly(A)-binding protein (44) are all O-GlcNAcylated; however, the functional consequence of these modifications remains to be established. O-GlcNAcylation of the eIF2-associated factor p67 prevents its degradation and stabilizes the protein to promote association with eIF2 (45). This association prevents eIF2α phosphorylation (45), which is a key mechanism responsible for the inhibition of translation initiation in response to cell stress. Thus, O-GlcNAcylation of proteins comprising the translational machinery likely represents a key signaling mechanism that is relatively unexplored in terms of its functional consequences on protein synthesis.
Regulation of 4E-BP1 function by O-GlcNAcylation is particularly appealing in light of the fact that other components of the insulin-signaling cascade have been reported to undergo regulation by similar modification (i.e. insulin receptor-β, insulin receptor substrate 1 (IRS-1), PDK1, and Akt (11, 46, 47)). In fact, elevation of intracellular O-GlcNAc concentrations promotes insulin resistance in multiple cell types (11, 15). Under hyperglycemic conditions, the Src homology 2 (SH2) domain-containing C terminus of IRS-1 is modified on multiple residues by O-GlcNAcylation, and these modifications appear to inhibit the insulin-signaling cascade (7). Similarly, O-GlcNAcylation of Akt has been shown to inhibit its phosphorylation at Thr308 leading to repressed kinase activity and thus insulin action (7). Thus, O-GlcNAcylation of 4E-BP1 represents another level of regulation whereby flux through the hexosamine biosynthetic pathway represses mediators of the insulin-signaling cascade.
When glucose enters the cell, it is phosphorylated to form glucose-6-phosphate in a reaction catalyzed by the enzyme hexokinase. The liver, which was the focus of the present study, contains glucokinase, a specialized isoform of hexokinase that is characterized by a low affinity for glucose, relative insensitivity to feedback inhibition by glucose-6-phosphate, and sigmoid kinetics displaying cooperative behavior for glucose (48). Adipocytes exposed to increasing amounts of glucose in the presence of insulin exhibit a 365% increase in glucose uptake, yet UDP-GlcNAc concentrations are only elevated by ∼35% (49). However, by working in conjunction with the high capacity/low affinity GLUT2 glucose transporter, glucokinase allows for greater glucose utilization by the liver under hyperglycemic conditions, thus enabling the organ to regulate whole body glucose levels. Therefore, high hepatic intracellular glucose levels, such as those seen in type 1 diabetes, would not limit the flux of glucose through the hexosamine biosynthetic pathway in the liver, as would potentially be the case in other tissues (i.e. adipose or muscle). As a result, the effects of hyperglycemia on changes in 4E-BP1 O-GlcNAcylation potentially have dramatic effects on protein synthesis in the liver of diabetic animals.
In addition to being modified by O-GlcNAcylation, 4E-BP1 in the liver was also post-translationally modified by N-terminal truncation. Under conditions of cellular stress, activation of p53 stimulates proteasome-dependent truncation of 4E-BP1 to produce an unphosphorylated isoform (tr4E-BP1) that interacts preferentially with eIF4E (19). Under normal conditions, p53 expression is low due to its phosphorylation on Thr155, which targets the protein for proteasomal degradation. However, under conditions that promote flux through the hexosamine biosynthetic pathway, phosphorylation of p53 on Thr155 is impaired by O-GlcNAcylation on neighboring Ser149, which blocks the interaction between p53 and mdm2, an E3 ubiquitin ligase that targets p53 for proteasomal degradation (21). In the present study, increased levels of tr4E-BP1 associated with eIF4E in the liver of diabetic mice in comparison to non-diabetic mice. Thus, conditions that favor increased O-GlcNAcylation potentially reduce 4E-BP1 phosphorylation by both promoting p53-mediated truncation and through competitive O-GlcNAcylation of residues at or around phosphorylation sites. It is unclear how p53 activation controls the proteasome-mediated cleavage of 4E-BP1; however, phosphorylation of 4E-BP1 appears to inhibit its truncation (19). In addition, recent findings that O-GlcNAcylation of host cell factor-1 promotes limited proteolytic cleavage of the protein (50) make it tempting to speculate that O-GlcNAcylation of 4E-BP1 could directly promote its truncation.
4E-BP1 O-GlcNAcylation and truncation represent a novel mechanism for selecting specific mRNAs for translation. Initiation factor eIF4E is essential for recognition of the 5′-m7GTP cap on mRNA. Thus, changes in the level or availability of eIF4E has differential effects on various mRNAs such that association with 4E-BPs prevents the translation of mRNA through a cap-dependent process. By increasing the association with eIF4E, O-GlcNAcylation of 4E-BP1 likely contributes to the pathogenesis of metabolic abnormalities associated with diabetes and makes further exploration of this modification an intriguing area for future research.
Supplementary Material
Acknowledgments
We thank Wendy Dunton for assistance with animal maintenance, phlorizin treatment, and blood glucose analysis; Sharon L. Rannels for performing liver perfusion experiments; and Chichun E. Sun for assistance with immunoprecipitations and Western blot analysis.
This work was supported by National Institutes of Health Grants DK13499 (to L. S. J.) and DK088416-01A1 (to M. D. D.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
- mTORC1
- mammalian target of rapamycin in complex 1
- GFAT
- glutamine-fructose-6-phosphate amidotransferase
- PUGNAc
- O-(2-acetamido-2-deoxy-d-glucopyranosylidenamino)N-phenylcarbamate
- WCL
- whole cell lysate.
REFERENCES
- 1. Ptushkina M., von der Haar T., Karim M. M., Hughes J. M., McCarthy J. E. (1999) EMBO J. 18, 4068–4075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. von der Haar T., Gross J. D., Wagner G., McCarthy J. E. (2004) Nat. Struct. Mol. Biol. 11, 503–511 [DOI] [PubMed] [Google Scholar]
- 3. Gingras A. C., Kennedy S. G., O'Leary M. A., Sonenberg N., Hay N. (1998) Genes Dev. 12, 502–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kimball S. R., Jefferson L. S., Fadden P., Haystead T. A., Lawrence J. C., Jr. (1996) Am. J. Physiol. 270, C705–709 [DOI] [PubMed] [Google Scholar]
- 5. Zeidan Q., Hart G. W. (2010) J. Cell Sci. 123, 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Buse M. G., Robinson K. A., Marshall B. A., Hresko R. C., Mueckler M. M. (2002) Am. J. Physiol. Endocrinol. Metab. 283, E241–250 [DOI] [PubMed] [Google Scholar]
- 7. Yang X., Ongusaha P. P., Miles P. D., Havstad J. C., Zhang F., So W. V., Kudlow J. E., Michell R. H., Olefsky J. M., Field S. J., Evans R. M. (2008) Nature 451, 964–969 [DOI] [PubMed] [Google Scholar]
- 8. Griffith L. S., Schmitz B. (1999) Eur. J. Biochem. 262, 824–831 [DOI] [PubMed] [Google Scholar]
- 9. Cheng X., Cole R. N., Zaia J., Hart G. W. (2000) Biochemistry 39, 11609–11620 [DOI] [PubMed] [Google Scholar]
- 10. Wells L., Vosseller K., Hart G. W. (2001) Science 291, 2376–2378 [DOI] [PubMed] [Google Scholar]
- 11. Vosseller K., Wells L., Lane M. D., Hart G. W. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 5313–5318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Snow D. M., Hart G. W. (1998) Int. Rev. Cytol. 181, 43–74 [DOI] [PubMed] [Google Scholar]
- 13. Zachara N. E., Hart G. W. (2002) Chem. Rev. 102, 431–438 [DOI] [PubMed] [Google Scholar]
- 14. Zachara N. E., Hart G. W., Cole R. N., Gao Y. (2002) Curr. Protoc. Mol. Biol. Chapter 17, Unit 17 16 [DOI] [PubMed] [Google Scholar]
- 15. McClain D. A., Lubas W. A., Cooksey R. C., Hazel M., Parker G. J., Love D. C., Hanover J. A. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 10695–10699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu K., Paterson A. J., Chin E., Kudlow J. E. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 2820–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Buse M. G. (2006) Am. J. Physiol. Endocrinol. Metab. 290, E1-E8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Constantinou C., Clemens M. J. (2007) Cell Death Differ 14, 576–585 [DOI] [PubMed] [Google Scholar]
- 19. Constantinou C., Elia A., Clemens M. J. (2008) Biol. Cell 100, 279–289 [DOI] [PubMed] [Google Scholar]
- 20. Constantinou C., Clemens M. J. (2005) Oncogene 24, 4839–4850 [DOI] [PubMed] [Google Scholar]
- 21. Yang W. H., Kim J. E., Nam H. W., Ju J. W., Kim H. S., Kim Y. S., Cho J. W. (2006) Nat. Cell Biol. 8, 1074–1083 [DOI] [PubMed] [Google Scholar]
- 22. Yoshinaga T., Nakatome K., Nozaki J., Naitoh M., Hoseki J., Kubota H., Nagata K., Koizumi A. (2005) Biol. Chem. 386, 1077–1085 [DOI] [PubMed] [Google Scholar]
- 23. Yoshioka M., Kayo T., Ikeda T., Koizumi A. (1997) Diabetes 46, 887–894 [DOI] [PubMed] [Google Scholar]
- 24. Oyadomari S., Koizumi A., Takeda K., Gotoh T., Akira S., Araki E., Mori M., Oyadomari S., Koizumi A., Takeda K., Gotoh T., Akira S., Araki E., Mori M. (2002) J. Clin. Invest. 109, 525–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kimball S. R., Horetsky R. L., Jefferson L. S. (1998) J. Biol. Chem. 273, 30945–30953 [DOI] [PubMed] [Google Scholar]
- 26. Kimball S. R., Jurasinski C. V., Lawrence J. C., Jr., Jefferson L. S. (1997) Am. J. Physiol. 272, C754–759 [DOI] [PubMed] [Google Scholar]
- 27. Kimball S. R., Siegfried B. A., Jefferson L. S. (2004) J. Biol. Chem. 279, 54103–54109 [DOI] [PubMed] [Google Scholar]
- 28. Flaim K. E., Liao W. S., Peavy D. E., Taylor J. M., Jefferson L. S. (1982) J. Biol. Chem. 257, 2939–2946 [PubMed] [Google Scholar]
- 29. Dennis M. D., Baum J. I., Kimball S. R., Jefferson L. S. (2011) J. Biol. Chem. 286, 8287–8296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sun H., Lu C. H., Shi H., Gao L., Yao S. Q. (2008) Nat. Protoc. 3, 1485–1493 [DOI] [PubMed] [Google Scholar]
- 31. Hong E. G., Jung D. Y., Ko H. J., Zhang Z., Ma Z., Jun J. Y., Kim J. H., Sumner A. D., Vary T. C., Gardner T. W., Bronson S. K., Kim J. K. (2007) Am. J. Physiol. Endocrinol. Metab. 293, E1687–1696 [DOI] [PubMed] [Google Scholar]
- 32. Barber A. J., Antonetti D. A., Kern T. S., Reiter C. E., Soans R. S., Krady J. K., Levison S. W., Gardner T. W., Bronson S. K. (2005) Invest Ophthalmol. Vis. Sci. 46, 2210–2218 [DOI] [PubMed] [Google Scholar]
- 33. Elia A., Constantinou C., Clemens M. J. (2008) Oncogene 27, 811–822 [DOI] [PubMed] [Google Scholar]
- 34. Cantin G. T., Yi W., Lu B., Park S. K., Xu T., Lee J. D., Yates J. R., 3rd. (2008) J. Proteome Res. 7, 1346–1351 [DOI] [PubMed] [Google Scholar]
- 35. Marshall S., Bacote V., Traxinger R. R. (1991) J. Biol. Chem. 266, 4706–4712 [PubMed] [Google Scholar]
- 36. McClain D. A. (2002) J Diabetes Complications 16, 72–80 [DOI] [PubMed] [Google Scholar]
- 37. Du X., Matsumura T., Edelstein D., Rossetti L., Zsengellér Z., Szabó C., Brownlee M. (2003) J. Clin. Invest. 112, 1049–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yki-Järvinen H., Daniels M. C., Virkamäki A., Mäkimattila S., DeFronzo R. A., McClain D. (1996) Diabetes 45, 302–307 [DOI] [PubMed] [Google Scholar]
- 39. Haltiwanger R. S., Grove K., Philipsberg G. A. (1998) J. Biol. Chem. 273, 3611–3617 [DOI] [PubMed] [Google Scholar]
- 40. Zeidan Q., Wang Z., De Maio A., Hart G. W. (2010) Mol. Biol. Cell 21, 1922–1936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Khidekel N., Ficarro S. B., Clark P. M., Bryan M. C., Swaney D. L., Rexach J. E., Sun Y. E., Coon J. J., Peters E. C., Hsieh-Wilson L. C. (2007) Nat. Chem. Biol. 3, 339–348 [DOI] [PubMed] [Google Scholar]
- 42. Teo C. F., Ingale S., Wolfert M. A., Elsayed G. A., Nöt L. G., Chatham J. C., Wells L., Boons G. J. (2010) Nat. Chem. Biol. 6, 338–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wells L., Vosseller K., Cole R. N., Cronshaw J. M., Matunis M. J., Hart G. W. (2002) Mol. Cell Proteomics 1, 791–804 [DOI] [PubMed] [Google Scholar]
- 44. Wang Z., Pandey A., Hart G. W. (2007) Mol. Cell Proteomics 6, 1365–1379 [DOI] [PubMed] [Google Scholar]
- 45. Datta R., Choudhury P., Bhattacharya M., Soto Leon F., Zhou Y., Datta B. (2001) Biochimie 83, 919–931 [DOI] [PubMed] [Google Scholar]
- 46. Ball L. E., Berkaw M. N., Buse M. G. (2006) Mol. Cell Proteomics 5, 313–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Klein A. L., Berkaw M. N., Buse M. G., Ball L. E. (2009) Mol. Cell Proteomics 8, 2733–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Massa M. L., Gagliardino J. J., Francini F. (2011) IUBMB Life 63, 1–6 [DOI] [PubMed] [Google Scholar]
- 49. Bosch R. R., Pouwels M. J., Span P. N., Olthaar A. J., Tack C. J., Hermus A. R., Sweep C. G. (2004) Endocrine 23, 17–24 [DOI] [PubMed] [Google Scholar]
- 50. Daou S., Mashtalir N., Hammond-Martel I., Pak H., Yu H., Sui G., Vogel J. L., Kristie T. M., Affar el B. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 2747–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.