Abstract
Glycogen synthase is a rate-limiting enzyme in the biosynthesis of glycogen and has an essential role in glucose homeostasis. The three-dimensional structures of yeast glycogen synthase (Gsy2p) complexed with maltooctaose identified four conserved maltodextrin-binding sites distributed across the surface of the enzyme. Site-1 is positioned on the N-terminal domain, site-2 and site-3 are present on the C-terminal domain, and site-4 is located in an interdomain cleft adjacent to the active site. Mutation of these surface sites decreased glycogen binding and catalytic efficiency toward glycogen. Mutations within site-1 and site-2 reduced the Vmax/S0.5 for glycogen by 40- and 70-fold, respectively. Combined mutation of site-1 and site-2 decreased the Vmax/S0.5 for glycogen by >3000-fold. Consistent with the in vitro data, glycogen accumulation in glycogen synthase-deficient yeast cells (Δgsy1-gsy2) transformed with the site-1, site-2, combined site-1/site-2, or site-4 mutant form of Gsy2p was decreased by up to 40-fold. In contrast to the glycogen results, the ability to utilize maltooctaose as an in vitro substrate was unaffected in the site-2 mutant, moderately affected in the site-1 mutant, and almost completely abolished in the site-4 mutant. These data show that the ability to utilize maltooctaose as a substrate can be independent of the ability to utilize glycogen. Our data support the hypothesis that site-1 and site-2 provide a “toehold mechanism,” keeping glycogen synthase tightly associated with the glycogen particle, whereas site-4 is more closely associated with positioning of the nonreducing end during catalysis.
Keywords: Carbohydrate Chemistry, Enzyme Kinetics, Glycogen, Glycogen Synthase, X-ray Crystallography
Introduction
Glycogen synthase was the first reported intracellular target of insulin, and the enzyme catalyzes the linear polymerization of glucose residues from activated sugar donor molecules to the nonreducing end of the glycogen chain. Recent structural studies have shown that the enzyme folds into two Rossmann fold-like domains, with a deep cleft in between harboring the active site (1–3). Although the basic fold is conserved between the prokaryotic, archaeal, and eukaryotic enzymes, there are multiple sequence insertions in the eukaryotic enzymes. The largest of these insertions (a long coiled-coil insert in the C-terminal domain) gives rise to their unique tetrameric arrangement as well as the structural plasticity necessary for the complex regulation of glycogen synthase activity in eukaryotes (3). Furthermore, a conserved arginine cluster present in the C-terminal region of the eukaryotic enzymes mediates the sensitivity to inhibition by phosphorylation and activation by glucose 6-phosphate (4, 5). In our recent structural studies, we demonstrated that the middle two arginine residues (Arg-583 and Arg-587)3 are necessary and sufficient to confer regulation by glucose 6-phosphate and that the first three arginine residues (Arg-580, Arg-581, and Arg-583) are required for full regulatory response to phosphorylation (3).
The yeast Saccharomyces cerevisiae possesses two genes encoding glycogen synthase, GSY1 and GSY2, which are differentially controlled, with the protein product of GSY2 contributing more to glycogen accumulation under most metabolic conditions (6). Unlike many other glycogen-associated proteins and enzymes, such as Stbd1 (7), laforin (8), type 1 protein phosphatase glycogen-targeting subunits (9), and AMP kinase (10), glycogen synthases from yeast or higher eukaryotes do not harbor a readily recognizable carbohydrate-binding module. In addition, very little is known about how glycogen synthase interacts with the nonreducing end during catalysis or stays so tightly associated with the glycogen particle while still maintaining a highly efficient level of catalysis. Early studies with rabbit muscle glycogen synthase using sugar acceptors with varying lengths and branching features demonstrated that the enzyme efficiency increases as longer and more branched oligosaccharides are used as acceptors (11). Based on these results, it was proposed that the enzyme has two distinct acceptor substrate-binding sites, a catalytic site and a polysaccharide-binding site, and occupancy of both sites is required for high catalytic efficiency (11). Consistent with this hypothesis, structural studies of the Escherichia coli (12) and Pyrococcus abyssi (13) glycogen synthases have identified oligosaccharide-binding sites in the N-terminal domain of the respective enzymes that impact catalysis on glycogen in both the archaeal and human enzymes (13).
To examine the mechanism by which eukaryotic glycogen synthases bind their substrates, we solved the structures of yeast Gsy2p in association with maltooctaose in both its basal (R580A/R581A/R583A) and activated (R589A/R592A with glucose 6-phosphate) state conformations. These structures reveal the presence of four distinct maltodextrin-binding sites on the surface of the enzyme. One of the maltodextrin-binding sites is similar in general location to sites observed in the P. abyssi and E. coli enzymes (12, 13) but is formed from structural elements unique to the eukaryotic enzymes (residues 117–121, 145–156, and 182–184). The remaining three sites (site-2 (residues 365–367 and 439–465), site-3 (residues 333–340 and 461–463) and site-4 (residues 208–211, 324, and 507–561)) are found on the surface of the C-terminal domain and have no known counterpart in the bacterial or archaeal enzymes. Site-1, site-2, and site-4 utilize structural elements derived from unique sequence insertions found in the eukaryotic enzymes. Sequence alignment of the eukaryotic enzymes shows that the residues providing the hydrophobic and polar interactions for maltodextrin binding are highly conserved among eukaryotic species (supplemental Fig. S1). Mutations to conserved residues within these sites decreased the catalytic efficiency toward glycogen as well as glycogen binding. Furthermore, studies in Δgsy1-gsy2 yeast cells demonstrated that the glycogen-binding mutants significantly reduced the net glycogen accumulation. Finally, we developed a new and highly sensitive assay that directly measures the ability of glycogen synthase to utilize small oligosaccharides, such as maltooctaose, as substrates and can distinguish between glycogen-binding sites from those that affect positioning of the nonreducing end for catalysis. Using this assay, we show that site-4 affects the ability of Gsy2p to utilize maltooctaose as a substrate and is implicated in acceptor end positioning during catalysis.
EXPERIMENTAL PROCEDURES
Protein Purification and Crystallization
A PCR-based site-directed mutagenesis approach was used for generating the mutant enzymes from the pET-28A-wild-type Gsy2p construct. His-tagged Gsy2p was expressed in E. coli BL21(DE3) cells, and the recombinant enzyme was purified as described previously (3) by affinity and ion exchange chromatographies. Crystals of the basal and activated state conformations of yeast Gsy2p R580A/R581A/R583A and R589A/R592A were prepared, cryoprotected, and frozen as described previously (3). The complexes with bound oligosaccharides were prepared in crystal soaking experiments using 50 mm maltooctaose (Carbosynth Ltd.).
Data Collection, Processing, Structure Solution, and Refinement
The data sets were collected at the Advanced Photon Source at beamline 23-ID operated by the General Medical Sciences and Cancer Institute Collaborative Access Team (GM/CA-CAT). The data sets were indexed, integrated, and scaled using the HKL2000 program suite (14). The structures were solved by the molecular replacement method using the program MOLREP as implemented in the CCP4 program package (15) using Protein Data Bank codes 3NAZ and 3NB0 as the search models for the basal and activated state conformations, respectively. Initial rigid-body and restrained refinements were performed with Refmac5 (16), and subsequent rounds of refinement utilized Phenix (17, 18).
Enzyme Assays and Kinetic Data Analysis
The activities of the purified Gsy2p enzymes were measured as described previously (19). Glycogen titration of the enzymes was performed using a fixed UDP-glucose concentration of 4.4 mm and nine different concentrations of glycogen ranging between 0.03 and 12 mg/ml in the absence and presence of 7.2 mm glucose 6-phosphate. The SigmaPlot software package (version 10) was used to determine the apparent Michaelis-Menten parameters by fitting to the following equation: v = (Vmax·[S])/(Km + [S]).
Maltooctaose Extension Assay
The reaction conditions for maltooctaose utilization by Gsy2p were as follows: 25 mm Tris-HCl (pH 7.6), 12.5 mm UDP-glucose, and 1.3–3.3 μm Gsy2p enzyme in a final reaction volume of 20 μl. The maltooctaose concentration was varied between 5 and 150 mm. The enzyme concentration and incubation time were optimized for each enzyme to ensure that maltononaose was the only product formed during the reaction. The enzymatic reactions were carried out at 30 °C for either 10 or 20 min and terminated by heating at 95 °C for 3 min, followed by cooling on ice for 5 min. After cooling, maltotriose was added to the reactions as an internal standard to control for injection volume variability, and the sample was subjected to centrifugation at 14,000 rpm for 1 min. For each substrate concentration, a control reaction was prepared in which an equivalent concentration of UDP was substituted for UDP-glucose. For consistency of analysis, prior to injection, the samples were diluted with water to give a final oligosaccharide concentration of ∼1 μm. All samples were analyzed using a Dionex ICS3000 high-performance anion-exchange chromatography system equipped with a PA1 column and monitored using pulse amperometric detection as described previously (20). All samples were filtered prior to injection. Eluent A consisted of 100 mm NaOH, and Eluent B consisted of 100 mm NaOH containing 1 m sodium acetate. The samples were eluted with a continuous gradient of 0–31.25% Eluent B over 25 min at a flow rate of 0.25 ml/min. To quantitate the reaction progress, a calibration curve for maltooctaose was prepared between 0.5 to 15 μm and analyzed under conditions identical to those used in the reaction. Reaction rates were determined by monitoring the decrease in maltooctaose as a function of time. All areas under the curve for maltooctaose were normalized to that of the internal standard.
Gsy2p Construct for Yeast Expression
The pJR1420-A vector (21) was used for expressing wild-type and mutant Gsy2p in yeast cells. pJR1420-A has the promoter and coding region of the GSY2 gene with the stop codon deleted cloned into pRS416 (22) containing the gene for enhanced GFP and a yeast terminator sequence from the UGP1 gene. The various maltodextrin-binding mutants were transferred from pET-28a into the pJR1420-A vector suitable for yeast expression by a three-step procedure. The pJR1420-A vector was digested with NotI and SpeI, and a fragment containing the GSY2 promoter and coding sequence was subcloned between the NotI and SpeI sites of pBluescript II SK(+), generating pBluescript-Gsy2. Each maltodextrin-binding mutant of GSY2 in pET-28a was digested with BstBI and NdeI, releasing a fragment of GSY2 spanning the region that contained the mutations of interest. These BstBI/NdeI fragments were then used to replace BstBI/NdeI fragments of pBluescript-Gsy2. The resulting GSY2 mutants were removed from pBluescript-Gsy2 by digestion with NotI and SpeI and cloned into pJR1420-A, which had been digested with NotI and SpeI. Each construct expressed wild-type Gsy2p or the maltodextrin-binding mutant fused at the C terminus to GFP. The yeast strain WW100, which is deficient for both the GSY1 and GSY2 genes (MATα leu2 trp1 ura3 gsy1::kanMX6 gsy2::natMX4), was used for the in vivo studies. Yeast cells were transformed with the wild-type or mutant Gsy2p-expressing plasmids and grown as described previously (21).
Glycogen Synthase Activity and Glycogen Accumulation Measurement in Yeast Cells
Yeast cells were lysed with glass beads, and the glycogen synthase activity in the cell lysate was determined by the filter paper method described previously under the standard assay conditions (19, 23). The glycogen accumulation in the cells was quantified by the method described previously (24). Yeast cells transformed with the vector alone were used as the negative control for activity measurement and glycogen quantification. The relative levels of expression of the various glycogen synthase constructs were determined by Western blot analysis of the yeast cell extracts with anti-Gsy2p and anti-GFP antibodies (Roche Applied Science). Yeast Hsp90 was used as a loading control.
RESULTS
Overall Structures
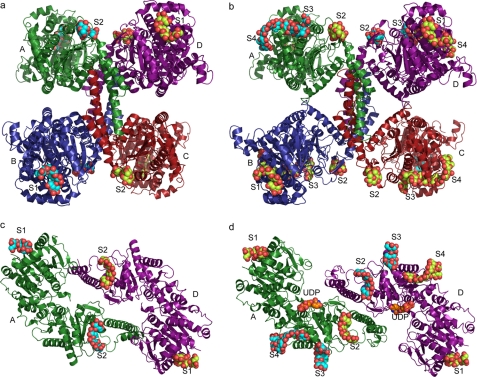
Two distinct crystal structures of yeast Gsy2p with bound maltodextrins were determined using maltooctaose as an analog of the linear chains of glycogen (Fig. 1 and Table 1). For these studies, we used non-phosphorylated mutant forms of Gsy2p that permit structure determination in a basal activity state (R580A/R581A/R583A) and in an activated state (R589A/R592A) (3). The first mutant is conformationally restricted and does not respond to glucose 6-phosphate as an activator or to inhibitory phosphorylation. In contrast, the R589A/R592A mutant exists in an inhibited state that can be fully activated by glucose 6-phosphate (3). We created complexes with the substrate analog in both the basal and activated states of the enzyme in an attempt to map how changes in conformation affect acceptor binding within the active site. The overall structures of these two activity states and the local structures within the maltodextrin-binding sites did not change substantially in response to substrate binding, demonstrating a root mean square deviation at their Cα positions of 0.34 and 0.36 Å to their respective uncomplexed structures (Protein Data Bank codes 3NAZ and 3NB0) (3). Surprisingly, rather than being bound in the active-site cleft, these substrate analogs bound to conserved surface crevices on the enzyme, and the number of available sites increased from two to four upon activation (supplemental Table S1). All of the sugar molecules bound in the basal and activated state conformations show a tight distribution of torsion angles and glycosidic angles (supplemental Table S2).
FIGURE 1.
Glycogen-binding sites in Gsy2p. a and c, ribbon diagram representation of the basal state conformation of the yeast Gsy2p tetramer (a) and dimer (c) (top view), highlighting the available glycogen-binding sites. The individual subunits are colored separately and labeled. The glycogen-binding sites are marked S1–S4, and the bound glycogen is represented in space-filling models. The cyan molecules represent the actual occupied sites, and the yellow molecules highlight the unoccupied sites. b and d, ribbon diagram representation of the activated conformation of the tetramer (b) and dimer (d) with similar coloring and labeling schemes. UDP binding is shown in the activated state dimer in a space-filling model and labeled. Structural alignments were utilized to place maltodextrins at all available binding sites to provide a representation of the full number of interaction surfaces available; see “Results” and supplemental Table S1 for site occupancies in the crystal structures.
TABLE 1.
Data collection and refinement statistics
| Basal (R580A/R581A/R583A) | Activated (R589A/R592A) | |
|---|---|---|
| Data collection | ||
| Space group | P21 | I222 |
| Cell dimensions | a = 96.6, b = 166.7, c = 121.1 Å; α = 90.0°, β = 103.2°, γ = 90.0° | a = 193.4, b = 205.3, c = 206.9 Å; α = 90.0°, β = 90.0°, γ = 90.0° |
| Resolution (Å) | 50.0-3.0 (3.1-3.0) | 50.0-2.8 (2.9-2.8) |
| Rmerge (%) | 8.2 (52.5) | 9.0 (60.3) |
| I/σ〈I〉 | 13.2 (2.0) | 13.7 (2.0) |
| Completeness (%) | 99.4 (94.7) | 99.8 (99.8) |
| Redundancy | 3.8 (3.1) | 5.0 (4.6) |
| Refinement | ||
| Resolution (Å) | 50.0-3.0 | 50.0-2.8 |
| No. of reflections | 73,079 | 96,658 |
| Rwork/Rfree (%) | 20.9/24.7 | 21.1/26.1 |
| No. of atoms | ||
| Protein | 19,742 | 20,617 |
| Ligand/ion | 260 | 333 |
| B-factors | ||
| Protein | 82.10 | 72.00 |
| Ligand/ion | 111.48 | 68.73 |
| r.m.s.d.a | ||
| Bond length (Å) | 0.008 | 0.007 |
| Bond angle | 1.15° | 1.01° |
| Ramachandran plot | ||
| Core | 86.1 | 85.6 |
| Allowed | 13.5 | 14.2 |
a r.m.s.d., root mean square deviation.
Maltodextrin Binding to the Basal State Conformation of Gsy2p
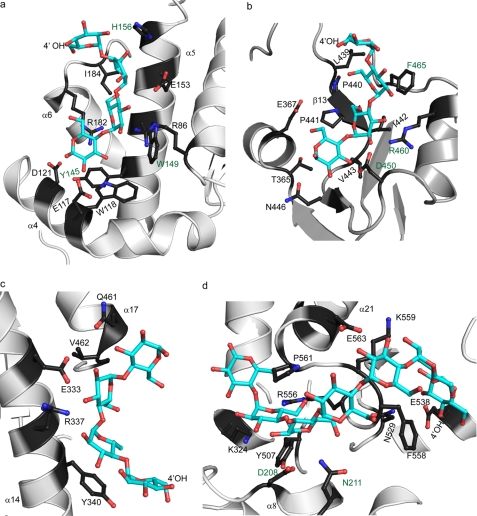
In the basal state conformation, we observed two oligosaccharide-binding sites in each subunit, and in each of these two sites, we found four ordered glucose residues bound along depressions on the surface of the enzyme (Fig. 1, a and c). Site-1 is occupied in subunits A and B. This site is not available in subunits C and D, as it is utilized in a crystal-packing contact. Site-1 is located on the surface of the N-terminal domain and is defined by helices α4, α5, and α6 (Figs. 1, a and c, and 2a). Helix α4 in Gsy2p is an element of secondary structure unique to eukaryotic enzymes and is adjacent to the 1′-end of the maltodextrin moiety. Helix α5 forms the base of the maltodextrin site and runs more or less parallel with the carbohydrate moiety. The side chains of Trp-118, Tyr-145, Trp-149, and Ile-184 form the hydrophobic contacts, and the side chains of Glu-117, Asp-121, Glu-153, His-156, Arg-182, and Arg-86 are positioned along the edges of the hydrophobic trough to provide hydrogen-bonding interactions.
FIGURE 2.
Protein-ligand interactions in the glycogen-binding sites of Gsy2p. a–d, ribbon representation of the secondary structural elements of the glycogen binding in Gsy2p in site-1–4, respectively. The bound polymer is represented in a cyan ball and stick model, and the 4′-hydroxyl group is labeled. The surrounding amino acids are represented in a ball and stick model and labeled. The residues mutated to alanine for the functional studies of the binding sites are shown in green.
Site-2 is located in a crevice near the subunit A/D and B/C interfaces and lies along the structural boundary between the conserved Rossmann fold domain and the unique helical elements that give rise to the subunit arrangement found in the eukaryotic enzymes (Fig. 1, a and c). Similar to the maltodextrin binding at site-1, only two of the available sites are occupied, each containing four ordered glucose residues in subunits A and B. The inner surface of site-2 is lined by a hydrophobic stretch of residues within the secondary structural element β13 comprising Leu-439–Val-443 and Phe-465 (Fig. 2b). The orientation of the maltodextrin moiety is essentially antiparallel to the direction of the β-strand. This hydrophobic trench is surrounded by Thr-365, Glu-367, Asn-446, Asp-450, and Arg-460, which are positioned for hydrogen-bonding interactions with the bound glucose polymer.
Maltodextrin Binding to the Activated State of Gsy2p
In the activated state of Gsy2p, we observed maltodextrins bound to site-1 and site-2 within subunits C and D as well as two additional sugar polymer-binding sites, designated site-3 and site-4 (Fig. 1, b and d). Site-3 is positioned adjacent to site-2 in the C-terminal domain and is bordered by helices α17 and α18 (Fig. 2c). In subunit A, site-3 has two ordered glucose residues visible, whereas in subunit D, this site contains four ordered glucose residues. The side chains of Tyr-340 and Val-462 are positioned on either side of the sugar polymer and provide the majority of the hydrophobic binding surface. In subunit D, the first and second glucose moieties stack against the side chain of Tyr-340, whereas the polar side chains of Gln-333, Arg-337, Gln-461, and Gln-463 are positioned to form hydrogen bonds with the hydroxyl groups of the glucose residues.
Unlike the other sites that are positioned on the surface of Gsy2p, site-4 is located within the interdomain cleft and is adjacent to the active site of the enzyme (Fig. 1d). In subunit A, site-4 has the longest ordered oligosaccharide with seven glucose residues (Fig. 2d). This extended binding pocket is defined by helix α21 and loop α21–β18 from the C-terminal Rossmann domain and helix α8 from the N-terminal domain, the latter of which is a unique structural element in the eukaryotic enzymes. The pyranose ring of Glc-2 in this polymer is stacked between the side chains of Tyr-507 and Pro-561, whereas Phe-558 stacks against Glc-6 and Glc-7 at the 4′-end of the polymer. The 2′-hydroxyl group of Glc-2 is located within hydrogen-bonding distance to the side chain of Lys-324. The side chains of Asp-208 and Asn-211 from helix α8 are positioned to hydrogen bond with the hydroxyl groups of Glc-3. The 1′-end of the oligosaccharide interacts with Arg-556, whereas the 4′-end is stabilized by interactions with the side chains of Glu-563, Lys-559, Asn-529, and Glu-538.
Kinetic Studies Using Glycogen
To study the influence of the sites on enzyme function, a sequence alignment of the yeast and mammalian glycogen synthase enzymes was generated (supplemental Fig. S1), and residues that made contacts critical for maltodextrin binding and are highly conserved across eukaryotes were selected for mutation to alanine. Because these sites possess multiple interactions across rather large surfaces, we chose to prepare multiple mutations within each site to ensure disruption of function. Using this strategy, we prepared the following mutations of Gsy2p for study: site-1 (W118A/W149A/H156A), site-2 (D450A/R460A/Y465A), site-3 (E333A/Y340A/Q461A), and site-4 (D208A/N211A/R556A). The site-4 triple mutant was not stable, was subject to extensive proteolysis, and had no detectable enzymatic activity. As our prior work had also demonstrated that mutations to residues 556–559 of Gsy2p were unstable (4), we prepared the D208A/N211A double mutant to study site-4. A similar but more intense protein stability problem was observed with the site-3 mutant. In this case, the triple alanine mutant and all possible combinations of double alanine mutants were not stable, suggesting that these amino acids serve a structural as well as functional role. Thus, wild-type Gsy2p and three stable maltodextrin-binding site mutants (site-1, site-2, and site-4) were used for all subsequent functional studies.
To determine the influence of glycogen concentration on enzymatic activity, we varied the concentrations of glycogen in both the absence and presence of 7.2 mm glucose 6-phosphate to determine the S0.5 for glycogen and overall catalytic efficiency toward glycogen (Table 2 and supplemental Fig. S3). All of the mutant enzymes exhibited increased S0.5 values, decreased Vmax values, and catalytic efficiencies that were reduced between 17- and 65-fold in the absence of glucose 6-phosphate and between 6- and 75-fold in the presence of glucose 6-phosphate (Table 2).
TABLE 2.
Kinetic properties of Gsy2p (glycogen extension)
| Enzyme |
Vmax for glycogen varied |
S0.5 for glycogen |
Vmax/S0.5 |
|||
|---|---|---|---|---|---|---|
| −Glc-6-P | +Glc-6-P | −Glc-6-P | +Glc-6-P | −Glc-6-P | +Glc-6-P | |
| min−1 | mg/ml | |||||
| WT | 1120 ± 20 | 1730 ± 10 | 0.29 ± 0.03 | 0.12 ± 0.01 | 3860 | 14,400 |
| Site-1 | 180 ± 20 | 1340 ± 30 | 1.73 ± 0.32 | 4.89 ± 0.08 | 100 | 270 |
| Site-2 | 220 ± 10 | 1070 ± 30 | 3.69 ± 0.41 | 5.57 ± 0.15 | 60 | 190 |
| Site-1/site-2 | NSa | NS | NS | NS | 4.5b | 4.5b |
| Site-4 | 780 ± 10 | 1450 ± 20 | 3.37 ± 0.12 | 0.60 ± 0.03 | 230 | 2420 |
a NS, non-saturable kinetics.
b Vmax/S0.5 values were estimated from the specific activity at 6.7 mg/ml glycogen.
Utilization of Maltooctaose as an Acceptor Substrate
To distinguish the contributions of the identified maltodextrin-binding sites to glycogen binding versus acceptor positioning at or near the active site, an assay was developed that could directly measure extension of small maltodextrin substrate analogs by glycogen synthase. The results from this assay compared favorably with the reported behavior toward maltodextrins using 14C incorporation (11). In contrast to its behavior toward glycogen as a substrate, mutation of site-2 had no demonstrable effect on maltooctaose utilization (Table 3). The site-4 mutant was least able to utilize maltooctaose as a substrate and did not show saturable behavior up to the maximal tested concentration of 150 mm, which precluded determination of Vmax or Km values. However, the Vmax/Km for maltooctaose could be estimated from the slope of this line (Table 3). The site-1 mutant showed intermediate behavior and exhibited an increased Km as well as a decreased Vmax for maltooctaose, resulting in a 4.5-fold lower catalytic efficiency toward maltooctaose (Table 3). Analysis of the product distributions following more extended incubations between the wild-type enzyme and maltooctaose gave results consistent with a distributive mode of catalysis (supplemental Fig. S4).
TABLE 3.
Kinetic properties of Gsy2p (maltooctaose extension)
| Enzyme | Vmax | Km | Vmax/Km |
|---|---|---|---|
| min−1 | m | m−1min−1 | |
| WT Gsy2p | 470 ± 29 | 0.044 ± 0.009 | 10,700 ± 2500 |
| Site-1 | 270 ± 20 | 0.110 ± 0.010 | 2400 ± 200 |
| Site-2 | 530 ± 33 | 0.046 ± 0.007 | 12,000 ± 1100 |
| Site-1/site-2 | 210 ± 20 | 0.067 ± 0.017 | 3200 ± 500 |
| Site-4 | NSa | NSa | 1400 ± 200b |
a NS, non-saturable kinetics.
b Vmax/Km was estimated from the linear slope of the Michaelis-Menten plot.
Glycogen Accumulation and Synthase Activity in Δgsy1-gsy2 Yeast
To study the effect of the maltodextrin-binding site mutants in cells, glycogen synthase-deficient yeast cell lines were complemented with various forms of wild-type and mutant Gsy2p enzymes. Cells expressing the individual site-1 and site-2 mutants accumulated between 2- and 6-fold less glycogen compared with wild-type enzyme-expressing cells, whereas expression of the site-4 mutant reduced glycogen accumulation by only 30% (Table 4). Cells transformed with the combined site-1/site-2 mutant possessed ∼40 times less glycogen than wild-type enzyme-transformed cells (Table 4). Consistent with the glycogen accumulation assays, the extracts from yeast cells expressing the combined site-1/site-2 mutant showed the lowest levels of glycogen synthase activity (Table 4). The differences in the amount of glycogen synthase activity cannot be accounted for by different levels of mutant protein expression (supplemental Fig. S5).
TABLE 4.
Yeast glycogen synthase activity and glycogen levels
| Construct | Yeast cell lysate GSa activity + Glc-6-P | Glycogen accumulation |
|---|---|---|
| nmol/min/mg | μg/107 cells | |
| Vector alone | 0.68 ± 0.33 | ND |
| WT | 19.5 ± 3.3 | 32.3 ± 4.5 |
| Site-1 | 3.25 ± 0.63 | 5.58 ± 1.28 |
| Site-2 | 1.88 ± 0.42 | 14.7 ± 2.7 |
| Site-1/Site-2 | 1.87 ± 0.34 | 0.81 ± 0.22 |
| Site-4 | 6.65 ± 1.69 | 22.9 ± 5.5 |
a GS, glycogen synthase; ND, not detectable.
DISCUSSION
Many enzymes acting on polysaccharides are characterized by the presence of a carbohydrate-binding module that facilitates the association between the enzyme and its glycan substrate (25). The data presented here are consistent with what has been found for the bacterial and archaeal glycogen synthases (12, 13) and glycogen phosphorylase (26) in that they integrate their carbohydrate-binding sites onto the surface of the enzyme. The physical and chemical features of the sites are comparable with the B-type carbohydrate-binding modules, which are characterized by the presence of a hydrophobic cleft or groove that accommodates at least two sugar moieties where additional points of contact are mediated through hydrogen bonding to the polar edges of the sugar molecules (25). Despite these physical and chemical similarities, there is no conservation of either the secondary structural elements or the amino acid sequences that contribute to glycan binding in glycogen synthase or carbohydrate-binding modules. It is an interesting aspect of functional evolution that the relative positioning of the respective N-terminal domain glycogen storage sites in glycogen phosphorylase, E. coli glycogen synthase, P. abyssi glycogen synthase, and site-1 of yeast Gsy2p is similar yet structurally distinct at the molecular level.
In both the P. abyssi and E. coli enzymes, all of the glycogen-binding sites are distributed on the surface of the N-terminal domain, where it was proposed, for the E. coli enzyme, that they efficiently uncouple substrate association from the domain movement necessary for catalysis (12). Unlike these enzymes, yeast Gsy2p has binding sites located on both the N- and C-terminal domains, and sequence insertions that form secondary structural elements unique to the eukaryotic enzymes contribute to these binding sites. The sequence conservation within the maltodextrin-binding sites suggests that these sites are present in all eukaryotic enzymes. Indeed, although the tyrosine residues identified to contribute to dextran binding in the P. abyssi enzyme (13) are not observed to directly bind maltodextrins in Gsy2p, these residues form the underlying structural foundation for site-1 in Gsy2p. This may explain why mutation of these tyrosine residues in human muscle glycogen synthase affects catalysis on glycogen and glycogen accumulation in cell culture (13). The mutational studies reported here for Gsy2p demonstrate that its dextran-binding sites, like the sites found in glycogen phosphorylase and glycogen synthase from P. abyssi, facilitate the association of the enzyme with the glycogen substrate and are necessary for efficient catalysis. These accessory sites also explain the observation that glycogen synthase remains bound to glycogen independent of its activity state because essentially all glycogen synthase is found in association with glycogen in vivo even when its activity ratio is reduced due to increased phosphorylation (27, 28). Efficient co-localization of enzyme and substrate would facilitate faster responses to the physiological signals that control glucose homeostasis and eliminate any lag period resulting from the reassociation of enzyme and substrate. Consistent with the impact of mutations within site-2 on glycogen association and accumulation, two mutations associated with glycogen storage disease type 0 in humans (T445M and H446D, corresponding to Thr-444 and His-445 in Gsy2p) (29) occur at amino acid positions in close proximity to the site-2 maltodextrin-binding site. Thus, the impaired glycogen synthesis in these patients may be due to reduced association between the enzyme and its substrate.
Although it is clear that site-1 and site-2 likely have primary functions directed toward maintaining association with glycogen, the role of site-4 is more complex. Unlike site-1–3, site-4 is located at the interdomain cleft and near the active site of Gsy2p. Tyr-507, which lies in the midst of this site (Fig. 2), is in a highly conserved stretch of residues (supplemental Fig. S1) that includes the putative active-site nucleophile Glu-509 (30). Mutations within site-4 were the least detrimental to catalysis using glycogen but the most detrimental to the enzyme's ability to utilize maltooctaose (Tables 2 and 3). Because glycogen is a heterogeneous substrate, it is not possible to directly compare the catalytic efficiencies toward the two substrates without some assumptions. However, if we assume that the average molecular mass of glycogen in the assay is ∼1.3 × 106 Da (11), then the S0.5 of the wild-type enzyme for glycogen could be estimated to be between 90 and 200 nm, giving a Vmax/S0.5 in excess of 1.0 × 108 m−1 s−1, which is very near the diffusion-limited condition for enzyme-catalyzed reactions. In contrast, the Vmax/Km of the wild-type enzyme for maltooctaose is ∼200 m−1 s−1, a reduction in catalytic efficiency of nearly 106. Clearly, glycogen synthase is well optimized to use glycogen as a substrate. It is likely that this is a function of the enzyme's ability to remain tightly associated with the polymer through multiple contacts that lie outside the active site coupled to a lower affinity binding site for the nonreducing end toward which catalysis is directed. This strategy solves the problem of rapid binding and release of actively extending chains and at the same time does not require complete dissociation from the polymer to reset the position of the acceptor end within the active site. It also increases the local concentration of nonreducing ends at or near the active site such that the lower affinity acceptor site does not negatively impact the overall efficiency of catalysis.
Recent work has suggested that the binding at these accessory sites forms the basis for processivity of glycogen synthase toward glycogen (13). Although this may be true, processivity toward a heterogeneous substrate such as glycogen is difficult to assess because there are many nonreducing ends within a single glycogen particle where catalysis can be directed without dissociation from the particle itself. If one uses this definition of processivity (namely catalysis limited to a single glycogen particle), then our data would lend further support to this hypothesis. However, it is less clear how many rounds of elongation occur toward any individual nonreducing end, where a more strict definition of processivity would prevail. At present, there are no unequivocal methodologies available to assess this definition of processivity toward glycogen. However, because our assay directly measures and quantitates the products from maltooctaose elongation, we can say that yeast glycogen synthase acts distributively toward maltooctaose, producing a Gaussian distribution of products with variable length as a function of time (supplemental Fig. S5). Therefore, any processivity directed toward glycogen must be a function of the additional points of contact between the enzyme and this complex substrate.
In conclusion, multiple studies by different groups have demonstrated that glycogen synthases possess glycogen association sites integrated into the surface of the enzyme. Our studies have shown that yeast Gsy2p has four highly conserved glycogen association sites, distinct from the actual catalytic site, that function to keep glycogen synthase localized to the glycogen polymer. We have shown that these sites are responsible for the enzyme's high catalytic efficiency toward this complex polymer and impact the level of glycogen that accumulates in cells. Mutations within site-4 only moderately affect catalysis on glycogen but severely affect utilization of maltooctaose as an alternative substrate. Consequently, site-4 may participate in positioning the 4′-acceptor end of the growing glycogen chain in the active site. The tight association conferred by this multipoint glycogen-binding mechanism may provide a functional advantage during signaling processes because the enzyme remains bound to glycogen independent of its activation status and thus does not require reacquisition of its substrate when cellular conditions are conducive for glycogen synthesis.
Supplementary Material
Acknowledgments
We thank Dr. Martin Schmidt (Des Moines University) for providing the anti-yeast Hsp90 polyclonal antibody. We thank the staff of the General Medical Sciences Cancer Institute Collaborative Access Team at the Advanced Photon Source at the Argonne National Laboratory. The Argonne National Laboratory is operated by the UChicago Argonne, LLC, for the United States Department of Energy Office of Biological and Environmental Research under Contract DE-AC02-06CH11357.
Footnotes
This work was supported, in whole or in part, by National Institutes of Health Grants R37-DK027221 and R01-NS056454 (to P. J. R.), R01-DK079887 (to T. D. H.), and R15-GM081810 (to W. A. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5 and Tables S1 and S2.
The atomic coordinates and structure factors (codes 3RSZ and 3RT1) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
We have adopted the residue numbering recommended by the Human Genome Organisation (HUGO), and numbering begins at the initiator methionine.
REFERENCES
- 1. Buschiazzo A., Ugalde J. E., Guerin M. E., Shepard W., Ugalde R. A., Alzari P. M. (2004) EMBO J. 23, 3196–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Horcajada C., Guinovart J. J., Fita I., Ferrer J. C. (2006) J. Biol. Chem. 281, 2923–2931 [DOI] [PubMed] [Google Scholar]
- 3. Baskaran S., Roach P. J., DePaoli-Roach A. A., Hurley T. D. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 17563–17568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pederson B. A., Cheng C., Wilson W. A., Roach P. J. (2000) J. Biol. Chem. 275, 27753–27761 [DOI] [PubMed] [Google Scholar]
- 5. Hanashiro I., Roach P. J. (2002) Arch. Biochem. Biophys. 397, 286–292 [DOI] [PubMed] [Google Scholar]
- 6. Farkas I., Hardy T. A., Goebl M. G., Roach P. J. (1991) J. Biol. Chem. 266, 15602–15607 [PubMed] [Google Scholar]
- 7. Jiang S., Heller B., Tagliabracci V. S., Zhai L., Irimia J. M., DePaoli-Roach A. A., Wells C. D., Skurat A. V., Roach P. J. (2010) J. Biol. Chem. 285, 34960–34971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang J., Stuckey J. A., Wishart M. J., Dixon J. E. (2002) J. Biol. Chem. 277, 2377–2380 [DOI] [PubMed] [Google Scholar]
- 9. Ceulemans H., Bollen M. (2004) Physiol. Rev. 84, 1–39 [DOI] [PubMed] [Google Scholar]
- 10. Hudson E. R., Pan D. A., James J., Lucocq J. M., Hawley S. A., Green K. A., Baba O., Terashima T., Hardie D. G. (2003) Curr. Biol. 13, 861–866 [DOI] [PubMed] [Google Scholar]
- 11. Larner J., Takeda Y., Hizukuri S. (1976) Mol. Cell. Biochem. 12, 131–136 [DOI] [PubMed] [Google Scholar]
- 12. Sheng F., Yep A., Feng L., Preiss J., Geiger J. H. (2009) Biochemistry 48, 10089–10097 [DOI] [PubMed] [Google Scholar]
- 13. Díaz A., Martínez-Pons C., Fita I., Ferrer J. C., Guinovart J. J. (2011) J. Biol. Chem. 286, 18505–18514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 15. (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 16. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 17. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 18. Terwilliger T. C., Grosse-Kunstleve R. W., Afonine P. V., Moriarty N. W., Zwart P. H., Hung L. W., Read R. J., Adams P. D. (2008) Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Thomas J. A., Schlender K. K., Larner J. (1968) Anal. Biochem. 25, 486–499 [DOI] [PubMed] [Google Scholar]
- 20. Tagliabracci V. S., Heiss C., Karthik C., Contreras C. J., Glushka J., Ishihara M., Azadi P., Hurley T. D., DePaoli-Roach A. A., Roach P. J. (2011) Cell Metab. 13, 274–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wilson W. A., Boyer M. P., Davis K. D., Burke M., Roach P. J. (2010) Can J. Microbiol. 56, 408–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sikorski R. S., Hieter P. (1989) Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hardy T. A., Huang D., Roach P. J. (1994) J. Biol. Chem. 269, 27907–27913 [PubMed] [Google Scholar]
- 24. Wang Z., Wilson W. A., Fujino M. A., Roach P. J. (2001) Mol. Cell. Biol. 21, 5742–5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Christiansen C., Abou Hachem M., Janecek S., Viksø-Nielsen A., Blennow A., Svensson B. (2009) FEBS J. 276, 5006–5029 [DOI] [PubMed] [Google Scholar]
- 26. Johnson L. N., Acharya K. R., Jordan M. D., McLaughlin P. J. (1990) J. Mol. Biol. 211, 645–661 [DOI] [PubMed] [Google Scholar]
- 27. Peng Z. Y., Trumbly R. J., Reimann E. M. (1990) J. Biol. Chem. 265, 13871–13877 [PubMed] [Google Scholar]
- 28. Suzuki Y., Lanner C., Kim J. H., Vilardo P. G., Zhang H., Yang J., Cooper L. D., Steele M., Kennedy A., Bock C. B., Scrimgeour A., Lawrence J. C., Jr., DePaoli-Roach A. A. (2001) Mol. Cell. Biol. 21, 2683–2694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Weinstein D. A., Correia C. E., Saunders A. C., Wolfsdorf J. I. (2006) Mol. Genet. Metab. 87, 284–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cid E., Gomis R. R., Geremia R. A., Guinovart J. J., Ferrer J. C. (2000) J. Biol. Chem. 275, 33614–33621 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.