Background: Fibroblasts can remodel the extracellular matrix by phagocytosis, but how the adhesion mechanisms are regulated is not well understood.
Results: Non-muscle myosin IIA provides a docking site for the actin-severing protein to localize at collagen adhesion sites and remodel actin filaments there.
Conclusion: Gelsolin binds to non-muscle myosin IIA at collagen adhesions and enables phagocytosis to occur in a calcium-dependent manner.
Significance: Dissection of the mechanisms by which collagen phagocytosis is mediated may provide new insights into fibrotic conditions under which phagocytosis is perturbed.
Keywords: Calcium, Collagen, Fibroblast, Integrin, Phagocytosis
Abstract
The formation of adhesion complexes is the rate-limiting step for collagen phagocytosis by fibroblasts, but the role of Ca2+ and the potential interactions of actin-binding proteins in regulating collagen phagocytosis are not well defined. We found that the binding of collagen beads to fibroblasts was temporally and spatially associated with actin assembly at nascent phagosomes, which was absent in gelsolin null cells. Analysis of tryptic digests isolated from gelsolin immunoprecipitates indicated that non-muscle (NM) myosin IIA may bind to gelsolin. Immunostaining and immunoprecipitation showed that gelsolin and NM myosin IIA associated at collagen adhesion sites. Gelsolin and NM myosin IIA were both required for collagen binding and internalization. Collagen binding to cells initiated a prolonged increase of [Ca2+]i, which was absent in cells null for gelsolin or NM myosin IIA. Collagen bead-induced increases of [Ca2+]i were associated with phosphorylation of the myosin light chain, which was dependent on gelsolin. NM myosin IIA filament assembly, which was dependent on myosin light chain phosphorylation and increased [Ca2+]i, also required gelsolin. Ionomycin-induced increases of [Ca2+]i overcame the block of myosin filament assembly in gelsolin null cells. We conclude that gelsolin and NM myosin IIA interact at collagen adhesion sites to enable NM myosin IIA filament assembly and localized, Ca2+-dependent remodeling of actin at the nascent phagosome and that these steps are required for collagen phagocytosis.
Introduction
Extracellular matrix synthesis and degradation are tightly coordinated to maintain connective tissue homeostasis. Collagen is the most abundant extracellular matrix protein of mammalian connective tissues (1) and is synthesized by fibroblasts, the predominant mesenchymal cells of soft connective tissue. Although collagen degradation by matrix metalloproteinases has been studied in depth (2), the critical regulatory steps of the intracellular collagen degradation pathway mediated by fibroblasts have not been defined.
Phagocytosis is essential for the uptake and degradation of microorganisms and damaged cells by specialized phagocytes (3, 4), but phagocytosis is also a critical process for the remodeling of connective tissue matrices by fibroblasts. Indeed, collagen phagocytosis by fibroblasts is critically involved in physiological remodeling of the extracellular matrix and in wound healing (5, 6). In collagen phagocytosis, following initial binding of collagen fibrils by fibroblasts, the fibril is cleaved extracellularly and is then internalized, after which it is degraded by lysosomal hydrolases including cathepsins (7–10). It is believed that the initial binding step of collagen phagocytosis may be a rate-limiting step in the degradation of collagen (8, 10–14).
The phagocytic process in many cell types, including fibroblasts, involves an initial attachment of the particle to cell surface receptors, which triggers recruitment of several families of adhesion proteins that interact with the actin cytoskeleton to mediate binding and internalization (15). For efficient collagen phagocytosis, actin rearrangements at collagen binding sites depend on the function of actin-severing and -capping proteins such as gelsolin (11, 13, 16). Gelsolin plays an important role in FcγR-mediated phagocytosis but not in complement receptor-mediated phagocytosis (17). Although a role for gelsolin in the initial collagen binding steps of α2β1 integrin-mediated phagocytosis has been shown (13), the potential roles of gelsolin-interacting binding proteins at collagen adhesion sites have not been examined.
Gelsolin, an actin-binding protein that can sever and cap actin filaments, nucleates filament assembly in a Ca2+-dependent manner (18, 19). At low [Ca2+]i, gelsolin is a compact, inactive structure consisting of six homologous repeat domains, G1–G6 (20, 21). On binding to Ca2+, gelsolin undergoes conformational changes to open the G1–G3 and G4–G6 latches (22, 23); these alterations expose actin binding sites on G1 and G4 and a filament side binding site on G2 (24, 25). Efficient severing of actin filaments by the N-terminal half of gelsolin (G1–G3) requires internal cooperation with the C-terminal half (G4–G6) (26). Notably, the reversibility of actin-gelsolin associations is achieved by reducing Ca2+ and phosphoinositides (27) or tropomyosin (28), which dissociates gelsolin from actin filaments in vitro.
The role of gelsolin in the initial binding step of collagen phagocytosis has been examined in fibroblasts derived from gelsolin null and matched background wild-type mice (13). The altered organization and reduced turnover of subcortical actin filaments in gelsolin null cells is thought to interfere with Ca2+ entry through integrin-gated channels; this results in greatly attenuated collagen binding and phagocytosis (13). In this context, treatment of cells with latrunculin B increases collagen bead binding (29), possibly by reducing interactions between cortical actin filaments and channel proteins to enhance Ca2+ entry as has been observed in other systems (30).
In addition to its role in activating gelsolin and other adhesion proteins, the net effect of increased Ca2+ on the actin cytoskeleton is to depolymerize actin filaments (31). Disassembly of actin filaments has been linked to myosin filament assembly (32), contraction, and changes in cell shape, all of which are needed for efficient phagocytosis. Myosin superfamily motor proteins interact with actin filaments to generate contractile forces that are critical for phagosome development (33). Myosin molecules are hexamers composed of myosin heavy chain dimers and two pairs of myosin light chains (34). In mammalian cells, phosphorylation on Ser-19 of the myosin II regulatory light chain is a commonly invoked mechanism for the regulation of myosin filament assembly in vivo, as in vitro studies show that light chain phosphorylation promotes filament assembly (35).
NMIIA2 is involved in complement receptor- but not FcγR-mediated phagocytosis, perhaps by controlling actin polymerization (36). NMIIA filaments may also coordinate phagocytosis (37) by depolymerizing (32) or fluidizing cortical actin (38). We showed earlier that NMIIA filaments localize to collagen adhesion sites and are required for binding and sequestering the small GTPase Rap1 in adhesions (12), which is important for regulating β1 integrin activation (39) and, accordingly, for collagen phagocytosis. As gelsolin is enriched in nascent collagen adhesions and plays a key role in mediating collagen phagocytosis by regulating actin assembly and β1 integrin activation (11, 13), we considered that gelsolin may interact with NMIIA to mediate myosin and actin filament assembly at the collagen binding site.
EXPERIMENTAL PROCEDURES
Reagents
Latex (2-μm diameter) beads were purchased from Polysciences (Warrington, PA). Purified bovine collagen solution was purchased from Advanced Biomatrix (Palo Alto, CA). Antibodies to β-actin (clone AC-15), myosin light chain, GAPDH, fluorescein isothiocyanate (FITC)-conjugated goat anti-mouse antibody and TRITC-phalloidin were obtained from Sigma. Immobilized protein G was obtained from Pierce. The antibody to NMHC II-A (BT-564) was from Biomedical Technologies, and the monoclonal antibody to NMHC II-B (CMII23) was from the Developmental Studies Hybridoma Bank (Iowa City, IA). Antibodies to myosin light chain (MLC) and phospho-MLC (P-Ser-19) were purchased from Cell Signaling (Danvers, MA). Heavy meromyosin IIA (1-1337 amino acids) was generously provided by Dr. J. R. Sellers (National Institutes of Health). Purified plasma gelsolin, purified skeletal muscle actin, and non-muscle actin purified from platelets were purchased from Cytoskeleton (Denver, CO). The antibody to gelsolin was prepared by immunization of a specific pathogen-free rabbit (New Zealand White strain) with purified mouse gelsolin. Immunization was carried out by subcutaneous injections in combination with adjuvant. Serum of prebleed (before first injection) and 1–3 bleeds was checked by ELISA to determine the titer (11). The myosin light chain kinase inhibitor ML-7, latrunculin B, jasplakinolide, and blebbistatin were purchased from (Calbiochem). Myosin light chain antibody and phospho-myosin light chain antibody were purchased from Cell Signaling. Ionomycin, fura2/AM, and BAPTA/AM were purchased from Invitrogen. Antibody to FLAG was purchased from Sigma.
Cell Preparation
Fibroblasts were obtained from either wild-type or Gsn− day 12 mouse fetuses and cultured as described (40). The embryos from which the fibroblasts were derived were genotyped by PCR of tail snips to confirm the deletion of gelsolin. Cells were grown in DMEM (Invitrogen) supplemented with 10% fetal calf serum and 1% antibiotics. The fibroblastic identity of the cells was verified by staining for intracellular vimentin and type I collagen as well as by the absence of desmin. To study the role of NMIIA in collagen phagocytosis we used embryonic stem (ES) cells. ES cells have been used extensively for studies of NMIIA function (41, 42) and provide complete ablation of NMIIA, whereas siRNA provides ∼70% knockdown of this protein (see “Results”). NMHC II-A null, NMHC II-B null, and wild-type (WT) embryonic stem cells were cultured as described previously (41).
Collagen Bead Binding
Collagen-coated latex beads (2-μm diameter) were applied to microbiological (i.e. non-tissue culture) dishes, dried down, and attached as described (43) followed by washing with PBS. The number of beads plated per dish was adjusted to produce final bead:cell ratios specific for each experiment. Cells were counted electronically, and the cell concentration was adjusted prior to plating cells on dishes containing collagen-coated beads. The plates were maintained at room temperature for 20 min to allow the cells to settle and subsequently were washed with fresh medium at 37 °C. Detached cells were removed by repeated washes.
In experiments to evaluate collagen bead internalization, FITC-collagen-coated beads were incubated with cells for timed incubation periods. Internalization was stopped by cooling on ice. Fluorescence from the extracellular beads was quenched by trypan blue, whereas internalized beads retained their bead-associated fluorescence (7), which was used to estimate the percentage of internalized beads by fluorescence microscopy.
Isolation of Collagen Bead-associated Proteins and Immunoprecipitation
Cell suspensions were allowed to attach to bead-coated dishes for 20 min. Floating cells were aspirated and replaced with media warmed to 37 °C to synchronize phagocytosis. Cells and collagen-coated latex beads were collected with a cell scraper into buffer (10 mm Tris, 1 mm NaF, and 10 mm iodoacetamide, pH 7.5) containing protease inhibitors (Sigma) and incubated on ice for 20 min before cells were passed through 25-gauge needle (five times). Lysates were centrifuged and fractionated into cytosolic (supernatant) and pellet fractions (membrane). Pellets were solubilized in extraction buffer (1% Triton X-100, 150 mm NaCl, 10 mm Tris-HCl, pH 7.2, 1 mm Na3VO4, 20 μg/ml aprotinin, and 1 μg/ml Pefabloc). Equal amounts of proteins were incubated with antibodies to gelsolin and NMIIA to form immunocomplexes that were captured on protein G-Sepharose beads (Pierce). Samples were boiled, separated on SDS-PAGE, and immunoblotted, and bands were quantified by scanning densitometry. To evaluate the myosin filament assembly at the collagen bead sites WT and gelsolin-deficient cells were incubated with collagen-coated beads. Triton-insoluble fractions were prepared by lysis in ice-cold Triton X-100 buffer (50 mm Tris-HCl, pH 7.4, 100 mm NaCl, 50 mm KCl, 5 mm MgCl2, 0.5% Triton X-100, 5 mm DTT, and 1 mm PMSF with protease inhibitors). After 5 min of incubation, soluble and insoluble proteins were separated by centrifugation (44).
Cells were incubated with BAPTA/AM (4 μm) for 45 min before and during treatment with collagen-coated beads. Cells were treated with ionomycin (2 μm) for 10 min and ML-7 (1 μm) for 30 min before and during incubation with collagen beads.
Gelsolin Constructs
Human full-length gelsolin cDNA (45) was provided by H. L. Yin. A primer pair (forward, 5′-GCGCAAGCTTGTGGTGGAACACCCCGAGTTCCTCAAGGCA-3′; reverse, 5′-GCGCGGATCCCTAAGACCAGTAATCATCATCCCAGCCAAG-3′) was designed to amplify full-length gelsolin (G1–G6, 2.151 kb, encoding gelsolin residues 26–742). A primer pair, (forward, 5′-GCGCAAGCTTGTGGTGGAACACCCCGAGTTCCTCAAGGCA-3′; reverse, 5′-GCGCGGATCCCTAGTCCCGCCAGTTCTTGAAGAACTGCTT-3′) was used to amplify the three N-terminal domains (G1–G3, 1.038 kb, encoding gelsolin residues 26–371 (23)), and a primer pair (forward, 5′-GCGCAAGCTTGACGATGGCACAGGCCAGAAACAGATCTGG-3′; reverse, 5′-GCGCGGATCCCTAAGACCAGTAATCATCATCCCAGCCAAG-3′) was used to amplify the three C-terminal domains (G4–G6, 0.987 kb, encoding gelsolin residues 414–742). The resulting PCR products were ligated into HindIII/BamHI sites (underlined sequences in the primers) of N3×FLAG-pCMV5c (constructed by Dr. Miriam Barrios-Rodiles) and sequenced (ACGT Corp., Toronto, Ontario, Canada).
The pEGFP-N1-Lifeact and pRuby-N1-Lifeact constructs (all in the pEGFP backbone from Clontech, Palo Alto, CA) were kindly provided by Roland Wedlich-Soldner (Cell Dynamics and Cell Patterning, Max Planck Institute of Biochemistry, Martinsried, Germany). Lifeact, a 17-amino acid peptide from the N terminus of the actin-binding protein Abp140, which binds actin filaments and localizes to actin patches and cables without interference to cellular processes, served as an actin filament binding marker. The peptide sequence for Lifeact is MGVADLIKKFESISKEE (46).
Gelsolin and NMIIA Binding Assay
To assess the binding of NM myosin IIA rods (residues 1338–1960) to GST-gelsolin full-length (G1–G6), proteins were dialyzed against assembly buffer overnight before binding assays were performed. Varying concentrations of GST-gelsolin Sepharose-bound proteins (6–14 μm) were incubated with assembled NM II-A rods (12 μm; 20 mm Tris, pH 7.5, 20 mm NaCl, 2 mm MgCl2, and 1 mm DTT) at 23 °C for 1 h in reaction buffer containing 20 mm Tris-HCl, pH 7.5, 20 mm NaCl, 2 mm MgCl2, and 1 mm DTT in the presence of Ca2+ (0.3 mm) or EGTA (2 mm). The Sepharose beads were washed three times and separated on SDS-PAGE.
Myosin Filament Extraction
For assessment of myosin filament assembly at collagen bead sites, we used immunofluorescence methods as described previously (47). Briefly, gelsolin WT and gelsolin null cells were permeabilized by cooling to 4 °C, washed with ice-cold PBS, and extracted for 30–90 s with ice-cold 10 mm Tris, pH 7.0, 60 mm KCl, 125 mm sucrose, and 0.05% Triton X-100. Cells were washed three times with ice-cold 10 mm Tris, pH 7.0, 30 mm KCl, 5 mm MgCl2, and 1 mm CaCl2 and fixed followed by immunostaining.
For immunoblotting, myosin filament-enriched samples associated with the collagen beads were prepared by cell lysis in ice-cold Triton X-100 (Triton X-100, 50 mm Tris-HCl, pH 7.4, 100 mm NaCl, 50 mm KCl, 5 mm MgCl2, 0.5% Triton X-100, and 5 mm DTT with protease and phosphatase inhibitors). After 5 min of incubation, soluble and insoluble proteins were separated by centrifugation, and Triton-insoluble components were suspended in 2× Laemmli sample buffer.
Transfection
We determined the functional relationship between gelsolin expression and collagen phagocytosis in fibroblasts by transfecting Gsn− cells with a gelsolin expression vector. Gsn− cells were transfected using FuGENE 6 transfection reagent (Roche Diagnostics). The optimum vector concentrations were determined by titration experiments. Cells were transfected and incubated for 48 h. To estimate transfection efficiency and to serve as a transfection control, pEGFPluc (Clontech) was used, and the numbers of fluorescent cells were counted.
Immunofluorescence, Confocal Total Internal Reflection Fluorescence (TIRF) Microscopy, and Fluorescence Recovery after Photobleaching
We determined the spatial distribution of gelsolin and myosin II. Cells were incubated with collagen-coated beads, fixed with 3% formaldehyde in PBS, permeabilized with 0.2% Triton X-100, and stained. The spatial distribution of staining around beads was determined by confocal microscopy (Leica, Heidelberg, Germany; ×40 oil immersion lens). Transverse optical sections were obtained at 0.5 μm nominal thickness using Leica software and analyzed with Adobe Photoshop. Co-localization was analyzed in 3–6 fields/cell depending on the number of bound beads by using the Image J plug-in, JACoP (48). The Pearson's correlation coefficient was expressed as the mean ± S.D.
We used TIRF microscopy to selectively illuminate fluorophores within ∼100 nm of the interface of collagen bead attachments to the cell surface. We observed the targeting of actin filaments to the bead sites close to the cell surface with red fluorescent protein-Lifeact (46) and myosin filaments with GFP-NMIIA (Addgene), both of which were co-transfected in gelsolin WT and null cells. Images were obtained simultaneously over a time course. Post-acquisition processing was used to measure fluorescence in a 2-μm square zone around attached beads of GFP-NMMIIA or RFP actin filaments.
For fluorescence recovery after photobleaching measurements, Gsn− and WT type cells were labeled directly with phycoerythrin-α2 integrin antibody for 30 min and washed in HEPES-buffered DMEM supplemented with 10 mm glucose, 1 mm Mg2+, and 1 mm Ca2+. Preparations were kept at room temperature to minimize the internalization of receptors. Fluorescently labeled receptors in small circular areas (4-μm2 bleach spots) were illuminated and photobleached (maximum intensity, 500 ms) with an argon laser at 488 nm. Fluorescence in the bleach spot was measured before bleaching and for 180 s after bleaching. The diffusion coefficient (D × 10−10cm2/s) and the mobile fractions (%R) were calculated (49, 50).
Intracellular Ca2+ Measurements
Cells were loaded with fura2/AM (3 μm) and plated on collagen-coated beads. The intracellular Ca2+ concentration ([Ca2+]i) in single, attached cells was estimated as described with a microscope-based ratio fluorimeter (51).
Statistical Analyses
For continuous variables, the means ± S.E. were computed, and differences between groups were evaluated by Student's unpaired t test or analysis of variance for multiple comparisons with statistical significance set at p < 0.05. Post hoc comparisons after analysis of variance were performed with Tukey's test, a statistical test that allows comparisons between different groups. For all experiments, at least three independent experiments were evaluated, each performed in triplicate.
RESULTS
Effect of Gelsolin and NMIIA on Collagen Bead Binding and Phagocytosis
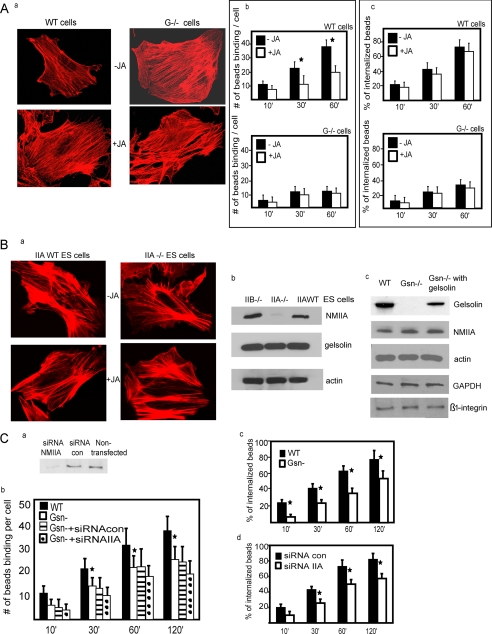
Gelsolin null cells exhibit much more prominent actin filaments than WT cells (40), which may be important in regulating collagen phagocytosis. We attempted to phenocopy the abundance of actin filaments in fibroblasts by treating gelsolin WT fibroblasts with jasplakinolide (JA), an agent that promotes actin assembly by inhibiting actin turnover (52). JA-treated WT cells exhibited more pronounced actin staining with phalloidin (Fig. 1A, a), which resembled gelsolin null cells that were stained with phalloidin. JA did not affect gelsolin expression levels in gelsolin WT cells or the levels of NMIIA in NMIIA WT cells (supplemental Fig. 1A) nor did it affect the interaction of NMIIA with actin filaments (supplemental Fig. 1B).
FIGURE 1.
A, a, gelsolin WT and gelsolin null cells with and without treatment with JA (500 nm) and stained with rhodamine-phalloidin. b, gelsolin WT and null cells were treated with JA and plated on collagen-coated beads. Data are means ± S.E. of bound collagen beads per cell (n = 100 cells/group). *, difference of p < 0.05. c, similarly, the % bead internalization was not different in the presence or absence of JA in gelsolin null cells and gelsolin WT cells. B, a, phalloidin-stained cells showed no difference in actin filament distribution in untreated or JA-treated cells. b, total cell lysates from NMIIB null, NMIIA null, and WT ES cells were immunoblotted for the indicated proteins. c, similarly, cell lysates from gelsolin WT, gelsolin null cells, and gelsolin null cells transfected with gelsolin cDNA show levels of indicated proteins. C, a, fibroblasts from gelsolin null mice transfected with NMIIA siRNA show >70% reduction of NMIIA protein levels compared with cells treated with non-targeted siRNA. b, the numbers of collagen beads binding per cell were significantly (p < 0.05) lower in gelsolin null cells and in gelsolin null cells transfected with siRNA NMIIA. c, in fibroblasts from WT mice treated with NMIIA or non-targeted siRNA as control, internalized beads were discriminated by quenching the fluorescence of extracellular bound beads with trypan blue. Fluorescent and quenched beads were counted in 60 cells/sample at each time point, and NMIIA knockdown cells showed reduced internalization for all time points (p < 0.02). d, collagen bead internalization assays on gelsolin WT and null cells.
Treatment with JA inhibited collagen bead binding at 30 and 60 min in WT cells (p < 0.05) but did not further reduce collagen binding in gelsolin null cells (p > 0.2; Fig. 1A, b). When we compared collagen bead binding in gelsolin WT cells and gelsolin null cells without JA treatment, at all times after the addition of collagen beads there was more bead binding in WT cells, whereas in the presence of JA we found significantly higher binding only at 60 min (supplemental Fig. 1C). The percentage of collagen bead internalization was not different in the presence or absence of JA when gelsolin null cells and gelsolin WT cells were analyzed separately (Fig. 1A, c). Notably, the percentage of collagen bead internalization was significantly higher in gelsolin WT compared with gelsolin null cells with or without JA (supplemental Fig. 1D).
We assessed actin filament distribution in NMIIA WT and NMIIA−/− embryonic stem cells. Phalloidin-stained cells showed no difference in actin filament distribution. JA-treated or untreated cells also showed no significant difference of staining (Fig. 1B, a), which may suggest differences in actin remodeling in ES cells compared with gelsolin WT fibroblast cells. Notably, JA did not affect the recruitment of GFP-NMIIA (supplemental Fig. 1E), actin filaments (supplemental Fig. 1F), or talin to bound collagen beads (supplemental Fig. 1G).
We assessed the expression levels of β-actin, β1 integrin, and NMIIA in the WT and gelsolin knock-out cells and the levels of β-actin, β1 integrin, and gelsolin in the NMIIA WT and NMIIA−/− embryonic stem cells (Fig. 1B, b and c). However, apart from the expected deletion of gelsolin and NMIIA, we found no differences.
Our previous data showed that phosphatidylinositol 4,5-bisphosphate (PIP2), which is important for the uncapping of gelsolin from actin filaments (31), is involved in regulating the internalization stages of collagen phagocytosis but not in the earlier collagen binding stage (11). Accordingly, we sought to determine what other molecules may associate with gelsolin and regulate actin assembly at the collagen binding site. Gelsolin was immunoprecipitated from cells that had been incubated with collagen beads or from untreated cells. The immunoprecipitates were analyzed by tandem mass spectrometry. These analyses identified 69 tryptic peptides in samples treated with collagen beads that matched sequences in mouse NMIIA compared with nine tryptic peptides in untreated samples (Table 1). These data indicate that gelsolin may associate with NMIIA in response to collagen binding.
TABLE 1.
LC-MS/MS analysis of gelsolin immunoprecipitates
| Protein name | GenBankTM accession no. | No. of peptides |
|
|---|---|---|---|
| Untreated | CCB-treateda | ||
| NMMHC IIA | 1143264 | 9 | 69 |
| Actin | 4501885 | 18 | 18 |
| Vimentin | 202368 | 10 | 15 |
| Myosin light chain | 71037403 | 4 | 8 |
| Supervillin | 1486910 | 1 | 2 |
| Flightless 1 | 1152849 | 0 | 1 |
| Myosin regulatory chain 10 | 15809016 | 0 | 2 |
| Mts1 | 199818 | 0 | 1 |
| Prothymosin α | 1487082 | 0 | 2 |
a CCB, collagen-coated bead.
We used siRNA to knock down NMIIA in fibroblasts obtained from gelsolin null mice in order to study the role of gelsolin and NMIIA in collagen binding. Protein levels of NMIIA were reduced by 70% compared with protein levels in cells transfected with control siRNA or non-transfected cells (Fig. 1C, a). In time course experiments (bead:cell ratio = 8:1), the number of collagen beads bound per cell was significantly lower (2.5-fold; p < 0.05) in gelsolin null cells and in gelsolin null cells treated with control siRNA compared with bead binding in wild-type cells (Fig. 1C, b). Gelsolin null cells transfected with NMIIA siRNA showed only small additional reductions in collagen binding, suggesting that gelsolin and NMIIA may regulate collagen binding through a common pathway.
In the process of collagen phagocytosis, binding is followed by collagen internalization and degradation in phagolysosomes by cathepsins (7). To assess collagen bead internalization, beads on the cell surface were distinguished from internalized beads with trypan blue, a method that quenches fluorescence of extracellular beads while the fluorescence of internalized beads is not affected (11). From 30–120 min there was a 30–40% reduction in bead internalization in gelsolin null cells, and similarly siRNA NMIIA cells showed a reduction in bead internalization (p < 0.05; Fig. 1C, c and d). There was no significant difference in the percentage of bead internalization between gelsolin null cells and NMIIA siRNA-treated cells (supplemental Fig. 1H).
Gelsolin and NMIIA Are Required for Actin Assembly at Collagen Bead Binding Sites
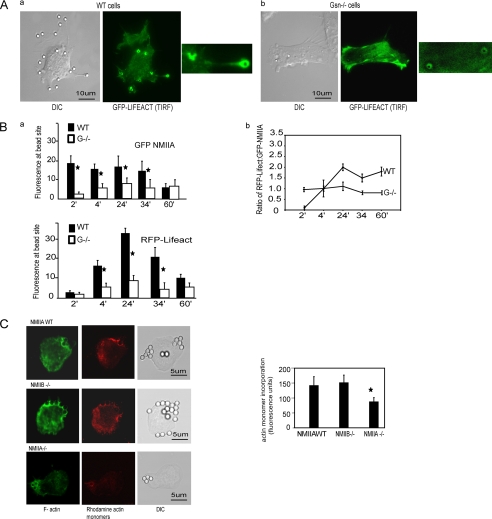
We examined the accumulation of actin at collagen bead binding sites in WT and gelsolin null cells that had been transfected with GFP-Lifeact, a 17-amino acid peptide that selectively binds actin filaments (46). TIRF microscopy of sites around bead peripheries showed a marked accumulation of actin around collagen beads in WT cells, which was not seen in gelsolin null cells (Fig. 2A).
FIGURE 2.
A, recruitment of F-actin (GFP-Lifeact) to bead binding sites imaged by TIRF microscopy in living gelsolin WT (a) and gelsolin null cells (b). Insets show fluorescence at the collagen-coated bead sites in gelsolin WT and null cells. DIC, differential interference contrast. B, a, WT and gelsolin null cells were co-transfected with RFP-Lifeact and GFP-NMIIA. Images were taken on live cells over 60 min. Histograms show mean fluorescence ± S.E. from regions of interest (3-μm diameter circles) around individual bead sites obtained from the images of 30 different cells at each time point. b, line graph shows the mean ± S.E. of the ratios of RFP-Lifeact to GFP-NMIIA fluorescence from around beads in 30 different cells for each time point. C, representative images of NMIIA WT, NMIIA B null, and NMIIA null cells showing the incorporation of rhodamine-labeled actin monomers in permeabilized cells to estimate free barbed ends. Cells were stained with Alexa 488 phalloidin to show F-actin. Histogram shows means ± S.E. of rhodamine fluorescence from 50 cells for each condition.
Gelsolin WT and null cells were transfected with RFP-Lifeact and GFP-NMIIA to examine the temporal and spatial relationships of actin and NMIIA accumulation at collagen bead binding sites. In gelsolin WT cells there was abundant GFP-NMIIA at the collagen bead binding sites during initial bead attachment, which remained constant until 34 min and then declined by 60 min after binding. In the absence of gelsolin, GFP-NMIIA fluorescence around beads was much lower than in WT cells (Fig. 2B, a). In WT cells actin filament fluorescence at collagen bead binding sites was very low at early times after bead attachment but increased sharply by 24 min and then, by 60 min, decreased by 4-fold (Fig. 2B, a). An examination of the mean ratios of RFP-Lifeact to GFP-NMIIA fluorescence at each bead site (n = 30 cells at each time point) revealed a large, time-dependent increase of the ratios of the relative abundance of actin filaments to NMIIA, which peaked at 24 min and declined thereafter (Fig. 2B, b). This pattern reflects large fluctuations in accumulations of actin filament at collagen adhesion sites. In gelsolin null cells, the ratios of actin filament to NMIIA fluorescence were not substantially altered over time after collagen bead binding.
We measured the effect of NMIIA on de novo incorporation of actin monomers into filaments around collagen-coated beads at 20 min after bead incubation, a time that was contemporaneous with maximal actin filament assembly. NMIIA WT cells or NMIIA null cells were incubated with collagen-coated beads, permeabilized with octyl glucoside, and then incubated with rhodamine labeled actin monomers (53–55). NMIIA-deficient cells showed ∼2-fold less incorporation of actin monomers compared with NMIIA WT and NMIIB−/− cells (p < 0.01; Fig. 2C), which is similar to previous observations of gelsolin-deficient cells that showed reduced actin monomer incorporation compared with WT type cells (11). These data indicate that NMIIA enhances actin monomer incorporation into actin filaments at collagen bead adhesion sites.
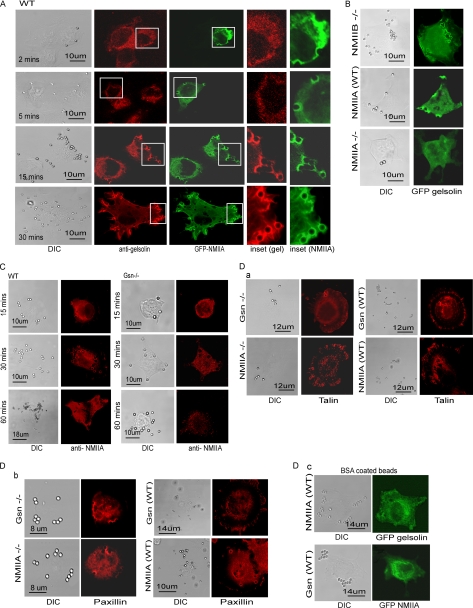
Targeting of NMIIA and Gelsolin to Collagen-coated Beads
Gelsolin WT cells were transfected with GFP-NMIIA and immunostained for gelsolin. NMIIA was rapidly recruited to collagen beads (within 2 min), whereas gelsolin was not detected at these early time points. By 15 and 30 min after collagen bead incubation, GFP-NMIIA and endogenous gelsolin were both localized to beads (Pearson's correlation coefficient = 0.88 ± 0.09; Fig. 3A). We determined whether NMIIA would be required for gelsolin accumulation at collagen bead binding sites. In NMIIA null cells GFP-gelsolin was not recruited to collagen bead sites (Fig. 3B, a), but gelsolin recruitment was similar in NMIIAWT and NMIIB−/− cells after 20 min of incubation with collagen-coated beads. These results were not because of differences in the protein expression levels of β-actin or gelsolin in NMIIA null cells, as WT cells and NMIIB−/− cells exhibited similar levels of expression of these proteins (Fig. 1B, b and c). Collectively these data show that NMIIA is enriched at collagen bead binding sites prior to gelsolin and that NMIIA is important for gelsolin enrichment at collagen binding sites.
FIGURE 3.
A, representative images showing distribution of GFP-NMIIA and endogenous gelsolin in WT cells. Insets show the targeting of gelsolin and NMIIA proteins to the developing phagosomes. B, NMIIB null, NMIIA WT, or non-muscle myosin IIA-deficient ES cells showing localization of GFP-gelsolin to collagen-coated beads after 20 min of incubation. A total of 20 cells were examined. These experiments were repeated three times. C, collagen-coated beads incubated with gelsolin WT and gelsolin null cells show enhanced localization of endogenous NMIIA to beads over time. D, a and b, gelsolin WT and gelsolin null cells and NMIIA WT ES cells and NMIIA null ES cells show an equivalent accumulation of talin and paxillin at collagen beads by immunostaining for talin (a) and paxillin (b). c, BSA-coated beads incubated with WT NMIIA and WT gelsolin cells transfected with GFP gelsolin and GFP NMIIA, respectively. A total of 25 cells were examined. DIC, differential interference contrast.
We examined the spatial distribution of endogenous NMIIA with respect to bound collagen beads in gelsolin WT and null cells. Over a period of 5–60 min, we observed increasing accumulations of NMIIA at attached collagen beads in WT cells that was not observed in gelsolin null cells (Fig. 3C). We also noted an increase of the number of bound beads per cell in WT compared with gelsolin null cells, which was consistent with the data shown in Fig. 1.
As deletion of either gelsolin or NMIIA from cells may have affected the structure of the actin cytoskeleton, which could in turn affect the localization of gelsolin and NMIIA adjacent to beads, we incubated gelsolin WT and gelsolin null cells and NMIIA WT ES cells and NMIIA−/− ES cells with collagen-coated beads and immunostained for talin and paxillin, proteins that are recruited in the early stages of adhesion formation (56). Talin and paxillin localized equivalently around collagen beads at 5 and 10 min after bead incubation in both cell types (Fig. 3D, a and b). Protein targeting to collagen-coated beads was β1 integrin-specific, as there was no recruitment of either myosin IIA or gelsolin to BSA-coated beads (Fig. 3D, c). Collectively these data indicate that reciprocal targeting of gelsolin and NMIIA to bead sites is required for enhanced collagen bead binding over time.
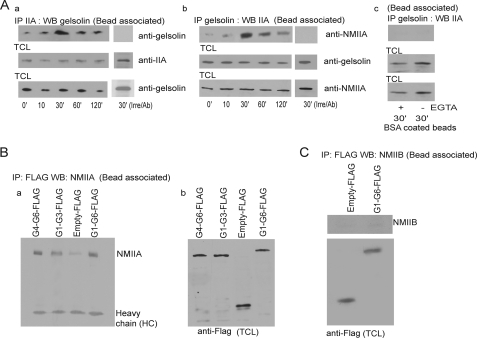
Gelsolin Associates with NMIIA at Collagen Bead Sites
As gelsolin and NMIIA apparently associated with each other in response to collagen-coated beads (Table 1) and were both enriched at collagen bead binding sites, we examined by biochemical methods a potential association between these proteins. Gelsolin immunoprecipitates from collagen bead-associated proteins were examined in cells collected at 0–120 min after collagen bead incubations. Compared with cells without beads, there was a 4-fold increase of gelsolin associated with NMIIA at 30 min compared with 0 min (Fig. 4A, a). This association was confirmed by immunoprecipitation of NMIIA, which showed a 5-fold increase of abundance of gelsolin in NMIIA immunoprecipitates at 30 min compared with 0 min (Fig. 4A, b). These associations were measured in the presence of 1 mm Ca2+; under low Ca2+ conditions the association was weak (data not shown). Control immunoprecipitations with irrelevant antibodies showed no gelsolin or NMIIA in the immunoprecipitates (Fig. 4A, a and b). To determine the specificity of the gelsolin/NMIIA associations in response to collagen bead binding, we performed the above experiment in response to BSA-coated beads in the presence or absence of Ca2+ (Fig. 4A, c).
FIGURE 4.
A, a and b, NMIIA or gelsolin immunoprecipitates of bead-associated proteins were immunoblotted for gelsolin and NMIIA, respectively. In separate experiments an irrelevant antibody was used for immunoprecipitation, and immunoprecipitates were immunoblotted with NMIIA and gelsolin. c, gelsolin immunoprecipitates of bead-associated proteins were immunoblotted for NMIIA in response to BSA-coated beads in the presence or absence of Ca2+. B, gelsolin null cells transfected with FLAG-tagged G1–G6, G1–G3, and G4–G6 constructs were incubated with collagen beads for 30 min. Bead-associated proteins immunoprecipitated (IP) with FLAG antibody were immunoblotted (WB, Western blot) for NMIIA (a). TCL, total cell lysates (b). These observations were made in four different experiments. C, gelsolin null cells transfected with FLAG-tagged G1–G6 and FLAG-tagged empty construct. Cells were incubated with collagen beads for 30 min, and bead-associated proteins were immunoprecipitated with FLAG antibody and then immunoblotted for NMIIB.
We prepared FLAG-tagged constructs of full-length gelsolin, the C-terminal half of gelsolin (G4–G6), and the N-terminal half of gelsolin (G1–G3) to assess the potential interactions with NMIIA in cells. Gelsolin null cells transfected with FLAG-tagged gelsolin constructs showed a slightly stronger interaction of NMIIA with the G4–G6 (the C-terminal half of gelsolin) than with G1–G3 (Fig. 4B). We probed the FLAG-tagged gelsolin pulldowns with antibody to NMIIB but found no immunoreactivity (Fig. 4C).
Gelsolin and NMIIA Are Required for Collagen-induced Ca2+ Flux
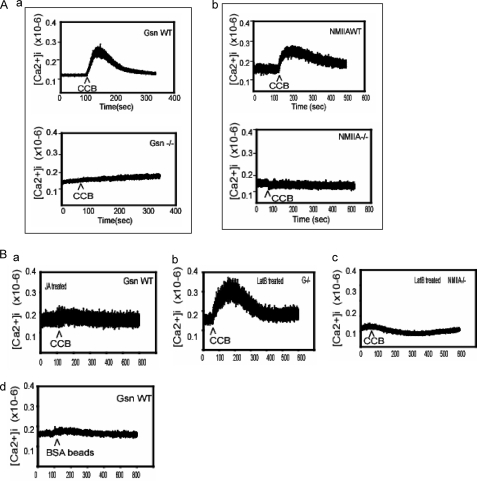
As gelsolin is a Ca2+-activated protein that enhances actin filament severing and promotes actin assembly (31), we examined Ca2+ flux in response to collagen beads. In WT cells, collagen-coated beads caused marked increases in [Ca2+]i, but there were only small increases in [Ca2+]i in gelsolin-deficient cells (Fig. 5A, a). These data are consistent with the idea that in gelsolin null cells the more densely polymerized cortical actin filament network inhibits the activation of Ca2+-permeable, integrin-gated plasma membrane channels (13). Experiments of a similar design with NMIIA-deficient ES cells or NMIIA WT ES cells also showed marked differences in Ca2+ fluxes in response to collagen bead binding (Fig. 5A, b). The means of the maximum [Ca2+]i increases above basal levels induced by collagen beads were: gelsolin null cells, 10 ± 5 nm; gelsolin WT cells, 130 ± 12 nm; NMIIA−/− cells, 12 ± 7 nm; NMIIA WT cells, 125 ± 27 nm (n = 5 cells/group).
FIGURE 5.
A, fura2/AM-loaded gelsolin (Gsn) WT and gelsolin null cells (a) and NMIIA WT and NMIIA null cells (b) showing Ca2+ fluxes in response to collagen-coated bead (CCB) binding measured by ratio fluorimetry. B, a and b, Ca2+ fluxes in response to collagen bead binding in gelsolin WT and null cells treated with JA (500 nm) and latrunculin B (LatB, 1 μm), respectively. c, intracellular calcium ion responses to collagen-coated beads in latrunculin B (1 μm)-treated NMIIA−/− cells. d, calcium responses were also measured in gelsolin WT cells after incubation with BSA-coated beads as controls. Data are means ± S.E. of bound collagen beads per cell (n = 100 cells/group).
Treatment of gelsolin WT cells with JA (1 μm, 15 min), an agent that stabilizes actin filaments and promotes actin assembly (52), inhibited Ca2+ flux in response to collagen binding, whereas gelsolin null cells treated with lantrunculin B (1 μm, 10 min) showed sharp increases in [Ca2+]i in response to collagen bead binding (Fig. 5B, a and b); but NMIIA−/− cells treated with lantrunculin B (1 μm, 10 min) showed no increase of [Ca2+]i in response to collagen bead binding (Fig. 5B, c). There was no calcium flux in response to BSA-coated beads in gelsolin WT cells (Fig. 5B, d). Notably, when gelsolin WT cells were treated with JA and incubated with or without ionomycin, there were more beads bound in the presence of ionomycin compared with samples without ionomycin treatment (supplemental Fig. 1I).
The mobility of β1 integrins in the plane of the plasma membrane is an important determinant of collagen phagocytosis because of the requirement for increased β1 integrin clustering for collagen binding (13). We considered that β1 integrin mobility could be impacted by NMIIA at collagen bead binding sites. Accordingly, we measured β1 integrin mobility and diffusion coefficients by fluorescence recovery after photobleaching. With this method we found no differences between NMIIA-deficient and NMIIA WT cells (percent mobile fraction: 38% in WT cells, 36% in NMIIA-deficient cells; diffusion coefficient (D): 2.52 × 10−10 cm2/s in WT cells, 2.36 × 10−10 cm2/s in NMIIA deficient-cells). Therefore NMIIA is unlikely to affect collagen bead binding-induced actin assembly because of its influence on β1 integrin mobility. Notably, we had shown earlier that the actin cytoskeletal elements that regulate β1 integrin mobility in the plane of the plasma membrane are functional in the absence of gelsolin (13).
Collagen Bead Binding Induces Phosphorylation of MLC
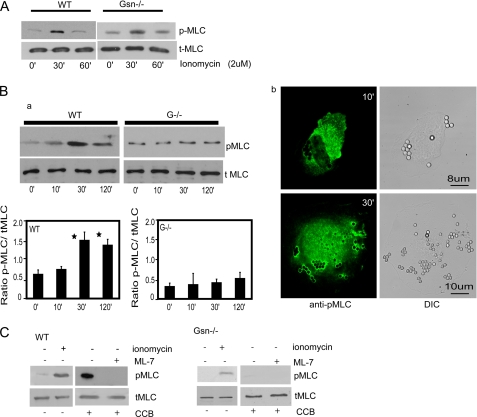
Increased [Ca2+]i is important for myosin light chain phosphorylation, which in turn is required for NMIIA filament formation in vitro (57). In view of our immunoprecipitation data showing an association between NMIIA and gelsolin (Fig. 4A), we examined collagen bead-induced myosin light chain phosphorylation. As expected, when WT or gelsolin null cells were treated with the calcium ionophore ionomycin (2 μm), there was prominent phosphorylation of the myosin light chain independently of gelsolin expression (Fig. 6A). Collagen bead incubations also induced marked, time-dependent phosphorylation of myosin light chain in WT cells (maximal time, 30 min), but phosphorylation was greatly attenuated in gelsolin null cells (Fig. 6B, a). WT cells that were immunostained with antibody to phospho-MLC showed prominent staining in the vicinity of bound collagen beads (Fig. 6B, b). When incubated with the myosin light chain kinase inhibitor ML-7, WT or gelsolin null cells showed a complete absence of myosin light chain phosphorylation after incubation with collagen beads (Fig. 6C). Taken together, these data suggest that the increased [Ca2+]i induced by collagen bead binding was required for phosphorylation of the myosin light chain, which may affect NMIIA filament formation (57).
FIGURE 6.
A, cells treated with ionomycin exhibit myosin light chain phosphorylation in WT and gelsolin null cells. B, a, bead-associated proteins collected from cells treated with collagen-coated beads show maximal phosphorylation of MLC at 30 min in WT cells but not in gelsolin-deficient cells. The histogram below shows ratios of quantification of blot densities as indicated. Data are mean ± S.E. of ratios. b, immunostaining shows localization of phospho-MLC (pMLC) at the bead sites after 10 and 30 min incubation with collagen-coated beads. C, in the presence of the MLC kinase inhibitor ML-7, gelsolin WT or null cells incubated with collagen beads show a complete block of MLC phosphorylation. The experiment was repeated three times. DIC, differential interference contrast; CCB, collagen-coated beads; tMLC, total myosin light chain.
Gelsolin Regulates Collagen-induced NMIIA Filament Assembly
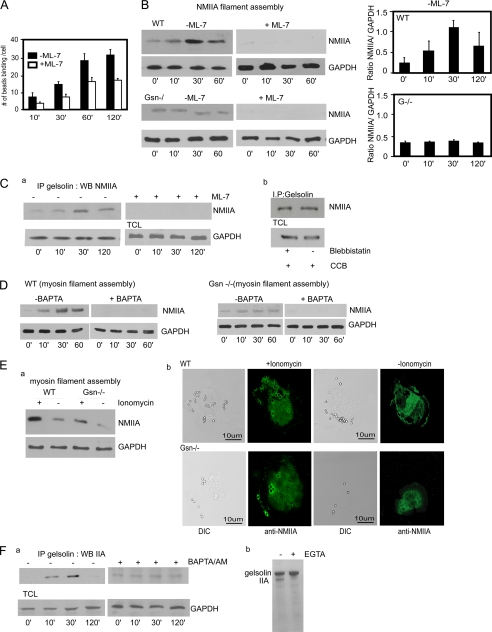
Because our data showed that [Ca2+]i-induced phosphorylation of the myosin light chain may mediate NMIIA filament assembly at the collagen bead binding site, we sought to determine whether gelsolin would be required for myosin filament assembly in response to collagen beads. We first examined whether inhibition of myosin light chain phosphorylation affects collagen bead binding. Treatment with ML-7 reduced collagen bead binding in WT cells (to 50% of controls, p < 0.05; Fig. 7A). In the absence of collagen beads, there were low levels of Triton X-100-insoluble (i.e. pelletable) NMIIA filaments. NMIIA filament assembly was increased after collagen bead incubation of gelsolin WT cells, but this increase was not detectable in gelsolin null cells (Fig. 7B). Even in the absence of collagen bead binding, we observed low levels of NMIIA filament accumulation, indicating the presence of sparse NMIIA filaments under unstimulated conditions. In response to collagen binding, ML-7 completely inhibited myosin filament assembly (Fig. 7B).
FIGURE 7.
A, gelsolin WT cells treated with ML-7 (25 μm) show reduced collagen binding (*, p < 0. 01 after 10 min). Data are mean ± S.E. numbers of collagen beads bound to cells (n = 75 cells/group). B, ML-7 treated or untreated gelsolin WT and null cells were incubated with collagen beads and probed with NMIIA antibody. Histograms are the ratios of quantification of blot densities as indicated. Data are mean ± S.E. of ratios. C, gelsolin WT cells treated with or without ML-7 (a) and with or without blebbistatin (b). Gelsolin immunoprecipitates (IP) from collagen-treated samples were probed with NMIIA antibody. CCB, collagen-coated beads; TCL, total cell lysates; WB, Western blot. D, cells loaded with the Ca2+ chelator BAPTA/AM and incubated with collagen beads inhibit myosin filament assembly in Triton-insoluble fractionation assay. E, a, ionomycin treatment of gelsolin null cells overcomes the block of myosin filament assembly in Triton-insoluble fractionation assay. b, ionomycin-treated gelsolin null cells show targeting of NMIIA to bead sites in collagen binding assays. DIC, differential interference contrast. F, a, in the presence or absence of BAPTA/AM, gelsolin immunoprecipitates after incubation with beads collagen were probed with NMIIA antibody. b, GST-Sepharose beads with bound gelsolin and NMIIA (residues 1338–1960) at a 2:1 molar ratio were incubated for 1 h in Ca2+- or EGTA-containing buffer followed by extensive washing. Bead-associated proteins were eluted and separated by SDS-PAGE. Gelsolin associated with NMIIA in the presence of Ca2+ but not in the presence of EGTA.
We examined whether the association of gelsolin and NMIIA required phosphorylation of the myosin light chain. Cells were treated with ML-7, and gelsolin immunoprecipitates were immunoblotted for NMIIA. After inhibition of myosin light chain kinase activity, there was no detectable association between gelsolin and NMIIA (Fig. 7C, a). When cells were treated with blebbistatin, an inhibitor of NMIIA motor activity (58), there was no loss of the association between gelsolin and NMIIA, suggesting that NMIIA motor activity was not required for the association between gelsolin and NMIIA (Fig. 7C, b).
As shown above and consistent with earlier data (59, 60), elevated [Ca2+]i mediates phosphorylation of the myosin light chain (61). We evaluated the Ca2+ dependence of myosin IIA assembly by treating cells with BAPTA/AM to dampen the increases of [Ca2+]i that were induced by collagen bead binding. BAPTA/AM-treated cells, which were incubated with collagen beads, showed minimal formation of NMIIA filaments, suggesting that increased [Ca2+]i is required for filament assembly (Fig. 7D). As a control, we treated gelsolin null cells with ionomycin and observed an increase of NMIIA filaments associated with collagen beads (Fig. 7E, a). Further, there was increased localization of NMIIA around collagen bead sites in ionomycin-treated gelsolin null cells (Fig. 7E, b), suggesting that Ca2+ is an important determinant of NMIIA assembly at collagen bead sites.
We examined whether the association of gelsolin with NMIIA required increased [Ca2+]i. Cells were treated with BAPTA/AM and gelsolin immunoprecipitates were immunoblotted for NMIIA. In the presence of BAPTA/AM there was no collagen-induced increase in the association between gelsolin and NMIIA (Fig. 7F, a). Collectively these data suggest that gelsolin associates with NMIIA more avidly in the presence of Ca2+ and that the interaction of gelsolin with NMIIA may be an important determinant of collagen bead binding. As gelsolin and NMIIA are both actin-binding proteins, we considered that the observed association between these proteins was mediated by actin. To determine whether the association between gelsolin and NMIIA is specific and direct, GST-Sepharose beads with bound gelsolin were incubated with NMIIA (residues 1338–1960). The proteins were incubated for 1 h followed by extensive washing in buffer. We found that gelsolin associated with NMIIA in the presence of Ca2+ but not in the presence of EGTA (Fig. 7F, b). Notably, the binding of gelsolin to NMIIA impacted the formation of NMIIA filaments as assessed by turbidimetry, whereas the gelsolin family protein flightless I (used here as a control) had no effect on NMIIA filament formation (supplemental Fig. 2).
DISCUSSION
Integrin-dependent collagen phagocytosis is a critical but largely unexplored pathway for maintaining the normal structure and function of soft connective tissues (e.g. uterus and normally functioning periodontium and in wound healing) (5). The process of collagen phagocytosis requires actin accumulation at nascent adhesions (7). We had shown previously that gelsolin (13) and non-muscle myosin IIA (12) are required for adhesion to collagen, but their potential functional interactions in regulating the early steps of phagocytosis were not defined. Here, using fibroblasts isolated from gelsolin null or WT mice and treated with or without NMIIA siRNA and NMIIA WT and null ES cells, as well as collagen-coated bead binding assays, we examined how Ca2+, along with gelsolin and NMIIA, interacts functionally at adhesion sites to regulate collagen phagocytosis. Our main finding was that gelsolin and NMIIA are temporally and spatially associated with the assembly of actin filaments at collagen binding sites. Our data indicate that NMIIA filaments tether gelsolin at collagen bead sites to promote actin remodeling at the nascent phagosome. At these sites, localized Ca2+ transients, which are induced by collagen binding and enhanced by gelsolin expression, promote myosin light chain phosphorylation. This process in turn enables NMIIA filament formation (Fig. 8) and enhanced gelsolin association. Collectively, the association of gelsolin with NMIIA filaments provides a feed-forward pathway that regulates the adhesion step in collagen phagocytosis. Notably, in specialized phagocytes such as granulocytes, gelsolin plays an important role in FcγR-mediated phagocytosis but not in complement receptor-mediated phagocytosis (17), whereas NMIIA is involved in complement receptor- but not FcγR-mediated phagocytosis (36). In contrast, the much slower process of β1 integrin-dependent phagocytosis of collagen fibrils by fibroblasts evidently requires both gelsolin and NMIIA.
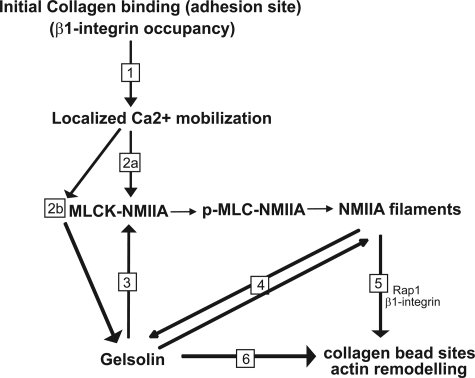
FIGURE 8.
Schematic diagram indicating the functional dependence of collagen phagocytosis on NMIIA interactions with gelsolin. 1, initial binding of collagen to β1 integrin at collagen bead adhesion sites. 2a, localized Ca2+ flux enhances phosphorylation of myosin light chain at serine 19 and increases NMIIA filament assembly. 2b, Ca2+ flux induces activation of gelsolin and MLCK, myosin light chain kinase. 3, gelsolin is required for MLC phosphorylation because the absence of [Ca2+]i in gelsolin null cells prevents the phosphorylation of NMIIA MLC required for myosin IIA filament assembly. 4, NMIIA filaments interact with gelsolin to provide anchorage for gelsolin at collagen bead binding sites. 5, NMIIA filaments provide Rap1 localization at bead sites and enable β1 integrin activation (12). 6, spatial localization of gelsolin by NMIIA filaments provides remodeling of actin filaments around bead binding sites, which is required for phagosome formation.
Cell substratum adhesion is regulated in part by the affinity of integrins for extracellular ligands (62), by integrin clustering, and by cytoskeletal reorganization (63, 64). Our data show that stabilization of actin filaments by jasplankinolide inhibited collagen bead binding and presumably integrin activation, as has been shown for other integrins (65). Conversely, and in agreement with this notion, actin depolymerization by latrunculin B promotes enhanced collagen bead binding (29). We found that compared with gelsolin null cells, WT cells exhibited enhanced actin filament accumulation and actin monomer incorporation into filaments at collagen binding sites. Taken together with previous data showing that gelsolin is a high abundance actin-severing protein (66, 67), the importance of gelsolin in collagen binding is likely dependent on its role in turnover of actin filaments. Consistent with this notion, we found that the ability of WT cells to remodel actin filaments was accompanied by much higher collagen bead binding than gelsolin null cells, indicating that actin remodeling at adhesion sites is important for the binding step of collagen phagocytosis. Similarly, supervillin, a peripheral membrane protein of the gelsolin family that binds actin filaments and myosin II, reorganizes the actin cytoskeleton to promote the formation of invadopodia for migration of COS-7 cells (68). Evidently, the ability to remodel actin filaments at adhesion sites is a critical function of migratory and phagocytic cells. It would seem that cells that more avidly phagocytose collagen (with gelsolin expression) are less likely to remodel collagen by traction.
Previous in vitro studies have demonstrated that in addition to its well described role in contraction, myosin II may be involved directly in actin depolymerization (32) and actin network treadmilling in cell migration (69). Our analyses of collagen bead binding and gelsolin/NMIIA co-localization at collagen binding sites indicate that these proteins may also be involved in the remodeling of actin filaments that occurs at nascent collagen adhesions. We found that at collagen bead binding sites, NMIIA is detected prior to the recruitment of gelsolin and actin assembly. Notably, we found a reduced but measureable amount of collagen bead binding in the absence of gelsolin. In this context, actin filaments can be remodeled by myosin family proteins (70, 71) and by other actin-binding proteins such as cofilin. However, the anchorage of gelsolin at collagen binding sites that is provided by NMIIA greatly increases collagen phagocytosis and enables site-specific actin remodeling (72–74). Conceivably, the actin depolymerizing activity of NMIIA cells (32) may either arise in part from gelsolin-mediated actin severing or be enhanced by gelsolin activity.
Collagen phagocytosis by fibroblasts is a relatively much slower process than the internalization of microbes by specialized phagocytes such as macrophages. Earlier we showed the requirement for gelsolin severing activity in the initial binding step of collagen phagocytosis (13) and the involvement of phosphoinositides in regulating gelsolin capping activity at the later internalization stage (11). Here we considered how gelsolin is sequestered at the collagen bead binding site. Initial screening experiments with tandem mass spectrometry pointed to NMIIA as a potential tethering molecule, which was supported by immunoprecipitation experiments demonstrating a Ca2+-dependent association between gelsolin and NMIIA. These data underline a new role for interactions between gelsolin and NMIIA that may be important for sequestering the actin severing activity of gelsolin to the nascent phagosome.
Previous data showed that the severing activity of gelsolin (22) and phosphorylation of NMIIA are controlled by Ca2+ (60). In this context, the Ca2+ regulation of gelsolin severing activity has been thought to be restricted to the C-terminal half of the molecule (24, 75). However, more recent crystallographic studies of the isolated C-terminal half of gelsolin indicate that the Ca2+ binding site on G4 is not required for actin binding (76). Conceivably, the increased [Ca2+]i that is induced by initial collagen binding may enhance the interaction of NMIIA with gelsolin, thereby localizing gelsolin severing activity to the adhesion site, which is needed for the actin filament assembly observed in macrophage (72) and fibroblast (13) phagosome formation.
Although NMIIA is required for gelsolin recruitment to collagen adhesions, gelsolin in turn is needed for NMIIA recruitment to collagen adhesions and for the assembly of myosin filaments. The importance of these functional interactions comes from our observation that gelsolin null cells transfected with NMIIA siRNA exhibited no further reductions of collagen binding compared with cells treated with control siRNA. These data suggest that gelsolin and NMIIA may act through a common pathway at bead sites to mediate collagen binding.
The immunoprecipitation data presented here showing that gelsolin associates with NMIIA does not distinguish between direct binding of these two proteins or an association that is mediated by other proteins such as actin filaments, which could bridge between gelsolin and NMIIA, possibly in a Ca2+-dependent manner. We have presented data here using purified NMIIA and gelsolin showing that the two proteins do indeed interact in a Ca2+-dependent manner. Further and more detailed in vitro studies using purified proteins are now under way to characterize the kinetics, affinity, and regulation of the binding of gelsolin to NMIIA.
During smooth muscle contraction, increased [Ca2+]i activates myosin light chain kinase and Ca2+/calmodulin-dependent protein kinase II, which in turn phosphorylates myosin light chain kinase (77). Phosphorylation of the myosin light chain on Ser-19 converts the folded myosin molecules into an extended, assembly-competent form (78, 79). In the context of collagen binding, our data suggest that gelsolin is required for enhanced NMIIA assembly into filaments. The Ca2+ influx generated by collagen bead binding in gelsolin WT cells enhances phosphorylation of the myosin light chain and myosin filament assembly, which evidently is a requirement for the association of gelsolin with NMIIA. We conclude, therefore, that NMIIA is required for gelsolin recruitment to collagen binding sites and that the mutual functional interdependence of gelsolin and NMIIA enables actin remodeling, which is a critical process in the binding step of collagen phagocytosis.
Supplementary Material
This work was supported by Canadian Institutes of Health Research Operating Grant MOP 36332 (to C. A. M.) and by Ontario Heart and Stroke Operating Grant T6022 (to C. A. M.). C. A. M. is also supported by a Canada Research Chair (Tier 1) in Matrix Dynamics.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1 and 2.
- NMIIA
- non-muscle myosin IIA
- NM
- non-muscle
- NMHC
- non-muscle myosin heavy chain
- Gsn
- gelsolin
- JA
- jasplakinolide
- MLC
- myosin light chain
- ES
- embryonic stem
- TIRF
- total internal reflection fluorescence
- TRITC
- tetramethylrhodamine isothiocyanate
- RFP
- red fluorescent protein.
REFERENCES
- 1. Pérez-Tamayo R. (1978) Am. J. Pathol. 92, 508–566 [PMC free article] [PubMed] [Google Scholar]
- 2. Murphy G., Nagase H. (2008) Mol. Aspects Med. 29, 290–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Caron E., Hall A. (1998) Science 282, 1717–1721 [DOI] [PubMed] [Google Scholar]
- 4. Krause K. H. (2000) Schweiz. Med. Wochenschr. 130, 97–100 [PubMed] [Google Scholar]
- 5. Everts V., van der Zee E., Creemers L., Beertsen W. (1996) Histochem J. 28, 229–245 [DOI] [PubMed] [Google Scholar]
- 6. McGaw W. T., Ten Cate A. R. (1983) J. Invest. Dermatol. 81, 375–378 [DOI] [PubMed] [Google Scholar]
- 7. Arora P. D., Manolson M. F., Downey G. P., Sodek J., McCulloch C. A. (2000) J. Biol. Chem. 275, 35432–35441 [DOI] [PubMed] [Google Scholar]
- 8. Lee H., Overall C. M., McCulloch C. A., Sodek J. (2006) Mol. Biol. Cell 17, 4812–4826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Melcher A. H., Chan J. (1981) J. Ultrastruct. Res. 77, 1–36 [DOI] [PubMed] [Google Scholar]
- 10. ten Cate A. R. (1972) J. Anat. 112, 401–414 [PMC free article] [PubMed] [Google Scholar]
- 11. Arora P. D., Chan M. W., Anderson R. A., Janmey P. A., McCulloch C. A. (2005) Mol. Biol. Cell 16, 5175–5190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Arora P. D., Conti M. A., Ravid S., Sacks D. B., Kapus A., Adelstein R. S., Bresnick A. R., McCulloch C. A. (2008) Mol. Biol. Cell 19, 5032–5046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arora P. D., Glogauer M., Kapus A., Kwiatkowski D. J., McCulloch C. A. (2004) Mol. Biol. Cell 15, 588–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Knowles G. C., McKeown M., Sodek J., McCulloch C. A. (1991) J. Cell Sci. 98, 551–558 [DOI] [PubMed] [Google Scholar]
- 15. Tjelle T. E., Lovdal T., Berg T. (2000) Bioessays 22, 255–263 [DOI] [PubMed] [Google Scholar]
- 16. Weeds A., Maciver S. (1993) Curr. Opin. Cell Biol. 5, 63–69 [DOI] [PubMed] [Google Scholar]
- 17. Serrander L., Skarman P., Rasmussen B., Witke W., Lew D. P., Krause K. H., Stendahl O., Nüsse O. (2000) J. Immunol. 165, 2451–2457 [DOI] [PubMed] [Google Scholar]
- 18. Yin H. L., Albrecht J. H., Fattoum A. (1981) J. Cell Biol. 91, 901–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yin H. L., Stossel T. P. (1979) Nature 281, 583–586 [DOI] [PubMed] [Google Scholar]
- 20. Kwiatkowski D. J., Janmey P. A., Yin H. L. (1989) J. Cell Biol. 108, 1717–1726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Burtnick L. D., Koepf E. K., Grimes J., Jones E. Y., Stuart D. I., McLaughlin P. J., Robinson R. C. (1997) Cell 90, 661–670 [DOI] [PubMed] [Google Scholar]
- 22. Kiselar J. G., Janmey P. A., Almo S. C., Chance M. R. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3942–3947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Burtnick L. D., Urosev D., Irobi E., Narayan K., Robinson R. C. (2004) EMBO J. 23, 2713–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Way M., Gooch J., Pope B., Weeds A. G. (1989) J. Cell Biol. 109, 593–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pope B., Way M., Weeds A. G. (1991) FEBS Lett. 280, 70–74 [DOI] [PubMed] [Google Scholar]
- 26. Selden L. A., Kinosian H. J., Newman J., Lincoln B., Hurwitz C., Gershman L. C., Estes J. E. (1998) Biophys. J. 75, 3092–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Janmey P. A., Stossel T. P. (1987) Nature 325, 362–364 [DOI] [PubMed] [Google Scholar]
- 28. Nyakern-Meazza M., Narayan K., Schutt C. E., Lindberg U. (2002) J. Biol. Chem. 277, 28774–28779 [DOI] [PubMed] [Google Scholar]
- 29. Segal G., Lee W., Arora P. D., McKee M., Downey G., McCulloch C. A. (2001) J. Cell Sci. 114, 119–129 [DOI] [PubMed] [Google Scholar]
- 30. Nishizaka T., Shi Q., Sheetz M. P. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 692–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Janmey P. A. (1994) Annu. Rev. Physiol. 56, 169–191 [DOI] [PubMed] [Google Scholar]
- 32. Haviv L., Gillo D., Backouche F., Bernheim-Groswasser A. (2008) J. Mol. Biol. 375, 325–330 [DOI] [PubMed] [Google Scholar]
- 33. Groves E., Dart A. E., Covarelli V., Caron E. (2008) Cell. Mol. Life Sci. 65, 1957–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Conti M. A., Adelstein R. S. (2008) J. Cell Sci. 121, 11–18 [DOI] [PubMed] [Google Scholar]
- 35. Scholey J. M., Taylor K. A., Kendrick-Jones J. (1980) Nature 287, 233–235 [DOI] [PubMed] [Google Scholar]
- 36. Olazabal I. M., Caron E., May R. C., Schilling K., Knecht D. A., Machesky L. M. (2002) Curr. Biol. 12, 1413–1418 [DOI] [PubMed] [Google Scholar]
- 37. Isik N., Brzostowski J. A., Jin T. (2008) Dev. Cell 15, 590–602 [DOI] [PubMed] [Google Scholar]
- 38. Humphrey D., Duggan C., Saha D., Smith D., Käs J. (2002) Nature 416, 413–416 [DOI] [PubMed] [Google Scholar]
- 39. Bos J. L., de Bruyn K., Enserink J., Kuiperij B., Rangarajan S., Rehmann H., Riedl J., de Rooij J., van Mansfeld F., Zwartkruis F. (2003) Biochem. Soc. Trans. 31, 83–86 [DOI] [PubMed] [Google Scholar]
- 40. Witke W., Sharpe A. H., Hartwig J. H., Azuma T., Stossel T. P., Kwiatkowski D. J. (1995) Cell 81, 41–51 [DOI] [PubMed] [Google Scholar]
- 41. Conti M. A., Even-Ram S., Liu C., Yamada K. M., Adelstein R. S. (2004) J. Biol. Chem. 279, 41263–41266 [DOI] [PubMed] [Google Scholar]
- 42. Even-Ram S., Doyle A. D., Conti M. A., Matsumoto K., Adelstein R. S., Yamada K. M. (2007) Nat. Cell Biol. 9, 299–309 [DOI] [PubMed] [Google Scholar]
- 43. Arora P. D., Fan L., Sodek J., Kapus A., McCulloch C. A. (2003) Exp. Cell Res. 286, 366–380 [DOI] [PubMed] [Google Scholar]
- 44. Dulyaninova N. G., House R. P., Betapudi V., Bresnick A. R. (2007) Mol. Biol. Cell 18, 3144–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kwiatkowski D. J., Stossel T. P., Orkin S. H., Mole J. E., Colten H. R., Yin H. L. (1986) Nature 323, 455–458 [DOI] [PubMed] [Google Scholar]
- 46. Riedl J., Crevenna A. H., Kessenbrock K., Yu J. H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T. A., Werb Z., Sixt M., Wedlich-Soldner R. (2008) Nat. Methods 5, 605–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Huang Y., Arora P., McCulloch C. A., Vogel W. F. (2009) J. Cell Sci. 122, 1637–1646 [DOI] [PubMed] [Google Scholar]
- 48. Bolte S., Cordelières F. P. (2006) J. Microsc. 224, 213–232 [DOI] [PubMed] [Google Scholar]
- 49. Axelrod D., Koppel D. E., Schlessinger J., Elson E., Webb W. W. (1976) Biophys. J. 16, 1055–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Jacobson K., Wojcieszyn J. (1984) Proc. Natl. Acad. Sci. U.S.A. 81, 6747–6751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Arora P. D., Bibby K. J., McCulloch C. A. (1994) J. Cell. Physiol. 161, 187–200 [DOI] [PubMed] [Google Scholar]
- 52. Bubb M. R., Spector I., Beyer B. B., Fosen K. M. (2000) J. Biol. Chem. 275, 5163–5170 [DOI] [PubMed] [Google Scholar]
- 53. Hartwig J. H. (1992) J. Cell Biol. 118, 1421–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hartwig J. H., Bokoch G. M., Carpenter C. L., Janmey P. A., Taylor L. A., Toker A., Stossel T. P. (1995) Cell 82, 643–653 [DOI] [PubMed] [Google Scholar]
- 55. Azuma T., Witke W., Stossel T. P., Hartwig J. H., Kwiatkowski D. J. (1998) EMBO J. 17, 1362–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nayal A., Webb D. J., Horwitz A. F. (2004) Curr. Opin. Cell Biol. 16, 94–98 [DOI] [PubMed] [Google Scholar]
- 57. Ip K., Sobieszek A., Solomon D., Jiao Y., Paré P. D., Seow C. Y. (2007) Cell. Physiol. Biochem. 20, 649–658 [DOI] [PubMed] [Google Scholar]
- 58. Straight A. F., Cheung A., Limouze J., Chen I., Westwood N. J., Sellers J. R., Mitchison T. J. (2003) Science 299, 1743–1747 [DOI] [PubMed] [Google Scholar]
- 59. Yumura S., Yoshida M., Betapudi V., Licate L. S., Iwadate Y., Nagasaki A., Uyeda T. Q., Egelhoff T. T. (2005) Mol. Biol. Cell 16, 4256–4266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ferrari M. B., Podugu S., Eskew J. D. (2006) Cell Biochem. Biophys. 45, 317–337 [DOI] [PubMed] [Google Scholar]
- 61. Watanabe T., Hosoya H., Yonemura S. (2007) Mol. Biol. Cell 18, 605–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Calderwood D. A. (2004) J. Cell Sci. 117, 657–666 [DOI] [PubMed] [Google Scholar]
- 63. Humphries M. J., McEwan P. A., Barton S. J., Buckley P. A., Bella J., Mould A. P. (2003) Trends Biochem. Sci. 28, 313–320 [DOI] [PubMed] [Google Scholar]
- 64. Hynes R. O. (2002) Cell 110, 673–687 [DOI] [PubMed] [Google Scholar]
- 65. Wang J., Chen H., Brown E. J. (2001) J. Biol. Chem. 276, 14474–14481 [DOI] [PubMed] [Google Scholar]
- 66. Lind S. E., Smith D. B., Janmey P. A., Stossel T. P. (1988) Am. Rev. Respir. Dis. 138, 429–434 [DOI] [PubMed] [Google Scholar]
- 67. Janmey P. A., Chaponnier C., Lind S. E., Zaner K. S., Stossel T. P., Yin H. L. (1985) Biochemistry 24, 3714–3723 [DOI] [PubMed] [Google Scholar]
- 68. Crowley J. L., Smith T. C., Fang Z., Takizawa N., Luna E. J. (2009) Mol. Biol. Cell 20, 948–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wilson C. A., Tsuchida M. A., Allen G. M., Barnhart E. L., Applegate K. T., Yam P. T., Ji L., Keren K., Danuser G., Theriot J. A. (2010) Nature 465, 373–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Pelham R. J., Chang F. (2002) Nature 419, 82–86 [DOI] [PubMed] [Google Scholar]
- 71. Wu J. Q., Pollard T. D. (2005) Science 310, 310–314 [DOI] [PubMed] [Google Scholar]
- 72. Defacque H., Bos E., Garvalov B., Barret C., Roy C., Mangeat P., Shin H. W., Rybin V., Griffiths G. (2002) Mol. Biol. Cell 13, 1190–1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Defacque H., Egeberg M., Antzberger A., Ansorge W., Way M., Griffiths G. (2000) Cytometry 41, 46–54 [PubMed] [Google Scholar]
- 74. Defacque H., Egeberg M., Habermann A., Diakonova M., Roy C., Mangeat P., Voelter W., Marriott G., Pfannstiel J., Faulstich H., Griffiths G. (2000) EMBO J. 19, 199–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chaponnier C., Janmey P. A., Yin H. L. (1986) J. Cell Biol. 103, 1473–1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Narayan K., Chumnarnsilpa S., Choe H., Irobi E., Urosev D., Lindberg U., Schutt C. E., Burtnick L. D., Robinson R. C. (2003) FEBS Lett. 552, 82–85 [DOI] [PubMed] [Google Scholar]
- 77. Tansey M. G., Luby-Phelps K., Kamm K. E., Stull J. T. (1994) J. Biol. Chem. 269, 9912–9920 [PubMed] [Google Scholar]
- 78. Craig R., Smith R., Kendrick-Jones J. (1983) Nature 302, 436–439 [DOI] [PubMed] [Google Scholar]
- 79. Smith R. C., Cande W. Z., Craig R., Tooth P. J., Scholey J. M., Kendrick-Jones J. (1983) Philos. Trans. R. Soc. Lond. B Biol. Sci. 302, 73–82 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.