Abstract
Mucus hypersecretion is a prominent feature of respiratory diseases, and MUC5B is a major airway mucin. Mucin gene expression can be affected by inflammatory mediators, including prostaglandin (PG) D2, an inflammatory mediator synthesized by hematopoietic PGD synthase (H-PGDS). PGD2 binds to either d-prostanoid receptor (DP1) or chemoattractant receptor homologous molecule expressed on T-helper type 2 cells (CRTH2). We investigated the mechanisms by which PGD2 induces MUC5B gene expression in airway epithelial cells. Western blot analysis showed that H-PGDS was highly expressed in nasal polyps. Similar results were obtained for PGD2 expression. In addition, we could clearly detect the expressions of both H-PGDS and DP1 in nasal epithelial cells but not CRTH2. We demonstrated that PGD2 increased MUC5B gene expression in normal human nasal epithelial cells as well as in NCI-H292 cells in vitro. S5751, a DP1 antagonist, inhibited PGD2-induced MUC5B expression, whereas a CRTH2 antagonist (OC0459) did not. These data suggest that PGD2 induced MUC5B expression via DP1. Pretreatment with extracellular signal-regulated kinase (ERK) inhibitor (PD98059) blocked both PGD2-induced ERK mitogen-activated protein kinase (MAPK) activation and MUC5B expression. Proximity ligation assays showed direct interaction between RSK1 and cAMP response element-binding protein (CREB). Stimulation with PGD2 caused an increase in intracellular cAMP levels, whereas intracellular Ca2+ did not have such an effect. PGD2-induced MUC5B mRNA levels were regulated by CREB via direct interaction with two cAMP-response element sites (−921/−914 and −900/−893). Finally, we demonstrated that PGD2 can induce MUC5B overproduction via ERK MAPK/RSK1/CREB signaling and that DP1 receptor may have suppressive effects in controlling MUC5B overproduction in the airway.
Keywords: DP1, MUC5B, Prostaglandin D2
Introduction
Mucus secretion in the airway, including the nasal and paranasal sinus cavities, is drained by the mucociliary transport system (1). Normal mucus secretion is essential for survival (2), but up-regulation of mucin gene expression is a major manifestation of chronic airway diseases such as allergic rhinitis, asthma, and cystic fibrosis (3, 4). Patients suffering from these diseases have pathological abnormalities in both the submucosal glands and surface epithelium, characterized by inflammation, increased mucus cell number, and increased airway mucus. Several classes of inflammatory mediators have been implicated in the process of mucus hypersecretion based on their ability to stimulate secretion from cultured cells and tissue explants (5). These inflammatory mediators are lipopolysaccharides (6), reactive oxygen species (7), and arachidonic acid metabolites (8, 9).
A total of 20 different mucin genes have been identified and subdivided into two groups, membrane-bound and secreted mucins, according to Human Genome Mapping conventions. The secreted mucins are MUC5AC, MUC5B, MUC6, and MUC19 (10–13). Mucins are primarily synthesized by two different cell types in the airway tract, namely, goblet cells and submucosal glandular cells. The major secreted mucins, MUC5AC and MUC5B, are highly expressed in the goblet cells of human airway epithelium and in the submucosal glands (14). Interestingly, differences in cell type-specific MUC5B gene expression are significantly associated with the pathogenesis of airway diseases (15), although little is known about the regulation of this gene.
Prostaglandins (PGs)2 are potent biologically active lipid mediators that are produced from arachidonic acid by almost every cell type (16) and are also known to regulate immune responses (17). One of them, prostaglandin D2 (PGD2), is thought to be involved in allergic reactions (18), and its actions are mediated via specific cell surface receptors coupled to G proteins, D prostanoid receptor 1 (DP1), and chemoattractant receptor homologous molecules expressed on Th2 cells (CRTH2/DP2) (19). Activation of DP receptor leads to a Gs-mediated increase in intracellular cAMP and agonist-induced Ca2+ flux (20). Moreover, PGD2 signaling through CRTH2 coupled with the Gi-type G protein leads to a decrease in cAMP, which subsequently stimulates intracellular Ca2+ in various cell types (21). Because increases in intracellular Ca2+ levels are often associated with immune-cell activation, the chemotactic effects of CRTH2 are in agreement with its reported signaling pattern (22). There are two isoforms of PGD synthase in the biosynthesis pathway. Hematopoietic PGD2 synthase (H-PGDS) contributes to the production of PGD2 in antigen-presenting cells and mast cells in diverse tissues (23, 24), whereas lipocalin-type PGDS is generally expressed in the central nervous system (25). In addition, it has been reported that mouse models of allergic pulmonary inflammation suggest an important pathogenetic role for PGD2 (26). Local antigen challenge also stimulates PGD2 production in the nasal mucosa of patients with allergic rhinitis (27). Thus, PGD2 seems to be an important chemical mediator in various allergic diseases.
A better understanding of PGD2-mediated activation of airway epithelial cells is potentially important for establishing a therapeutic strategy for allergic inflammation, but the precise effects of PGD2 on airway epithelial cells and receptor usage are not fully understood. In this study we investigated the mechanisms by which PGD2 induces MUC5B gene expression in airway epithelial cells. We found that the DP1 receptor played a critical role in PGD2-induced MUC5B gene expression in the airway. In addition, we observed that H-PGDS protein was highly expressed in nasal polyps tissues compared with the level in normal nasal mucosa. The level of PGD2 was also increased in nasal polyp tissues in both allergic and non-allergic patients. The DP1 receptor, but not the CRTH2-receptor, was highly expressed in human primary nasal epithelial cells. Our results showed a critical role of extracellular signal-regulated kinase (ERK1/2) mitogen-activated protein kinase (MAPK) in PGD2-induced MUC5B gene expression in airway epithelial cells. Furthermore, p90 ribosomal S6 protein kinase 1 (RSK1) and cAMP response element (CRE)-binding protein (CREB) were found to be required for PGD2-induced MUC5B gene expression. Fluorescent in situ proximity ligation assays of NCI-H292 cells demonstrated that RSK1 can directly bind to CREB in the nucleus. PGD2 did not directly induce an increase in intracellular Ca2+ levels. In addition, analysis of the transcriptional activities of MUC5B promoter regions showed that both CRE sites in the MUC5B promoter (−921/−914 and −900/−893) played an essential role in PGD2-induced MUC5B gene expression. Together these findings suggest new insights into the molecular mechanisms by which PGD2 induces MUC5B gene expression in airway epithelial cells.
EXPERIMENTAL PROCEDURES
Materials
PGD2, the PGD2-MOX EIA kit, and anti-DP1 receptor rabbit polyclonal antibody were purchased from Cayman Chemical (Ann Arbor, MI). Forskolin and 3-isobutyl-1-methylxanthine were purchased from Sigma. 8-Bromo-cAMP was purchased from ENZO life science (Plymouth Meeting, PA). S5751 (DP1-specific antagonist) and OC0459 (CRTH2-specific antagonist) were obtained from TAIHO Pharmaceutical Co., Ltd (Saitama, Japan). PD98059 was purchased from Calbiochem. Anti-prostaglandin D synthase murine monoclonal antibody was obtained from Osaka Bioscience Institute. Anti-CRTH2 goat polyclonal antibody and anti-β-actin mouse monoclonal antibody were purchased from Santa Cruz Biotechnology (Santa Cruz, CA); anti-phospho-p44/42 MAP kinase (Thr-202/Tyr-204), anti-p44/42 MAPK (ERK1/2), anti-phospho-p38 MAPK (Thr-180/Tyr-182), anti-phospho-SAPK/JNK (Thr-183/Tyr-185), anti-SAPK/JNK, anti-phospho-RSK1 (Ser-380), anti-RSK1/2/3, anti-phospho-CREB (Ser-133), and anti-CREB antibodies were purchased from Cell Signaling Technology (Beverly, MA). siERK1 (catalogue number sc-29307), siERK2 (catalogue number sc-35335), and negative control small Interfering RNA (siRNA; catalogue number sc-37007) were purchased from Santa Cruz Biotechnology. siDP1 (GenBankTM accession number NM_000953) and siCRTH2 (GenBankTM accession number NM_004778) were synthesized by Bioneer (Daejeon, Korea). siRSK1 and siCREB were synthesized by Invitrogen. An additional siRNA negative control was also utilized (Stealth, catalogue number 12935–300; Invitrogen). Fura-2 acetoxymethyl ester (Fura-2/AM) was purchased from Teflabs (Austin, TX).
Cell Culture
We obtained written permission from subjects and approval from the Institutional Review Board at Yonsei University College of Medicine for the use of specimens. Normal human nasal epithelial (NHNE) cells were cultured as previously described (28, 29). In brief, epithelial cells from turbinates were treated with 1% Pronase (Type XIV protease, Sigma) for 18–20 h at 4 °C. To remove fibroblasts, endothelial cells, and myoepithelial cells, isolated cells were placed in a plastic dish and cultured for 1 h at 37 °C. Isolated epithelial clusters were divided into single cells through incubation with 0.25% trypsin, EDTA. Passage-2 NHNE cells were seeded in 0.5 ml of culture medium onto 24.5-mm, 0.45-μm pore size Transwell clear (Costar) culture inserts. Cells were cultured in a 1:1 mixture of bronchial epithelial cell growth medium (Clonetics):Dulbecco's modified Eagle's medium (Invitrogen) containing all listed supplements as previously described (28). Cultures were grown while submerged, and the culture medium was changed on the first day and every other day thereafter. The human lung mucoepidermoid carcinoma cell line NCI-H292 was purchased from the American Type Culture Collection (catalogue number CRL-1848, Manassas, VA) and cultured in RPMI 1640 (Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen) in the presence of penicillin-streptomycin at 37 °C in a 5% CO2-humidified chamber.
Western Blot Analysis
Nasal polyps were homogenized in cell extraction buffer (Invitrogen). NCI-H292 cells were grown to confluence in six-well plates. After treatment with 1 μm PGD2, cells were lysed with cell extraction buffer (Invitrogen), and the total amount of protein was quantitated using the Pierce BCA Protein Assay kit (Thermo Rad Laboratories, Hercules, CA). Equal amounts of total protein (25 μg) were resolved via 10% SDS-polyacrylamide gel electrophoresis followed by electrophoretic transfer to a polyvinylidene difluoride membrane (PVDF, Immobilon; Millipore Corp., Bedford, MA) at 250 mA for 90 min at 4 °C. The PVDF membrane was then blocked with 5% skim milk in Tris-buffered saline (TBS, 50 mm Tris-Cl, pH 7.5, 150 mm NaCl) for 2 h at room temperature followed by overnight incubation with primary antibodies in TBST (0.5% Tween 20 in TBS). The membrane was washed with 1× TBST for 10 min and then probed for 2 h with secondary antibodies conjugated to horseradish peroxidase. The membrane was exposed to high performance autoradiography film (Fuji XR film, Fuji Film Corp., Tokyo, Japan) and visualized using the ECL system (Santa Cruz, CA).
RNA Extraction and RT-PCR Analysis
Total cellular RNA was extracted from NCI-H292 cells using RNA TRIzol reagent (Invitrogen) according to the manufacturer's protocol. The extracted RNA was treated with amplification grade deoxyribonuclease I (Invitrogen) before cDNA synthesis. DNase I-treated RNA (3 μg) was converted to cDNA with the addition of GeneAmp® RNA PCR kit components (Moloney murine leukemia virus reverse transcriptase, RNase inhibitor, and random hexamers; Applied Biosystems Inc., NJ) in a total volume of 25 μl. After reverse transcription, RT-PCR was performed using a MyCyclerTM thermal cycler system (Bio-Rad) for 35 cycles using previously described amplification conditions (30). All PCR primers were synthesized by Bioneer. The primer sequences were as follows: MUC5B forward (5′-GCTGCTGCTACTCCTGTGAGG-3′ (and reverse (5′-AGGTGATGTTGACCTCGGTCTC-3′); β2-microglobulin (β2M) forward (5′-TCGCGCTACTCTCTCTTTCT-3′) and reverse (5′-GCTTACATGTCTCGATCCCACTTAA-3′). The PCR products were visualized using 2% agarose gel electrophoresis and ethidium bromide staining.
Real-time PCR Analysis
Primers and probes were designed using the Perkin Elmer Life Science Primer Express® software purchased from PE Biosystems (Foster City, CA). Real-time RT-PCR was performed in a 96-well format using the PE Biosystems ABI PRISM® 7300 sequence detection system (Foster City, CA) with TaqMan® Universal PCR master mix (Applied Biosystems, Branchburg, NJ) and according to the conditions recommended in the manufacturer's protocol. Reactions were performed in a final volume of 25 μl containing 3 μg of cDNA (reverse transcription mixture), oligonucleotide primers at a final concentration of 800 nm, and 200 nm TaqMan hybridization probe. The probe for real-time PCR was labeled with carboxyfluorescein (FAM) at the 5′-end and with the quencher carboxytetramethylrhodamine (TAMARA) at the 3′-end. The following primers and probes were used: MUC5B forward (5′-TCTACCTGGACAACCACTACTGC-3′) and reverse (5′-TGGTG ACAGTGAGGACGATATCC-3′) and TaqMan probe (FAM-CTGCCACTGCCGCTGCCGCC-TAMARA). β2M endogenous control reagents were included as normal controls with the following primers and probes: β2M forward (5′-CGCTCCGTGGCCTTAGC-3′) and reverse (5′-GAGTACGCTGGATAGCCTCCA-3′) and TaqMan probe (FAM-TGCTCGCGCTACTCTCTCTTTCTGGC-TAMARA). Samples were analyzed in triplicate using PCR with an initial 10-min denaturation at 95 °C followed by 40 cycles of 15 s at 95 °C and 1 min at 60 °C. The relative quantity of MUC5B mRNA was determined using a comparative threshold method, and the results were normalized against β2M as an internal control.
Immunohistochemistry and Double Immunocytofluorescence Staining
We performed immunohistochemical staining for human hematopoietic-type prostaglandin D synthase (Osaka Bioscience Institute, Japan). Paraffin-embedded sections (4 mm) were de-paraffinized and hydrated using standard protocols, and antigen retrieval was performed by heating a slide-mounted section in a microwave oven in 0.01 m sodium citrate buffer, pH 6.0. The slide was then cooled to room temperature and washed with 0.01 m phosphate-buffered saline (PBS). Nonspecific endogenous peroxidase activity was blocked via treatment with 3% hydrogen peroxidase for 10 min. After washing, sections were incubated overnight with the primary antibodies at 4 °C. Negative controls were stained with purified mouse non-immune IgG at the same concentration as the primary antibody. Biotinylated link antibody and streptavidin/peroxidase were applied for 10 min followed by the addition of a solution of diaminobenzidine (Dako Corp., Carpinteria, CA) as a substrate chromogen. Finally, the sections were lightly counterstained with hematoxylin/eosin and then mounted.
For double immunocytofluorescence staining, cells were washed three times with PBS and treated with 1% Pronase (Type XIV protease, Sigma) for 18–20 h at 4 °C. The cells were then washed with PBS and separated into single cells by incubation in 0.05% trypsin/EDTA. Next, the cells were washed with PBS, and 200 μl of the cell suspension (1 × 104 cells/ml) was added to a slide funnel and fixed in cold methanol-acetone solution for 5 min. The slides were washed 3 times with PBS and then blocked with blocking solution (10% normal goat serum, Jackson ImmunoResearch Laboratories Inc., West Grove, PA) for 1 h. The cells were then washed with PBS and permeabilized in 0.1% Triton® X-100, PBS for 20 min. MUC5AC protein was detected using the MUC5AC monoclonal antibody (1:100, Santa Cruz). Prostaglandin D2 receptor proteins were detected via incubation with the DP1 rabbit polyclonal antibody (1:100, Cayman Chemical, CA) and CRTH2 goat polyclonal antibody (1:100, Santa Cruz Biotechnology) for 24 h at 4 °C followed by several washes in PBS. Incubation was repeated with an appropriate fluorescein isothiocyanate and Texas Red-conjugated secondary antibody (1:100, Jackson ImmunoResearch), and cell nuclei were stained with 4,6-diamidino-2-phenylindole. Coverslips were mounted onto slides with Vectashield Mounting Medium (Vector Laboratories, Inc., Burlingame, CA) and examined using a Zeiss LSM 510 confocal microscope (Carl Zeiss, Inc.).
Measurement of PGD2 Levels
Tissues (nasal polyps, nasal mucosa) were homogenized in ice-cold methanol (MeOH), and the resulting homogenate was incubated for about 30 min on ice to extract the lipid fraction. The homogenate was centrifuged at 3000 × g for 10 min at 4 °C, and the supernatant was collected in new clean tubes. To collect any remainder of the lipid fraction, MeOH was added to the residue and vortexed thoroughly. The precipitated proteins were removed via centrifugation, and the supernatant was evaporated to dryness under nitrogen (N2). PGD2 in the residue was derivatized to PGD2-methoxime. Next, the homogenate was centrifuged at 3000 × g for 10 min at 4 °C, after which the combined supernatant was concentrated at reduced pressure under N2 with centrifugation. Next, the dried residue was dissolved in ice-cold 1 m acetate buffer, pH < 4.0. Purification of arachidonic acid metabolite was performed with an SPE C18 cartridge (Sep-Pak C18 Vac, Cayman Chemicals). The eluted fraction was concentrated at reduced pressure under N2 via centrifugation. The dried residue was dissolved in ice-cold EIA buffer (included with the EIA kit). Concentrations of PGD2 in the purified samples were quantified using a prostaglandin D2-Mox enzyme immunoassay kit according to the manufacturer's directions (Cayman Chemical).
Measurement of MUC5B Concentrations
Enzyme-linked immunosorbent assay (ELISA) was used to determine the MUC5B protein levels. Cell lysates from human NCI-H292 airway epithelial cells and NHNE were prepared in phosphate-buffered saline (PBS). In brief, the cells were then washed three times with cold PBS. Cells were collected after adherent cells were digested by trypsin and then centrifuged. The homogenate was centrifuged at 5000 × g for 5 min at 4 °C, and the supernatant was then collected in new clean tubes. Then physical methods for cell cleavage were undertaken (cells were sonicated, then repeatedly frozen and thawed). Next, the homogenates were centrifuged for 5 min at 5000 × g, and the supernatant was collected in new tubes. Concentrations of MUC5B levels in the samples were quantified using a MUC5B enzyme immunoassay kit according to the manufacturer's directions (Uscn Life Science Inc.). The absorbance was read at 450 nm.
Transient Transfection and Luciferase Analysis
MUC5B gene constructs containing promoter deletions and CRE site point mutations were reported in our previous study (30). Deletion mutants covering the promoter regions of MUC5B were generated through PCR (30). NCI-H292 cells were seeded (2.5 × 105 cells/well) in six-well tissue culture plates with plasmid DNA (1 μg/well) 24 h before transfection using FuGENE6 Transfection Reagent (Roche Applied Science) according to the manufacturer's recommendations. After transfection, cells were maintained in RPMI media containing 0.2% serum for 24 h before treatment with 1 μm PGD2. Cells were harvested 24 h after treatment and assayed for luciferase activity using a dual-luciferase reporter assay system (Promega, Madison, WI) in which the activity value was normalized to that of a β-galactosidase expression vector. Luciferase activity in cell extracts (20 μl) was measured on a Berthold 9501 luminometer using luciferase assay reagent. Each plasmid was assayed in duplicate in at least three separate experiments. To confirm that the luciferase activity of each construct was induced by PGD2, the activity of each construct was also measured in the absence of PGD2.
siRNA Treatment
Cells were plated in 6-well plates 24 h before transfection with siERK1/2, siRSK1, siCREB, and negative control siRNA using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. After transfection, cells were maintained in 0.2% serum RPMI media for 16–18 h before treatment with PGD2 and then harvested. The efficiency of siRNA knockdown was determined using Western blot analysis with the relevant antibodies.
cAMP Detection Assay
NCI-H292 cells were plated in 6-well plates 1 day before treatment. Cells were incubated with DMSO or PGD2 for various times at 37 °C. 3-Isobutyl-1-methylxanthine (1 mm) was added to the culture 10 min before treatment to prevent degradation of cAMP. Cells were then lysed through three rounds of freezing and thawing and centrifuged at 10,000 × g for 10 min. Supernatants were then collected, and cAMP concentrations were determined using a cAMP detection kit according to the manufacturer's protocol (Cayman Chemical).
Measurement of Calcium Ion Concentration
NCI-H292 cells were seeded onto glass coverslips in 35-mm dishes (2.5 × 105 cells/coverslip) and cultured for 1 day. The cells were incubated with 5 μm Fura-2/AM for 40 min at room temperature and washed with bath solution (140 mm NaCl, 5 mm KCl, 1 mm MgCl2, 10 mm HEPES, 1 mm CaCl2, 10 mm glucose, 310 mosm, pH 7.4). The coverslips were transferred to a perfusion chamber, and the cells were continuously perfused with prewarmed (37 °C) bath solution. Fura-2-AM-loaded cells were mounted onto the stage of an inverted microscope (Nikon, Tokyo, Japan) for imaging. The cells were illuminated at 340 and 380 nm, and the emitted fluorescent images at 510 nm were collected with a CCD camera. The images were digitized and analyzed using the MetaFluor system (Universal Imaging Co., Downingtown, PA). The fluorescence ratio (340/380) was used as a measure of the free calcium ion concentration [Ca2+]i, and fluorescence images were obtained at 3-s intervals.
Chromatin Immunoprecipitation (ChIP) Assay
For ChIP assays, chromatin was isolated as previously described (31). In brief, ∼2 × 109 NCI-H292 cells in 150-mm dishes were treated with PBS containing 1% formaldehyde for 10 min and washed 3 times with ice-cold PBS. Cells were then harvested into PBS and collected via centrifugation. The cells were resuspended in 1 ml of cell lysis buffer (5 mm KOH, pH 8.0/85 mm KCl, 0.5% Nonidet P-40) and incubated for 10 min on ice. After a short spin (5 min at 5000 rpm), the pellets were resuspended in nuclear lysis solution (50 mm Tris, pH 8.1, 10 mm EDTA, 1% SDS) containing protease inhibitors and sonicated to break the chromatin into fragments with average lengths of 0.3 to 1.5 kb. After centrifuging at 5000 rpm for 5 min, the nuclear pellets were resuspended in ChIP dilution buffer (1.2 mm EDTA, 16.7 mm Tris-HCl, pH 8.1, 0.01% SDS, 1.1% Triton X-100, 167 mm NaCl) containing protease inhibitors. The assay used 2 μg of anti-phospho-CREB (Ser-133) antibody and 2 μg of rabbit IgG as a negative control. Primers used for ChIP analysis included the following: CRE (−956 to −753), 5′-ACGCGTGAGGTATTGCAGCGCGGACG-3′, 5′-GGTTGAGGAAGCAGCTGCTC-3′; negative controls (−3960 to −3761, upstream of the CRE element; non-CRE), 5′-CCAGGCACTGGCTCTGAGA-3′, 5′-GAGGGTCCCATCGTGTGAC-3′.
In Situ Proximity Ligation Assay
To detect the interaction between CREB and RSK1, we utilized the DuoLinkTM in situ proximity ligation assay (PLA) (Olink Bioscience, Uppsala, Sweden) according to the manufacturer's protocol. NCI-H292 cells, cultured on glass coverslips, were rinsed with PBS and fixed with 4% paraformaldehyde for 20 min at room temperature followed by a 10 min of incubation with 0.1% Triton X-100 solution in PBS and an additional 30 min of incubation in blocking solution. Cells were then immunolabeled with primary antibodies (1:100) for 1 h at 4 °C. A monoclonal antibody to CREB (Santa Cruz Biotechnology) and polyclonal antibody (Cell Signaling Technology) to RSK1 were used to detect protein-protein interactions under confocal microscopy at 60× magnification. The secondary antibodies with attached PLA probes were supplied in the Duolink kit, and a fluorescence signal was measured to indicate that the two proteins were separated by <40 nm (Olink Bioscience).
Statistical Analysis
Quantified RT-PCR signals were normalized to β2M levels and presented as the mean ± S.D. Student's t test was used to determine the significances of differences in expression data. Statistical analysis was performed using the program GraphPad Prism 4 (GraphPad Software, Inc., La Jolla, CA). Differences were considered to be significant at values less than p < 0.05.
RESULTS
Both H-PGDS and PGD2 Proteins Are Highly Expressed in Human Nasal Polyps
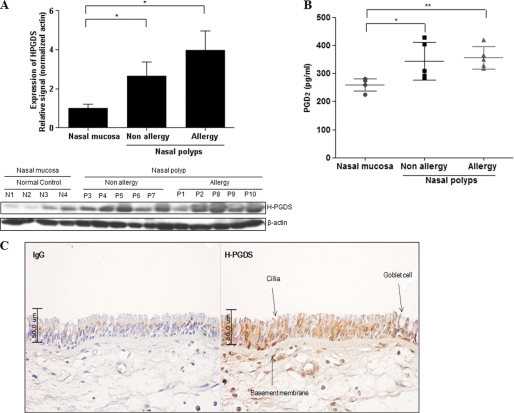
To determine whether H-PGDS is expressed in human nasal polyp tissues, we first examined H-PGDS expressions in normal nasal mucosa and nasal polyp tissue using Western blot analysis. The expression of H-PGDS in nasal polyp tissues was similar in patients from both allergic and non-allergic groups; however, the expression in nasal polyp tissues was significantly greater (more than 3-fold) than that in nasal mucosa (Fig. 1A). A similar result was obtained for PGD2 expression (Fig. 1B). We showed that H-PGDS expression was characteristic of nasal polyp tissues with a high secretion of PGD2. Next, we used immunohistochemistry to investigate whether H-PGDS was located in human nasal epithelial cells (Fig. 1C) and found that H-PGDS was highly expressed throughout the epithelium of nasal polyp tissue. These data suggested that PGD2 plays an important role in the inflammation response. DP1 receptor but not CRTH2 receptor is highly expressed in human primary nasal epithelial cells.
FIGURE 1.
H-PGDS protein and PGD2 are expressed in human nasal polyps. A, the expression of H-PGDS in homogenates of tissue specimens from 4 healthy subjects and 10 subjects with human nasal polyps is shown. Total cell lysates were prepared for Western blot analysis. Values above the figures represent the relative density of the bands normalized to β-actin. Results are representative of three independent experiments. Data are expressed as the mean ± S.D. *, p < 0.05 according to Wilcoxon's rank sum test for control versus non-allergic group and control versus allergic group. B, we measured the PGD2 level in human nasal polyps using a PGD2-MOX ELISA kit. PGD2 levels were standardized according to the concentration of total protein for each sample. Each bar represents the median concentration of PGD2. Significance was determined by the Mann-Whitney U test. The results are expressed as pg/mg of protein and are the mean ± S.D. of 4 healthy subjects and 10 subjects with nasal polyps. *, p < 0.05 versus healthy subjects. **, p < 0.05 versus healthy subjects. C, strong H-PGDS immune-reactivity is observed in human primary nasal epithelial cells. The negative control (IgG; without H-PGDS antibody) shows no immunoreactivity (200×).
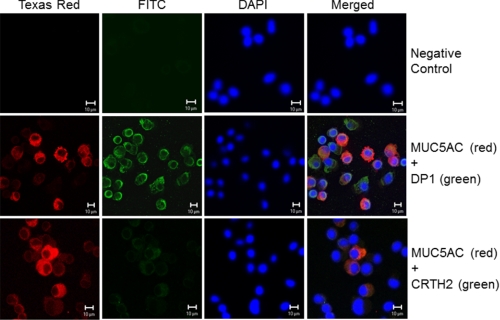
Next we investigated the cellular localization of PGD2 receptors (DP1 and CRTH2) in human primary nasal epithelial cells using double immunocytofluorescence staining. MUC5AC was used as a goblet cell marker (Fig. 2, red) (32), whereas DP1 (green staining) was detected on the surfaces of goblet cells as yellow objects in the merged image, indicating double immunofluorescence of DP1 and MUC5AC in both goblet and non-goblet cells of human primary nasal epithelia. In contrast, CRTH2 receptor was barely expressed. No staining was detected in control experiments in which primary antibodies against DP1, CRTH2, and MUC5AC were replaced with purified IgG (Fig. 2, upper panel).
FIGURE 2.
PGD2 receptors are expressed in human primary nasal epithelial cells. Cellular localization of PGD2 receptors (DP1 and CRTH2) and MUC5AC proteins in human nasal epithelial cells was analyzed using double-immunocytofluorescence staining with antibodies against MUC5AC and PGD2 receptors (DP1 and CRTH2). Immunostaining of MUC5AC (red) and DP1 (green) is shown. Merged images show co-localization of MUC5AC and DP1 in yellow (middle panels). Immunostaining of MUC5AC (red) and CRTH2 (green) is shown. Overlays of the images show co-localization of MUC5AC and CRTH2 in yellow (lower panels). No staining was detected when both primary antibodies were omitted (upper panels) (Bars indicate 10 μm).
PGD2 Induces MUC5B Expression in Both NHNE Cells and Human Lung Mucoepidermoid Carcinoma (NCI-H292) Cells
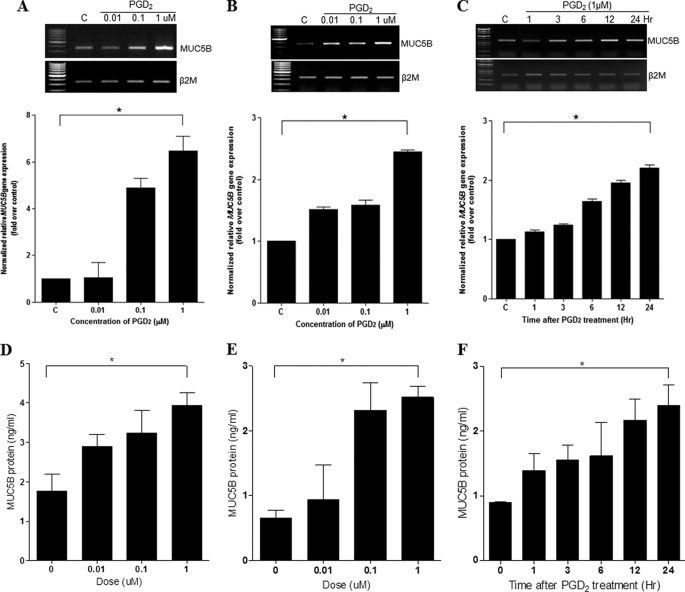
To determine whether PGD2 could induce mucin gene expression in NHNE and NCI-H292 cells, we performed real-time quantitative PCR on cells that had been treated with various concentrations of PGD2 (0.01, 0.1, and 1 μm) for 24 h. The mRNA levels of secreted mucin genes in the cell lysates, such as MUC5B, 6, 8, and 5AC, were determined and compared using RT-PCR (data not shown). Mucin gene expression was significantly induced by 0.01 μm PGD2. Interestingly, some secreted mucin genes in NCI-H292 cells, such as MUC6, -8, and -5AC, did not exhibit any increase due to PGD2 (data not shown). However, one of the major secreted gel-forming mucin genes (MUC5B) was increased in both cells (Fig. 3A, NHNE, greater than 6-fold induction; Fig. 3B, NCI-H292 cells, greater than 2-fold induction). Because the characteristics of NCI-H292 cells were similar to those of NHNE cells, we used NCI-H292 cells in subsequent experiments.
FIGURE 3.
PGD2 induces MUC5B expression in NHNE and human lung mucoepidermoid carcinoma (NCI-H292) cells. A, NHNE cells were pretreated apically and basolaterally with PGD2 at the indicated concentrations for 24 h. Cell lysates were harvested for real-time quantitative RT-PCR. B, NCI-H292 cells were treated with PGD2 (0.01, 0.1, and 1 μm) for 24 h. Cell lysates were harvested for real-time quantitative RT-PCR. C, cells were treated with PGD2 (1 μm) for 1, 3, 6, 12, and 24 h, and total RNA were isolated. MUC5B expression relative to that of β2M was determined using real-time quantitative RT-PCR. D, we measured MUC5B levels in NHNE using a MUC5B ELISA kit. MUC5B levels were standardized according to the concentration of total protein for each sample. NHNE cells were pretreated apically and basolaterally with PGD2 at the indicated concentrations for 24 h. E, NCI-H292 cells were treated with PGD2 (0.01, 0.1, and 1 μm) for 24 h. Cell lysates were harvested for ELISA. F, NCI-H292 cells were treated with PGD2 (1 μm) for 1, 3, 6, 12, and 24 h. The mean values of MUC5B estimated by ELISA in cell lysates and its subgroups are expressed as ng of standard reactivity/ml of diluted resolubilized mucin sample. Significance was determined by the Mann-Whitney U test. Data are expressed as the mean ± S.D. of triplicate cultures. At least three separate experiments were performed for each measurement. C, untreated control; *, p < 0.05 versus untreated control.
To investigate whether PGD2 induced MUC5B gene expression in a time-dependent manner, we performed RT-PCR after treating NCI-H292 cells with 1 μm PGD2 for various times (1, 3, 6, 12, and 24 h). We observed a time-dependent increase in expression of MUC5B in NCI-H292 cells over a 24-h time period (greater than 2-fold increase) (Fig. 3C).
Next, ELISA analysis was performed to investigate the effect of PGD2 on MUC5B protein expression in airway epithelial cells. We observed that PGD2 can induce MUC5B protein in a dose-dependent manner in both cells (Figs. 3, D, NHNE, and E, NCI-H292 cells; greater than 2-fold induction). These results show that the MUC5B expression was increased at all PGD2 dose and peaked at 1 μm PGD2 for 24 h. A concentration of 1 μm PGD2 was used for all subsequent experiments.
PGD2-induced MUC5B Gene Expression Is Dependent on the DP1 Receptor in NCI-H292 Cells
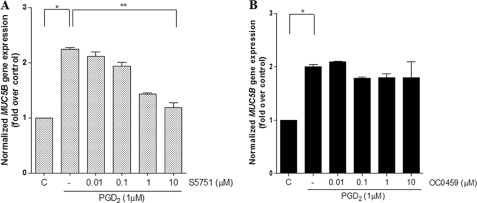
To identify which PGD2 receptor was involved in PGD2-mediated MUC5B gene expression, cells were treated for 24 h with the DP1 antagonist, S5751 (33, 34), or the CRTH2/DP2 antagonist, OC0459 (35). PGD2-dependent MUC5B gene expression in NCI-H292 cells was significantly inhibited by S5751 in a dose-dependent manner, whereas OC0459 had very little effect (Fig. 4, A and B). The increase in MUC5B gene expression by PGD2 treatment (2.2-fold induction) was decreased by 20% (1.8-fold) and 47% (1.18-fold) upon the addition of OC0459 and S5751, respectively. These results suggest that the DP1 receptor likely mediates PGD2-induced MUC5B gene expression in NCI-H292 cells.
FIGURE 4.
DP1 is required for PGD2-induced MUC5B gene expression in NCI-H292 cells. A, cells were treated with S5751 (DP1 antagonist, final concentration 0.01–10 μm) for 1 h before stimulation with 1 μm PGD2. B, cells were treated with OC0459 (CRTH2 antagonist, final concentration 0.01–10 μm) for 1 h before stimulation with 1 μm PGD2. Cells were subsequently stimulated with 1 μm PGD2 for 24 h. MUC5B expression relative to that of β2M was determined using real-time quantitative RT-PCR. Values represent the mean ± S.D. The results represent three independent experiments. C, untreated control; *, p < 0.05 versus untreated control; **, p < 0.001 versus PGD2 treatment alone.
ERK1/2 Is Essential for PGD2-induced MUC5B Gene Expression
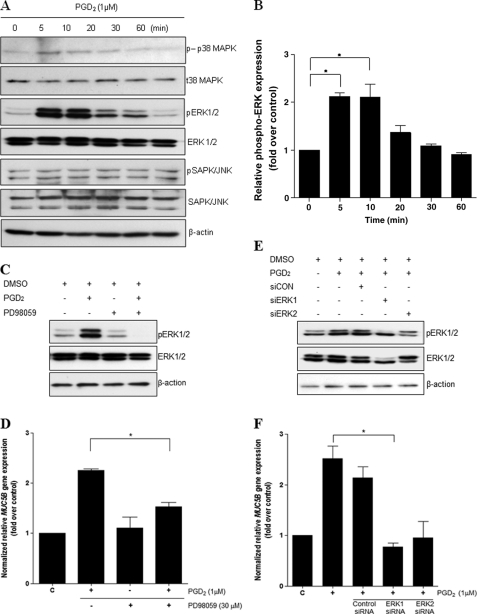
To investigate which extracellular ERK MAPK signaling pathway in NCI-H292 cells was activated by PGD2, we performed Western blot analysis using specific antibodies. Activation of ERK1/2 MAPK increased after 5 min of stimulation with PGD2 and then gradually decreased (Fig. 5A). Western blotting experiments indicated that p38 MAPK and SAPK/JNK (stress-activated protein kinase/Jun amino-terminal kinase) were not activated by PGD2 treatment (Fig. 5A) and thus did not appear to be involved in PGD2-induced MUC5B gene expression. To further examine the role of ERK1/2 MAP kinase, we pretreated cells with 30 μm PD98059, an ERK1/2 inhibitor, for 1 h before treatment with PGD2 and subsequent analysis of protein expression (Fig. 5C). PGD2 treatment significantly increased ERK1/2 activity, whereas pretreatment with PD98059 completely attenuated the effect of PGD2. Moreover, PD98059 completely inhibited PGD2-induced MUC5B mRNA expression (Fig. 5D). For further confirmation of these findings, cells were transfected with siRNAs specific for ERK1 or ERK2 MAPK. Interestingly, transfection with ERK1/2 siRNA specifically reduced PGD2-induced MUC5B gene expression compared with the effects of control siRNA (Fig. 5F). These results suggest that phosphorylation of ERK1/2 MAPK is required for PGD2-induced MUC5B gene expression in NCI-H292 cells.
FIGURE 5.
PGD2 induces MUC5B gene expression via ERK MAPK signaling. A, confluent cells were treated with PGD2 (1 μm) for 5, 10, 20, 30, and 60 min, and cell lysates were harvested for Western blot analysis. B, quantitation of the p-ERK/ERK ratio is shown. Representative Western blots using phospho-specific antibodies show transient activation of ERK1/2, with a maximum effect at 5 min. ERK1/2, total ERK1/2. C, cells were pretreated for 1 h with PD98059 (30 μm) and stimulated for 5 min with PGD2 (1 μm) before collection of cell lysates for Western blot analysis. D, confluent cells were pretreated for 1 h with PD98059 (30 μm) and stimulated for 24 h with PGD2 (1 μm) before collection of total RNA for real-time quantitative RT-PCR. E, after transfection with siERK1 (40 nm), siERK2 (40 nm), or siRNA-negative control (40 nm), cells were stimulated with PGD2 (1 μm) for 5 min followed by analysis of proteins using Western blotting. F, transfected cells were stimulated with PGD2 (1 μm) for 24 h before the collection of total RNA for real-time quantitative RT-PCR. MUC5B expression relative to that of β2M was determined using real-time quantitative PCR. The figures are representative of at least three independent experiments. Data are expressed as the mean ± S.D. C, untreated control; *, p < 0.05 versus PGD2 treatment alone.
DP1 Receptor Is Involved in PGD2-induced MUC5B Expression
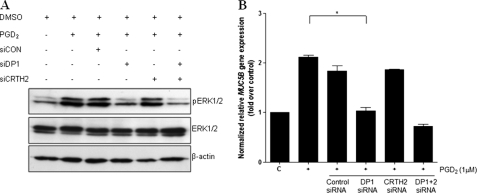
Next, we examined the role of PGD2 receptors in the signal transduction pathway leading to MUC5B gene expression in airway epithelial cells by transfecting NCI-H292 cells with 50 nm siRNA against DP1 or CRTH2. Cells were treated with PGD2 24 h after transfection and subsequently harvested for Western blot analysis. DP1 and CRTH2 siRNAs suppressed intracellular DP1 and CRTH2 mRNA levels in NCI-H292 cells, respectively (data not shown). PGD2 treatment initially caused a 2.12-fold increase in MUC5B gene expression. We found that the addition of DP1 siRNA decreased expression by 48.0% (1.03-fold compared with 2.12-fold), whereas the addition of CRTH2 siRNA only inhibited MUC5B gene expression by 87.7% (1.86-fold compared with 2.12-fold) (Fig. 6B). These results suggest that the DP1 receptor plays an important role in PGD2-induced MUC5B gene expression.
FIGURE 6.
DP1 is essential for PGD2-induced MUC5B expression through ERK signaling in NCI-H292 cells. A, cells were transiently transfected with constructs expressing siRNA for DP1, CRTH2, DP1 + DP2, or control siRNA. Cells were serum-starved and treated with 1 μm PGD2 for 5 min, after which cell lysates were harvested for Western blot analysis with anti-ERK1/2. B, cells were serum-starved and treated with 1 μm PGD2 for 24 h, after which cell lysates were harvested for real-time quantitative RT-PCR. MUC5B expression was determined relative to that of β2M using real-time quantitative PCR. C, untreated control; *, p < 0.05 versus PGD2 treatment alone.
DP1-specific siRNA treatment also decreased the levels of p-ERK1/2 (Fig. 6A), whereas CRTH2-specific siRNA had no effect. These results indicated that DP1 receptor is an essential mediator of ERK MAPK signaling for PGD2-induced MUC5B expression in NCI-H292 cells.
PGD2-induced MUC5B Gene Expression Is Mediated by RSK1
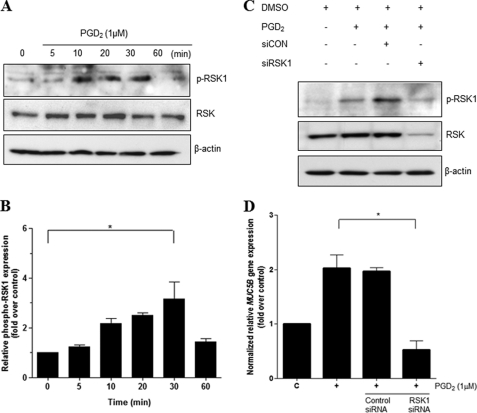
The p90 ribosomal S6 kinases (RSK) are a family of serine/threonine kinases that are activated downstream of the Ras/MAPK pathway. RSK phosphorylates multiple signaling effectors to play an essential role in a number of cellular functions, including regulation of gene expression through the phosphorylation of transcriptional regulators such as c-fos (36) and CREB (37, 38). We found that phosphorylation of RSK1 peaked 30 min after PGD2 stimulation and was decreased by 60 min (Fig. 7A). Densitometric analysis of Western blots showed a significant increase in p-RSK1 at 30 min (Fig. 7B).
FIGURE 7.
RSK1 is essential for PGD2-induced MUC5B gene expression. A, confluent and quiescent cells were stimulated with PGD2 (1 μm) in a time-dependent manner for up to 1 h followed by harvesting of cell lysates and Western blot analysis using anti-phospho-RSK1. β-Actin served as an internal control. B, quantitation of the p-ERK/ERK ratio is shown. The results of Western blot analyses are representative of three separate experiments. C, cells were transiently transfected with RSK1 siRNA constructs. Transfected cells were serum-starved overnight before treatment with PGD2 (1 μm) for 30 min, after which cell lysates were harvested for Western blot analysis using specific antibodies. D, 24 h after transfection, cells were treated with PGD2 (1 μm) and harvested for real time quantitative RT-PCR. MUC5B expression was determined relative to β2M by real-time quantitative PCR. Figures represent three independent experiments. Data are expressed as mean ± S.D. C, untreated control; *, p < 0.05 versus PGD2 treatment alone.
To determine whether RSK1 plays an important role in PGD2-induced MUC5B gene expression, we performed an RSK1 knockdown study. Cells were transfected with 40 nm siRNA against RSK1 or a negative control siRNA and treated with PGD2 24 h after transfection followed by harvesting for Western blot analysis. RSK1-specific siRNA treatment decreased the levels of p-RSK1 compared with the negative siRNA control, which had no effect (Fig. 7C). Knockdown of RSK1 led to an attenuation of MUC5B gene expression according to quantitative real-time RT-PCR (Fig. 7D). These results suggest that PGD2-induced MUC5B gene expression was mediated via phosphorylation of RSK1.
CREB but Not Ca2+ Signaling Is Required for PGD2-induced MUC5B Gene Expression
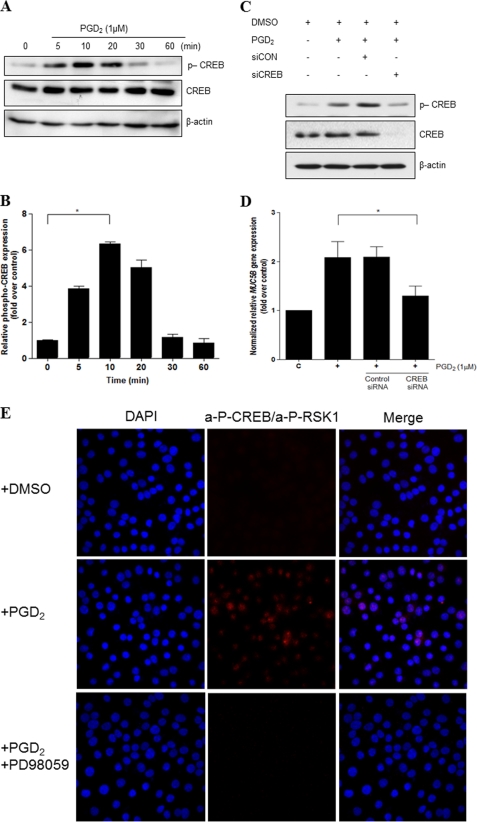
As RSK1 is a mediator of cytokine-induced CREB phosphorylation at Ser-133 and CREB was previously shown to mediate the transcriptional regulation of some mucins, we investigated whether CREB can be activated by PGD2. Cells were treated with PGD2 for various times up to 1 h followed by measurement of CREB activation through phosphorylation monitoring of Ser-133 (p-CREB) in Western blots. As shown in Fig. 8A, PGD2 induced CREB activation in a time-dependent manner: The activation of CREB was observed 5 min after PGD2 treatment and was decreased at 30 min. Densitometric analysis of the Western blots showed a significant increase in p-CREB at 10 min (Fig. 8B). To examine the function of activated CREB in transcriptional regulation of the MUC5B gene by PGD2, CREB was knocked-down with RNAi. Cells were treated with PGD2 24 h after transfection and harvested for Western blot analysis. As expected, CREB-specific siRNA caused a reduction in the level of p-CREB. Furthermore, CREB knockdown led to a decrease in the level of MUC5B gene expression as assessed by quantitative real-time RT-PCR (Fig. 8D).
FIGURE 8.
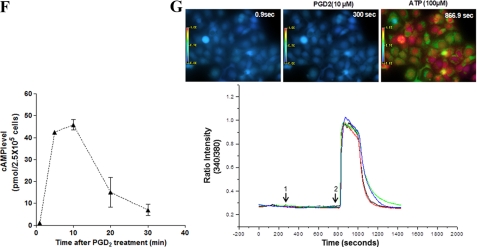
CREB mediates PGD2-induced MUC5B gene expression independent of Ca2+ signaling. A, confluent and quiescent cells were treated with PGD2 (1 μm) for 5, 10, 20, 30, and 60 min, and cell lysates were processed for Western blot analysis using phospho-specific antibodies. β-Actin was used as an internal control. B, quantitation of the p-CREB/CREB ratio is shown. The results of Western blot analyses are representative of three separate experiments. C, cells were transfected with siCREB (40 nm) or siRNA-negative control (40 nm) and stimulated with PGD2 (1 μm) for 10 min before Western blot analysis. D, cells were serum-starved overnight, stimulated with PGD2 (1 μm) for 24 h, and analyzed using real time quantitative RT-PCR. MUC5B expression was determined relative to that of β2M using real-time quantitative PCR. The figures are representative of three independent experiments. Data are expressed as the mean ± S.D. C, untreated control; *, p < 0.05 versus PGD2 treatment alone. E, fluorescence images are shown to indicate interaction between two proteins. Staining with primary anti-CREB goat polyclonal and anti-RSK1 rabbit polyclonal antibodies was followed by species-specific secondary antibodies with PLA probes. PLA signaling (red dots) was detected in NCI-H292 cells using confocal microscopy (60×). F, cells were serum-starved overnight and then treated with the indicated concentrations of PGD2 for 1 h, after which cAMP production was measured. The values shown are the means ± S.D. of experiments performed in triplicate. G, shown are fluorescence changes in response to PGD2 in NCI-H292 cells (10 μm). PGD2 in DMSO was added to Fluo-2/AM-labeled cells (arrow 1), and ATP (100 μm) was added after 8 min (arrow 2). The mean basal level of [Ca2+]i was measured for 1 min before any treatment. Blue areas represent basal [Ca2+]i, and green and red areas represent an increase from base line. [Ca2+]i versus time is shown for 10 cells (left). Colored lines show the trace of [Ca2+]i mobilization in individual cells (right). Results are representative of two independent experiments.
Furthermore, it has been reported that RSKs can phosphorylate CREB in the nucleus (39). To test if RSK1 can directly bind to CREB in NCI-H292 cells, we performed an in situ PLA assay. Cells were pretreated with PD98049 (ERK inhibitor) for 1 h before treatment with PGD2. We observed that the PLA signals visualizing RSK1 and CREB interactions were very low in non-treated cells but were strongly induced by PGD2 treatment. In contrast, reduction of the PLA signal (shown in red) indicated that inhibition of ERK greatly attenuated the interaction of RSK1 with CREB. This result suggests that RSK1 can bind to CREB directly, and significant downstream mediators of ERK MAPK signaling in PGD2 induced MUC5B gene expression (Fig. 8E).
DP1 is coupled to the G-protein subunit, Gαs, the activation of which results in elevated intracellular cAMP levels. If the effects of PGD2 on airway epithelial cells are mediated via DP1 signaling, PGD2 treatment of cells should result in increased intracellular cAMP levels compared with those in the controls. Indeed, we found that PGD2 treatment increased cAMP levels in NCI-H292 cells, peaking within 10 min of incubation and then decreasing over at least 30 min (Fig. 8F).
Binding of PGD2 to the DP1 receptor has also been shown to elicit an increase in the concentration of intracellular Ca2+ in certain cell types (40). Next, we tested the ability of PGD2 to mobilize Ca2+ in airway epithelial cells. As shown in Fig. 8G, PGD2-treated cells failed to induce a significant Ca2+ influx in NCI-H292 cells. In addition, there was no change in Ca2+ influx when the cells were treated with forskolin either in the absence or presence of 3-isobutyl-1-methylxanthine (data not shown).
Another study has reported that intracellular Ca2+ is dependent on CREB (41). In our study, PGD2 treatment did not directly trigger any significant increase in the intracellular Ca2+ concentration in NCI-H292 cells despite the involvement of CREB in PGD2-induced MUC5B gene expression. We also examined the effect of PGD2 on [Ca2+]i in A549 and BEAS-2B cells and obtained similar results (data not shown). These findings provide evidence that CREB plays an important role in PGD2-induced MUC5B gene expression, although Ca2+ signaling is not involved in this process.
ERK MAPK Signaling Is Required for Transcriptional Activation of PGD2-induced MUC5B Gene via Recruitment for Phospho-CREB on the CRE of MUC5B Gene
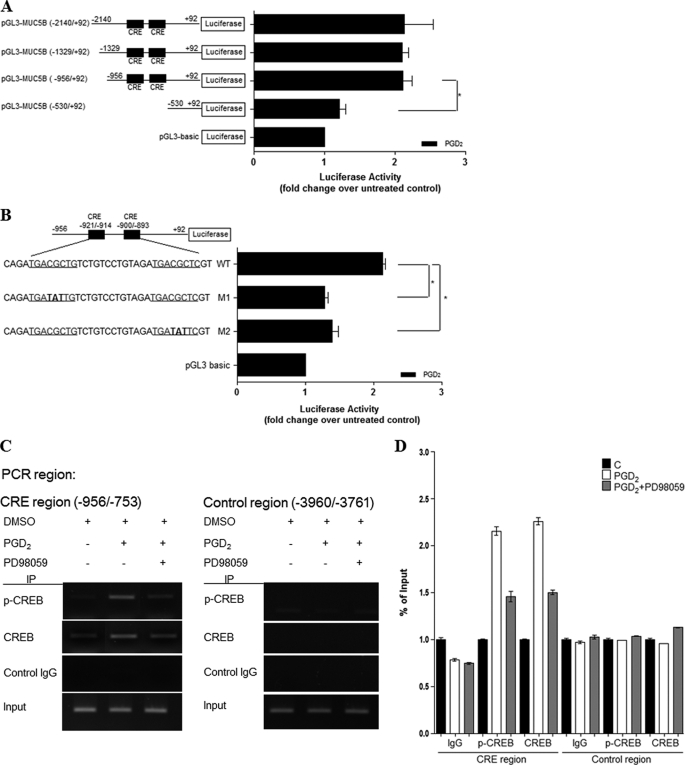
Our previous data showed that PGD2 induced CREB phosphorylation and CREB-dependent MUC5B transcription in NCI-H292 cells. To identify the PGD2-responsive region in the MUC5B promoter, cells were transiently transfected with MUC5B deletion mutants and treated with PGD2 for 24 h. We had already constructed multiple MUC5B promoter deletions in previous studies (30, 42). As shown in Fig. 9A, PGD2-induced luciferase activities of the −2140/+92, −1329/+92, and −956/+92 regions of the MUC5B promoter (2.2-fold) were enhanced compared with those in the −530/+92 regions, which indicates that the −956/−530 region of the MUC5B promoter may be critical for the response to PGD2.
FIGURE 9.
PGD2 enhances MUC5B promoter activity via cis-acting CRE regulatory motifs. A, cells were transiently transfected with various MUC5B promoter luciferase reporter constructs and stimulated with PGD2 (1 μm) for 24 h. Luciferase activity was measured in PGD2-treated and untreated cells. *, p < 0.05 versus −530/+92 reporter construct. B, cells were transfected with MUC5B promoter constructs containing mutated CRE sites as indicated. Cells were treated with PGD2 (1 μm) for 24 h and then harvested. The luciferase activities shown have been corrected for transfection efficiency using the β-galactosidase activity of the cell lysates. Data are expressed as the mean ± S.D. of experiments performed in triplicate cultures. *, p < 0.05 versus wild-type treated with PGD2 (1 μm). C, cells were stimulated with PGD2 (1 μm) for 20 min. Cross-linked protein-DNA complexes were immunoprecipitated using CREB antibody or normal rabbit IgG that either amplified the CREB binding flanking region in MUC5B promoter (CRE site) or a region farther upstream that did not contain a CREB-binding site (Non-CRE site). The immunoprecipitated chromatin was analyzed with PCR using primers specific to the indicated site. Input chromatin represents a portion of the sonicated chromatin before immunoprecipitation. Control IgG (C), goat anti-rabbit IgG (negative control for ChIP). D, the results were analyzed using real-time PCR and are shown as the percentage of input. The results are shown as means ± S.D. calculated from three independent experiments.
The CREB pathway may directly affect PGD2-induced transcriptional activation of the MUC5B gene if the two CRE sites within the −956/−530 region of the MUC5B promoter (5′-TGACGCTG-3′ at −921/−914 and 5′-TGACGCTC-3′ at −900/−893) act as cis elements. We, therefore, investigated whether CRE activation was required for PGD2-induced MUC5B transcription by performing site-directed mutagenesis of the CREB-binding site (Fig. 9B). Both CRE mutant constructs M1 and M2 showed decreased MUC5B transcriptional activity compared with that of the wild type (Fig. 9B), suggesting that both CRE sites in the regulatory region of the MUC5B promoter were involved in the PGD2-dependent up-regulation of MUC5B transcriptional activity.
We further confirmed this conclusion via ChIP assay. To this end, cells were first treated with or without PGD2 for 20 min and processed for ChIP using anti-phospho-CREB and anti-CREB antibodies. Because the region from −956 to −753 contains putative CRE binding sites, we performed ChIP assays using specific primers (−956/−753) for PCR. We also used the region −3960/−3761 as a negative control. After purification of DNA in the immunoprecipitate, the abundance of genomic DNA containing a promoter was determined through PCR amplification using sequence-specific primers. Anti-rabbit IgG, the negative control, did not yield any signals, indicating assay specificity. Input chromatins were also used in these assays to indicate that equal amounts of cell lysates were used. The ChIP assays clearly showed that phosphorylated CREB bound directly to the MUC5B promoter (−953/−753), and PD98059 treatment (ERK inhibitor) induced the dissociation of p-CREB from the CRE of the MUC5B gene (Fig. 9, C and D). However, no binding of CREB was detected regardless of PD98059 treatment. We also obtained similar results in NHNE cells (data not shown). Based on these results, we concluded that p-CREB protein is strongly bound to the chromatin regions of CRE binding sites of the MUC5B promoter region with PGD2 treatment and that CREB plays an important role in ERK MAPK signaling for PGD2-induced MUC5B gene expression in airway epithelial cells.
DISCUSSION
Mucus overproduction is related to several pathological features of respiratory diseases such as asthma (3) and chronic obstructive pulmonary disease (4). Excessive mucus in the airways has been linked to an increase in the frequency and duration of infection, a decline in lung function, and increased morbidity and mortality in respiratory diseases. Mucus is produced by goblet cells and submucosal glands in the large airways but only by goblet cells in the small airways (2). In asthma and chronic obstructive pulmonary disease patients, the major mucin components of airway mucus secretions are MUC5AC and MUC5B, both of which contribute to the viscoelastic properties of the mucus. However, the molecular mechanisms underlying the up-regulation of MUC5B gene expression due to inflammatory stimuli remains poorly understood. Here, we investigated the mechanism of MUC5B gene expression up-regulation by PGD2 in airway epithelial cells.
Prostaglandins are among the major mediators of airway inflammation and include human airway trypsin (43) and reactive oxygen species (7), all of which increase MUC5AC expression in vitro. PGD2 levels exhibit marked changes in various pathological conditions such as asthma (16) and inflammatory bowel disease (44). However, an increase in PGD2 level can have either deleterious or protective effects. In this study we observed H-PGDS and PGD2 expressions in nasal polyp tissue and normal mucosa. Because the levels of PGD2 and H-PGDS were much greater in nasal polyp tissue, we sought to ascertain whether an increased level of PGD2 might lead to increased MUC5B gene expression. We used PGD2 as a stimulant in airway epithelial cells, which led to an increase in MUC5B expression in airway epithelial cells in a time- and dose-dependent manner.
One of the PGD2 receptors, DP1, was expressed in nasal mucosa in epithelial goblet cells, serous glands, vascular endothelium, epithelium, and in some of the infiltrating inflammatory cells including eosinophils (45). We found that H-PGDS and DP1 were highly expressed in human primary nasal epithelial cells. CRTH2 was also expressed, but only weakly. Thus, PGD2 is likely involved in multiple aspects of allergic inflammation via its dual receptor systems, DP1 and CRTH2, and seems to be an important mediator in allergic diseases.
Recently, Van Hecken et al. (46) reported that a DP1 receptor antagonist, MK-0524, potently inhibits PGD2-induced nasal blockage. In the present study we also demonstrated that MUC5B gene expression was induced by PGD2 in airway epithelial cells and that this induction could be completely suppressed by the DP1 receptor antagonist, S-5751. These findings suggest that PGD2 acts as a key factor of MUC5B gene expression via the DP1 receptor in human airway epithelial cells.
We also showed that ERK MAPK/RSK1/CREB activation was required for PGD2-induced MUC5B gene expression. Several studies have demonstrated that multiple MAP kinases are involved in signal transduction pathways downstream of a variety of inflammatory mediators (47). Transcription of specific genes is induced by the phosphorylation and activation of various transcription factors (48); however, little is known about the specific effect of PGD2 on human airway epithelial cells. Thus, we examined the role of MAP kinases in PGD2-dependent signal transduction systems. Our previous work on MUC5AC gene expression showed that cytokines such as interleukin-1β and tumor necrosis factor-α activate at least two MAP kinases, ERK and p38 MAPK (14). These findings also suggest that ERK MAPK may be an important signaling molecule in mucin gene expression, although the molecules involved in the signaling of ERK MAP kinase for PGD2-induced MUC5B gene expression have yet to be identified.
One substrate of ERKs is RSKs. Upon activation, both ERKs and RSKs translocate to the nucleus, where RSKs can phosphorylate CREB at Ser-133 (39). We have previously shown that RSK1 and CREB are important downstream mediators of ERK MAPK activation in estrogen receptor-induced MUC5B expression (30). However, E2 (estradiol) does not directly bind to the DP1 receptor but triggers the signaling pathway by interacting with ER α (estrogen receptor α). It is only that E2 and PGD2 have common molecules (RSK1, CREB) that are involved in ERK MAPK downstream signaling.
In addition, CREB is a known regulator of the expressions of the mucin genes MUC2, MUC5AC, and MUC5B on the p15 arm of chromosome 11 (11p15) (49). Recently, Koo and co-workers (50) reported that CREB activation is required for PGF2α-induced MUC5AC overproduction. Activation of CREB plays a critical role in the signaling of inflammatory prostaglandins. We have now investigated whether RSK1 can be directly interaction with CREB in NCI-H292 cells. The interaction between RSK1 and CREB was significantly amplified in nucleus by PGD2 treatment. However, inhibition of ERK MAPK pathway greatly decreased the interaction of RSK1 and CREB. Our results indicate that CREB activation is involved in the downstream signaling of ERK MAPK and RSK1 for PGD2-induced MUC5B gene expression.
Like cAMP, Ca2+ functions as a second messenger in a large number of cellular processes (51). It is well established that transient Ca2+ fluxes regulate gene transcription and expression (52). Furthermore, generation of Ca2+ fluxes by intact bacteria and flagella stimulates transcription of the NF-κB-dependent genes, MUC2 and IL-8 (53, 54). In airway epithelial cells, Ca2+ fluxes travel from cell to cell and can also serve to amplify Ca2+-activated proinflammatory signals (55). In hippocampal neurons, coupling of Ca2+ influx and intracellular Ca2+ mobilization pathways to CREB activation has been observed (56).
However, although astrocytes are activated by cAMP, they lack the intracellular Ca2+ signaling cascades required to induce CREB-dependent gene transcription (41). This can be explained by the report that CREB-dependent genes may also be activated by the MAPK and phosphoinositide 3-kinase signaling pathways independent of Ca2+ signaling (57). Likewise, Shaywitz and Greenberg (58) have demonstrated that a variety of kinase signaling pathways can differentially contribute to Ca2+ influx-mediated phosphorylation of CREB in pheochromocytoma (PC12) cells. Notably, a recent paper reported that treatment of human NK cells with PGD2 does not directly increase intracellular Ca2+ (59). Our data showed that PGD2 does not induce changes in the intracellular Ca2+ content in NCI-H292 cells, and we observed similar results in both BEAS-2B and A549 cells. Together, these findings suggest that cAMP may not be involved in Ca2+-signaling responses, depending on the type, which may reflect different mechanisms of Ca2+ signaling.
CREB activation requires phosphorylation at Ser-133 to promote formation of an active transcriptional complex that includes recruitment of CREB-binding protein (CBP), p300, and other transcriptional co-activators to CREs in the promoters of target genes (60, 61). Another study (62) has revealed that co-transfection of Sp1 with fragment 1896 (−956/−1) of the MUC5B promoter leads to an increase in MUC5B expression in LS173T and Caco-2 cells, suggesting that Sp1 may be another important transcription factor in MUC5B gene expression. In addition, Choi et al. (30) reported that CRE sites play an important role in 17β-estradiol-induced MUC5B gene expression. The results of our study show that independent point mutations of each CRE site (−921 to −914 and −900 to −893) in the MUC5B promoter resulted in significant suppression of luciferase activity in NCI-H292 cells, indicating that both of these sites were critical for PGD2-induced MUC5B gene expression. The region (−956/−753) of MUC5B promoter was involved in the response to PGD2, and activation of CREB was important for MUC5B gene expression. We found that the PGD2-induced phosphorylation of CREB had DNA binding and transcriptional activities in the −956/−753 region. These results suggest that both the CRE motif and activation of the CREB signaling pathway are important in the up-regulation of MUC5B gene expression by inflammatory mediators.
In conclusion, our data indicate that H-PGDS was highly expressed in human nasal polyp tissues compared with that in human normal nasal mucosa and that PGD2 was also increased in human nasal polyp tissues. We found that one of the PGD2 receptors, DP1, was expressed in human primary nasal epithelial cells, whereas the CRTH2 receptor was barely detectable. Our data demonstrated that PGD2 induces MUC5B gene expression in airway epithelial cells through the DP1 receptor. Consistently, the DP1 antagonist S5751 significantly suppressed PGD2-induced MUC5B expression, whereas a CRTH2 antagonist did not. Furthermore, the activation of the ERK MAPK/RSK1/CREB signaling pathway, but not intracellular Ca2+, was shown to be involved in PGD2-induced MUC5B gene expression. We also showed that RSK1 can interact directly with CREB. Two CRE sites in the MUC5B promoter (−921 to −914 and −900 to −893) appeared to play a vital role in MUC5B expression in NCI-H292 cells. In addition, the activation of CREB is an important factor of cellular processes, indicating that that CREs in the MUC5B promoter might play an active role in these processes through their binding with CREB. Taken together, these observations provide new insights into the signaling mechanism of MUC5B gene expression in airway epithelial cells.
Acknowledgment
We thank Dr. Yoshihiro Urade (Dept. of Molecular Behavioral Biology, Osaka Bioscience Institute, Japan) for providing the anti-PGD2 synthase mAb.
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Education, Science, and Technology (2011-0001168).
- PGD2
- prostaglandin (PG) D2
- H-PGDS
- hematopoietic prostaglandin D synthase
- DP1
- d-prostanoid receptor
- CRTH2
- chemoattractant receptor homologous molecule expressed on T-helper type 2 cells
- RSK1
- p90 ribosomal S6 protein kinase 1
- CRE
- cAMP-response element
- CREB
- CRE-binding protein
- FAM
- carboxyfluorescein
- TAMARA
- carboxytetramethylrhodamine
- β2M
- β2-microglobulin
- PLA
- proximity ligation assay
- NHNE
- normal human nasal epithelial.
REFERENCES
- 1. Ali M. S., Pearson J. P. (2007) Laryngoscope 117, 932–938 [DOI] [PubMed] [Google Scholar]
- 2. Rogers D. F. (1994) Eur. Respir. J. 7, 1690–1706 [PubMed] [Google Scholar]
- 3. Yuta A., Ali M., Sabol M., Gaumond E., Baraniuk J. N. (1997) Am. J. Physiol. 273, L1203–L1207 [DOI] [PubMed] [Google Scholar]
- 4. Voynow J. A., Gendler S. J., Rose M. C. (2006) Am. J. Respir. Cell Mol. Biol. 34, 661–665 [DOI] [PubMed] [Google Scholar]
- 5. Borchers M. T., Carty M. P., Leikauf G. D. (1999) Am. J. Physiol. 276, L549–L555 [DOI] [PubMed] [Google Scholar]
- 6. Binker M. G., Binker-Cosen A. A., Richards D., Oliver B., Cosen-Binker L. I. (2009) Biochem. Biophys. Res. Commun. 386, 124–129 [DOI] [PubMed] [Google Scholar]
- 7. Kim H. J., Ryu J. H., Kim C. H., Lim J. W., Moon U. Y., Lee G. H., Lee J. G., Baek S. J., Yoon J. H. (2010) Am. J. Respir. Cell Mol. Biol. 43, 349–357 [DOI] [PubMed] [Google Scholar]
- 8. Enss M. L., Wagner S., Schmidt-Wittig U., Heim H. K., Beil W., Hedrich H. J. (1997) Prostaglandins Leukot. Essent. Fatty Acids 56, 93–98 [DOI] [PubMed] [Google Scholar]
- 9. Marom Z., Shelhamer J. H., Steel L., Goetzl E. J., Kaliner M. (1984) Prostaglandins 28, 79–91 [DOI] [PubMed] [Google Scholar]
- 10. van de Bovenkamp J. H., Hau C. M., Strous G. J., Büller H. A., Dekker J., Einerhand A. W. (1998) Biochem. Biophys. Res. Commun. 245, 853–859 [DOI] [PubMed] [Google Scholar]
- 11. Ho S. B., Roberton A. M., Shekels L. L., Lyftogt C. T., Niehans G. A., Toribara N. W. (1995) Gastroenterology 109, 735–747 [DOI] [PubMed] [Google Scholar]
- 12. Chen Y., Zhao Y. H., Kalaslavadi T. B., Hamati E., Nehrke K., Le A. D., Ann D. K., Wu R. (2004) Am. J. Respir. Cell Mol. Biol. 30, 155–165 [DOI] [PubMed] [Google Scholar]
- 13. Keates A. C., Nunes D. P., Afdhal N. H., Troxler R. F., Offner G. D. (1997) Biochem. J. 324, 295–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Song K. S., Lee W. J., Chung K. C., Koo J. S., Yang E. J., Choi J. Y., Yoon J. H. (2003) J. Biol. Chem. 278, 23243–23250 [DOI] [PubMed] [Google Scholar]
- 15. Yuan-Chen Wu D., Wu R., Reddy S. P., Lee Y. C., Chang M. M. (2007) Am. J. Pathol. 170, 20–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hata A. N., Breyer R. M. (2004) Pharmacol. Ther. 103, 147–166 [DOI] [PubMed] [Google Scholar]
- 17. Harris S. G., Padilla J., Koumas L., Ray D., Phipps R. P. (2002) Trends Immunol. 23, 144–150 [DOI] [PubMed] [Google Scholar]
- 18. Lewis R. A., Soter N. A., Diamond P. T., Austen K. F., Oates J. A., Roberts L. J., 2nd. (1982) J. Immunol. 129, 1627–1631 [PubMed] [Google Scholar]
- 19. Hirata M., Kakizuka A., Aizawa M., Ushikubi F., Narumiya S. (1994) Proc. Natl. Acad. Sci. U.S.A. 91, 11192–11196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sawyer N., Cauchon E., Chateauneuf A., Cruz R. P., Nicholson D. W., Metters K. M., O'Neill G. P., Gervais F. G. (2002) Br. J. Pharmacol. 137, 1163–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Spik I., Brénuchon C., Angéli V., Staumont D., Fleury S., Capron M., Trottein F., Dombrowicz D. (2005) J. Immunol. 174, 3703–3708 [DOI] [PubMed] [Google Scholar]
- 22. Tilley S. L., Coffman T. M., Koller B. H. (2001) J. Clin. Invest. 108, 15–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Urade Y., Ujihara M., Horiguchi Y., Ikai K., Hayaishi O. (1989) J. Immunol. 143, 2982–2989 [PubMed] [Google Scholar]
- 24. Urade Y., Ujihara M., Horiguchi Y., Igarashi M., Nagata A., Ikai K., Hayaishi O. (1990) J. Biol. Chem. 265, 371–375 [PubMed] [Google Scholar]
- 25. Blödorn B., Mäder M., Urade Y., Hayaishi O., Felgenhauer K., Brück W. (1996) Neurosci Lett. 209, 117–120 [DOI] [PubMed] [Google Scholar]
- 26. Naclerio R. M., Meier H. L., Kagey-Sobotka A., Adkinson N. F., Jr., Meyers D. A., Norman P. S., Lichtenstein L. M. (1983) Am. Rev. Respir. Dis. 128, 597–602 [DOI] [PubMed] [Google Scholar]
- 27. Matsuoka T., Hirata M., Tanaka H., Takahashi Y., Murata T., Kabashima K., Sugimoto Y., Kobayashi T., Ushikubi F., Aze Y., Eguchi N., Urade Y., Yoshida N., Kimura K., Mizoguchi A., Honda Y., Nagai H., Narumiya S. (2000) Science 287, 2013–2017 [DOI] [PubMed] [Google Scholar]
- 28. Yoon J. H., Gray T., Guzman K., Koo J. S., Nettesheim P. (1997) Am. J. Respir. Cell Mol. Biol. 16, 724–731 [DOI] [PubMed] [Google Scholar]
- 29. Yoon J. H., Moon H. J., Seong J. K., Kim C. H., Lee J. J., Choi J. Y., Song M. S., Kim S. H. (2002) Differentiation 70, 77–83 [DOI] [PubMed] [Google Scholar]
- 30. Choi H. J., Chung Y. S., Kim H. J., Moon U. Y., Choi Y. H., Van Seuningen I., Baek S. J., Yoon H. G., Yoon J. H. (2009) Am. J. Respir. Cell Mol. Biol. 40, 168–178 [DOI] [PubMed] [Google Scholar]
- 31. Choi K. C., Oh S. Y., Kang H. B., Lee Y. H., Haam S., Kim H. I., Kim K., Ahn Y. H., Kim K. S., Yoon H. G. (2008) Biochem. J. 411, 19–26 [DOI] [PubMed] [Google Scholar]
- 32. Kim J. H., Chun Y. S., Lee S. H., Mun S. K., Jung H. S., Lee S. H., Son Y., Kim J. C. (2010) Am. J. Ophthalmol 149, 45–53 [DOI] [PubMed] [Google Scholar]
- 33. Arimura A., Yasui K., Kishino J., Asanuma F., Hasegawa H., Kakudo S., Ohtani M., Arita H. (2001) J. Pharmacol. Exp. Ther. 298, 411–419 [PubMed] [Google Scholar]
- 34. Yasui K., Asanuma F., Hirano Y., Shichijo M., Deguchi M., Arimura A. (2008) Eur. J. Pharmacol. 578, 286–291 [DOI] [PubMed] [Google Scholar]
- 35. Armer R. E., Ashton M. R., Boyd E. A., Brennan C. J., Brookfield F. A., Gazi L., Gyles S. L., Hay P. A., Hunter M. G., Middlemiss D., Whittaker M., Xue L., Pettipher R. (2005) J. Med. Chem. 48, 6174–6177 [DOI] [PubMed] [Google Scholar]
- 36. Chen R. H., Abate C., Blenis J. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 10952–10956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shimamura A., Ballif B. A., Richards S. A., Blenis J. (2000) Curr. Biol. 10, 127–135 [DOI] [PubMed] [Google Scholar]
- 38. Carriere A., Ray H., Blenis J., Roux P. P. (2008) Front. Biosci. 13, 4258–4275 [DOI] [PubMed] [Google Scholar]
- 39. Xing J., Ginty D. D., Greenberg M. E. (1996) Science 273, 959–963 [DOI] [PubMed] [Google Scholar]
- 40. Boie Y., Sawyer N., Slipetz D. M., Metters K. M., Abramovitz M. (1995) J. Biol. Chem. 270, 18910–18916 [DOI] [PubMed] [Google Scholar]
- 41. Murray P. D., Kingsbury T. J., Krueger B. K. (2009) Glia 57, 828–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen Y., Zhao Y. H., Wu R. (2001) Am. J. Respir. Cell Mol. Biol. 25, 409–417 [DOI] [PubMed] [Google Scholar]
- 43. Chokki M., Eguchi H., Hamamura I., Mitsuhashi H., Kamimura T. (2005) FEBS J. 272, 6387–6399 [DOI] [PubMed] [Google Scholar]
- 44. Hatoum O. A., Gauthier K. M., Binion D. G., Miura H., Telford G., Otterson M. F., Campbell W. B., Gutterman D. D. (2005) Arterioscler. Thromb. Vasc. Biol. 25, 2355–2361 [DOI] [PubMed] [Google Scholar]
- 45. Nantel F., Fong C., Lamontagne S., Wright D. H., Giaid A., Desrosiers M., Metters K. M., O'Neill G. P., Gervais F. G. (2004) Prostaglandins Other Lipid Mediat. 73, 87–101 [DOI] [PubMed] [Google Scholar]
- 46. Van Hecken A., Depré M., De Lepeleire I., Thach C., Oeyen M., Van Effen J., Laethem T., Mazina K., Crumley T., Wenning L., Gottesdiener K. M., Deutsch P., Clement P., Lai E., de Hoon J. N. (2007) Eur. J. Clin. Pharmacol. 63, 135–141 [DOI] [PubMed] [Google Scholar]
- 47. Voynow J. A. (2002) Paediatr Respir. Rev. 3, 98–103 [DOI] [PubMed] [Google Scholar]
- 48. Whitmarsh A. J., Davis R. J. (1996) J. Mol. Med. 74, 589–607 [DOI] [PubMed] [Google Scholar]
- 49. Vincent A., Perrais M., Desseyn J. L., Aubert J. P., Pigny P., Van Seuningen I. (2007) Oncogene 26, 6566–6576 [DOI] [PubMed] [Google Scholar]
- 50. Chung W. C., Ryu S. H., Sun H., Zeldin D. C., Koo J. S. (2009) J. Immunol. 182, 2349–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ghosh A., Greenberg M. E. (1995) Science 268, 239–247 [DOI] [PubMed] [Google Scholar]
- 52. Mellström B., Savignac M., Gomez-Villafuertes R., Naranjo J. R. (2008) Physiol. Rev. 88, 421–449 [DOI] [PubMed] [Google Scholar]
- 53. McNamara N., Gallup M., Khong A., Sucher A., Maltseva I., Fahy J., Basbaum C. (2004) FASEB J. 18, 1770–1772 [DOI] [PubMed] [Google Scholar]
- 54. Pena J., Fu Z., Schwarzer C., Machen T. E. (2009) Infect Immun. 77, 2857–2865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Isakson B. E., Olsen C. E., Boitano S. (2006) Respir. Res. 7, 105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lam B. Y., Zhang W., Ng D. C., Maruthappu M., Roderick H. L., Chawla S. (2010) J. Neurochem. 112, 1065–1073 [DOI] [PubMed] [Google Scholar]
- 57. Schinelli S., Zanassi P., Paolillo M., Wang H., Feliciello A., Gallo V. (2001) J. Neurosci. 21, 8842–8853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shaywitz A. J., Greenberg M. E. (1999) Annu. Rev. Biochem. 68, 821–861 [DOI] [PubMed] [Google Scholar]
- 59. Chen Y., Perussia B., Campbell K. S. (2007) J. Immunol. 179, 2766–2773 [DOI] [PubMed] [Google Scholar]
- 60. Anjum R., Blenis J. (2008) Nat. Rev. Mol. Cell Biol. 9, 747–758 [DOI] [PubMed] [Google Scholar]
- 61. Kawasaki H., Eckner R., Yao T. P., Taira K., Chiu R., Livingston D. M., Yokoyama K. K. (1998) Nature 393, 284–289 [DOI] [PubMed] [Google Scholar]
- 62. Van Seuningen I., Pigny P., Perrais M., Porchet N., Aubert J. P. (2001) Front. Biosci. 6, D1216–D1234 [DOI] [PubMed] [Google Scholar]