Background: RNA processing is altered when ELL2, a transcription elongation factor, is induced during differentiation to plasma cells.
Results: Knocking down ELL2 by siRNA reduces IgH secretory mRNA and histone H3K4 and H3K79 methylations, marks of open chromatin.
Conclusion: Opening downstream chromatin favors proximal poly(A) sites and exon skipping, thereby modifying gene expression.
Significance: Differentiation often favors downstream poly(A) sites, but ELL2 reverses this.
Keywords: Chromatin Histone Modification, Gene Transcription, Polyadenylation, RNA Processing, Transcription Elongation Factors, Immunoglobulin
Abstract
In plasma cells, immunoglobulin heavy chain (IgH) secretory-specific mRNA is made in high abundance as a result of both increased promoter proximal poly(A) site choice and weak splice-site skipping. Ell2, the eleven-nineteen lysine rich leukemia gene, is a transcription elongation factor that is induced ∼6-fold in plasma cells and has been shown to drive secretory-specific mRNA production. Reducing ELL2 by siRNA, which reduced processing to the secretion-specific poly(A) site, also influenced the methylations of histone H3K4 and H3K79 on the IgH gene and impacted positive transcription factor b (pTEFb), Ser-2 carboxyl-terminal phosphorylation, and polyadenylation factor additions to RNA polymerase II. The multiple lineage leukemia gene (MLL) and Dot1L associations with the IgH gene were also impaired in the absence of ELL2. To investigate the link between histone modifications, transcription elongation, and alternative RNA processing in IgH mRNA production, we performed chromatin immunoprecipitation on cultured mouse B and plasma cells bearing the identical IgH γ2a gene. In the plasma cells, as compared with the B cells, the H3K4 and H3K79 methylations extended farther downstream, past the IgH enhancer to the end of the transcribed region. Thus the downstream H3K4 and H3K79 methylation of the IgH associated chromatin in plasma cells is associated with increased polyadenylation and exon skipping, resulting from the actions of ELL2 transcription elongation factor.
Introduction
Alternative processing of the immunoglobulin heavy chain (IgH) mRNA regulates antibody production, an important component of the adaptive immune response. When B cells differentiate into plasma cells with their high levels of IRF-4, blimp-1, and ELL2 2 (eleven-nineteen lysine-rich leukemia gene 2), both the IgH mRNA is extensively polyadenylated at the secretory-specific site (sec-mRNA) and the splice between CH3 and M1 is skipped, precluding use of the downstream membrane exons (1). In addition, IgH sec-mRNA abundance is increased at least 10-fold, leading to large amounts of mature mRNA and subsequently secreted Ig protein (2). Transcriptional elongation is an important regulator for RNA splicing (3, 4). Changes in mRNA splicing can occur as a consequence of changes in histone H3 methylations (5, 6), which are themselves linked to transcription elongation. How these impact the alternative splicing and polyadenylation of IgH mRNA processing is an important question.
Ell2 closely resembles Ell1, a transcription elongation factor and fusion partner with MLL, the multiple lineage leukemia gene (7). ELL2, is induced 6-fold in plasma cell differentiation and has been shown to have a central role in directing efficient alternative mRNA processing, influencing both proximal poly(A) site choice and exon skipping, as well as IgH alternative processing (1). The molecular context in which ELL2 acts to influence RNA processing remains unclear, but some information is emerging. In 293 HEK cultured cells, ELL1, -2, and -3 were shown to purify together in two distinct entities, either in the super elongation complex (SEC) or in another complex containing KIAA0947 and NARG2 proteins (8). The SEC is composed of positive transcription factor b (pTEFb), a protein heterodimer of cdk9 and cyclin T, as well as many of the previously identified MLL fusion partners that can perturb gene expression (8). ELL and ELL-associated factor (EAF) have also been implicated in interactions with pTEFb and some of the same MLL fusion partner components of SEC in HIV-mediated Tat-TAR regulation of mRNA synthesis; ELL2 alone was implicated in one study (9), whereas ELL1 and -2 were described in another (10). The SEC or Tat link the transcription elongation elements to the polymerase-associated factor (paf1) and polyadenylation factors; thus their assembly may represent a crucial control point for gene regulation. In contrast, the KIAA0947-NARG2 components isolated with ELL and EAF in 293 cells are less well characterized. Both NARG2 and KIAA0947 were found as neuronal-associated transcripts and show decreased expression in more differentiated cells (11, 12).
The appearances of methylated forms of histone H3, modified at lysine 4, lysine 36, or lysine 79 (aka H3K4me2/3, H3K36me3, and H3K79me2,3) are signals of gene activation (13). These are linked to serine phosphorylation of the carboxyl-terminal domain (CTD) of RNA polymerase II (RNAP-II), reviewed in Ref. 14. How H3 methylations relate to changes in RNA processing of the nascent transcripts and their distribution on genes undergoing alternative polyadenylation is not understood. The methylation of histone H3K4 is brought about by yeast SET1 and Complex Proteins Associated with Set1 or the MLL protein in mammalian cells at sites including the Hoxa7 and Hoxa9 genes, hallmarks of multiple lineage leukemia, aka MLL (15). The H3K36me3 mark has been associated with CTD Ser-5 and Ser-2 phosphorylation and recruitment of splicing factors to exons (16); H3K36 methylations can modulate alternative splicing by recruiting polypyrimidine tract-binding protein to suboptimal exons (5). H3K79 methylations are brought about by Dot1L (13), where conversion of monomethyl into di- and trimethyl is correlated with the transition from low to high level gene transcription. The multisubunit Dot1 complex (Dot-com) includes MLL partners: Enl, Af9/Mllt3, Af17/Mllt6, and Af10/Mllt10 (17). Although ENL is also present in the SEC, two separate complexes, namely SEC and Dot-com, were found (17). In another study, ENL was shown to independently associate with Dot1L to methylate Lys-79 (18). In the MLL-AF4 fusion-induced leukemia, a complex of MLL, SEC components, and Dot1L was found (19). Thus the associations of ELL2, the SEC, Dot-com, and the polyadenylation factors that occur for these H3-lysine modifications are still unclear, but elucidating this sequence of events would aid in understanding the assembly of the transcription elongation complex in general and in elucidating IgH mRNA alternative processing in B cells versus plasma cells in particular.
To relate histone modifications in the IgH locus to ELL2 and mRNA processing changes, we used cultured mouse B and plasma cells bearing the identical rearranged IgH γ2a gene. We found that the quantity and distribution of Lys-4, Lys-36, and Lys-79 methylations on the IgH gene differed when comparing B cells and plasma cells. In plasma cells with their high ELL2 levels, histone H3K4 and H3K79 methylations extended past the enhancer all the way to the end of the gene; in contrast, in B cells, there was an abrupt transition to Lys-4 and Lys-79 methylation-poor histones in B cells after the enhancer. The expressions of many factors involved in the SEC and transcription elongation are increased in the plasma cells, but ELL2 is the most elevated. We found that reducing ELL2 by siRNA influences the H3K4 and H3K79 methylations but did not inhibit RNA polymerase binding or Ser-5 phosphorylation, early initiation events. Although siRNA inhibition of ELL2 significantly alters processing at the secretion-specific poly(A) site, it also influences pTEFb and polyadenylation factor additions to the RNAP-II, MLL, and Dot1L associations with the gene. Thus ELL2 is important for the histone modifications accompanying transition from membrane-specific to secretory IgH mRNA expression.
EXPERIMENTAL PROCEDURES
Chromatin Immunoprecipitations
The culturing and isolation of mouse splenic B cells plus and minus LPS and the A20 B cell line and plasma cell lines AxJ and J558L have been described (1). The AxJ line (2) was made as a hybridoma fusion of the A20 line and J558L, which lacks its own heavy chain gene but retains the plasma cell phenotype; the expressed IgH V genes in A20 and AxJ are identical (20). The chromatin immunoprecipitation (ChIP) experiments were conducted as described previously (1, 20) using the ChIP-ITTM express kit from Active Motif (Carlsbad, CA) revised for magnetic beads coated with protein G (catalogue number 53014). Reverse cross-linking of the protein from the DNA was performed using NaCl and RNase at 67 °C overnight. Proteinase K digestion was increased to 20 μg/reaction at 37 °C for 2 h. DNA was then isolated on QIAquick® PCR purification kit (Qiagen, Gaithersburg, MD).
Antibodies for ChIP
Antibodies were used at 1–3 μg per ∼3 × 105 cell equivalents of fixed and sheared chromatin in a 100-μl volume with the Active Motif protein G-coupled magnetic beads: anti-RNAP-II, N-20 sc 899X (Santa Cruz Biotechnology, Santa Cruz, CA); anti-cdk9, H-169 sc 8338X (Santa Cruz Biotechnology); anti-cyclin T1 H245 sc 10750 (Santa Cruz Biotechnology); anti-H3, catalog number 1791 (Abcam, Cambridge, UK); anti-H3K36 me3, ab9050-100 (Abcam); anti-H3K4 me1, 8895 (Abcam); anti-H3K4 me2, 07-030 (Upstate Biotech Millipore) and ab32356 (Abcam); anti-H3K4 me2,3, 6000-100 (Abcam); anti-H3K4 me3, 39160 (Active Motif) and 07-473 (Millipore); anti-H3K79 me1, 2886-100 (Abcam); anti-H3K79 me2 ab3594 (Abcam); anti-H3K79 me3, 2621-100 (Abcam); anti-MLL, A300-086A (Bethyl Laboratories, Montgomery, TX); anti-Dot1L, A300-953A (Bethyl Laboratories); and normal rabbit serum from unimmunized animals as a negative control. Anti-Ser-2 and Ser-5 phosphorylated CTD of RNAP-II (Covance) were described previously (1, 20); these are IgM antibodies and require a secondary anti-IgM for maximum immunoprecipitation.
Primers for qPCR
Primers for the qPCR on the chromatin immunoprecipitated DNA for IgM and IgG heavy chains used here have been described (1, 20). For the gastrin promoter, we used 5′-CGG CAG TCA CTG GGC TGT GG-3′ forward and 5′-AGA GGG GGA GAG ACG GGG CT-3′ reverse. Primers used in the quantification of the mRNAs in the B cells versus plasma cells (γ2a heavy chains) and the transfected MPC11 γ2b gene, used in Ref. 1, are shown in Table 1, as are all the probes used in quantitative RT-PCRs.
TABLE 1.
Primers for RT-qPCR
| mRNA | Primers for RT-qPCR forward (top) and reverse (5′ to 3′) |
|---|---|
| 7SK RNA | CGCTCCATGTGCGTCCCTCC |
| AGGGGAGCGCAGCTACTCGT | |
| Af9 | GCAGCAGACCACTCCTGCCAC |
| CCCTGCCATTCCCTCTTTGTGCT | |
| Aff1 aka af4 | AGCGCCCAGGTCCTCCTCAG |
| GCTGCTGGCACTTCTGGGGG | |
| Aff4 | TCCTGGTGTACTCCTTGGCATTGT |
| TCGCAACAGTCTAACTTTGGCCCT | |
| Brd4 | TATGCCTGGCCTTTCTACAAGCCT |
| ATCAGCACCAAATTCCTGGGCATC | |
| Cdk8 | GAACCAGGACAGCGCCCACG |
| TCCCATGCTGCTCTGGGGCT | |
| Cdk9 | AGAATAGCCAGCCCAACCGATACA |
| ACGAGTCCACATCTCTGCCATGAT | |
| Cyclin C1 and C2 | TGTGCAGGACATGGGCCAGGA |
| GCGATCATGAACGGAGGGTACAGC | |
| Cyclin T1 | CAACCAACAGCCTGCATTTGACCA |
| AGGTCACAGTGGCGTCAACATACT | |
| Dot1L | GTGCCTACAGCAGCCACGGG |
| TGGGAGTGCTCCTCGCCCTC | |
| Eaf1 | CCGGGCAAGTGCAAGGCCAT |
| TGCCAACCTGTAGTTCTCCTTCACA | |
| Eaf2 | AAACCTGCTTCTAATGATACTTCTTGTG |
| ACTGTGACTGGTGGAGGTGAACCT | |
| ELL1 | GGTGTGATGGTTGCAAGATGGCG |
| CTCACGCGCCCACACGACAA | |
| ELL2 | AGCGTCTTTGCTGGAACAC |
| CTGGCCAGGTTACAGTGAGA | |
| ENL | AACACCCGCAAGGCCAACCC |
| GAGGGAGCGGCTTCTGGGGA | |
| HEXIM | AAAGTTCGACGAGAAGCAGAGCCT |
| ATCCATGAGGAACTGCGTGGTGTT | |
| Hprt | GAGACAGCCGCATCTTCTTGT |
| CCACAGGACTAGAACACCTGCTAA | |
| Irf4 | AGGATTGTTCCAGAGGGAGCCAAA |
| ATGGGATTTCTGGGTGTGACTGGT | |
| IgG2b sec | AAAAATTACTACCTGAAGAAGACC |
| TTTTTTTTTTTTTTTTTTTTACATGGTACC | |
| IgG2b mb | GGTCTGAAAAATTACTACCTGAAGAAG |
| CTTTACCTTGAAGAGTGTGACAG | |
| Kiaa0947 | GCCTGGACACTGGGTCCCCA |
| GTGGCTTCCTCCTTGGGGG | |
| MLL | GCAGGCGTCCCTCAAGCAGG |
| GGAGGCTTGGCAGCCTGACG | |
| Narg2 | AGGCCTTCGTAGGCGGGCA |
| AACTCAGGGTCCCAAGCGGCT | |
| Nsd1 | CACCAGGGGCAGGGCTTTGG |
| CCCCCAGCAGACCAGCAAGC | |
| poly(A) CstF2 | GATCCTGAGATTGCGCTGAAA |
| GGCTGAGGGTTGCCTGAAA |
siRNA for ELL2
The cloning of oligonucleotide inserts into pSilencer hygro 2.1-U6 (Ambion/Invitrogen) was previous described (1). The cloned DNA produces an siRNA driven by the U6 promoter. The clone with the maximal inhibition of ELL2 protein synthesis was CM482 (1), which contains the insert: 5′-Tcgagg CCA GTA GCT ACA CTG CTT CTT CAA GAG AGA AGC AGT GTA GCT ACT GGT TTT TTAC GCG TA-3′ and the complementary strand 5′-Agc ttac gcg taa aaaa CCA GTA GCT ACA CTG CTT CTC TCT TGA AGA AGC AGT GTA GCT ACT GGC C-3′. The siell2 sequence is not present in the transfected ELL2 cDNA clone used in these studies CM565. The ELL2 mRNA is decreased, and the expression of the secretory-specific Ig mRNA is reduced 5-fold with this siell2 (1). Another oligonucleotide clone of pSilencer hygro 2.1-U6, designated, CM 501, had no effect on ELL2 protein expression (1) and was used as a control (nonsense siRNA). The oligonucleotide sequence of CM 501 is 5′-Tcgagg TAT ATC GCT ATC GTC TCC TTT CAA GAG AAG GAG ACG ATA GCG ATA TAT TTT TTA CGC GTA-3′, and its complement if 5′-Agc ttac gcg taa aaaa TAT ATC GCT ATC GTC TCC TTC TCT TGA AAG GAG ACG ATA GCG ATA TAC C-3′. Transfections were performed with Lipofectamine 2000TM (Invitrogen) according the manufacturer's directions. Cells were incubated for 48 h before chromatin was isolated as detailed above.
Statistics
All experiments were repeated a minimum of three times with different biological replicates. The values for actin are indicated and served as an internal control. The error bars on the graphs indicate ± S.E. Samples in a given set were compared by two-way analysis of variance using the Prism 5TM/GraphPad Program with a Bonferroni post test. Significance is indicated by *, p ≤ 0.05, **, p ≤ 0.01, ***, p ≤ 0.001 and ****, p ≤ 0.0001.
RESULTS
Plasma Cell Lines Express Higher Levels of Elongation Factors, Especially ELL2
Our rationale for these studies was that changes in mRNA processing to IgH secretion in plasma cells might be linked to changes in multiple elongation factors and subsequent histone modifications both in amount and distribution across the gene. To study this, we used the A20 mouse cell line, which is representative of the B cell where sec and membrane (mb) IgH mRNA are about equal (sec = mb mRNA). The AxJ line, a plasma cell (where sec ≫ mb mRNA), was made by fusing A20 and a non-IgH-producing plasma cell line J558L (2). Each line is rapidly growing and expresses the identical IgH γ2a Vh gene (V-J2) with identical transcription start sites (20).
We examined the expression of transcription elongation factor mRNAs in these B cells and plasma cells. In Fig. 1, data are tabulated using quantitative RT-PCR, where values in each cell are normalized to hypoxanthine-guanine phosphoribosyltransferase. The mRNAs are grouped into four sets representing: ELL1 and -2 and its associated factors; the pTEFb components and associated genes; potential MLL fusion partners and other factors associated with them; and the histone methylases, etc. ELL2 expression is significantly increased (∼6-fold) in the plasma cells, the highest increase seen among any of these factors (Fig. 1A). Meanwhile for ELL1, the mRNA level is not altered with differentiation to plasma cells, also confirmed by protein expression (data not shown). There is a significant (∼2-fold) increase in the mRNAs for subunits of, or genes associated with, pTEFb (Fig. 1B) including cyclin T; spt5, the suppressor of yeast Ty homologue; the 7SK, a small but highly abundant RNA; and HEXIM. HEXIM protein and 7SK RNA have been shown to regulate pTEFb (21).
FIGURE 1.
The mRNA expression pattern in plasma cells favors transcriptional elongation. The mRNA expression in the B cell line (white bars) was compared with that of the plasma cell line (dark gray bars) using dT priming for cDNA synthesis followed by quantitative PCR. Expression of hypoxanthine-guanine phosphoribosyltransferase (HPRT) was set as 1 in each cell type, and the expression relative to hypoxanthine-guanine phosphoribosyltransferase was plotted on the x axis. Note the differences in scales between the groups of mRNA. A, contains mRNAs for the eleven-nineteen lysine-rich leukemia gene 1 or 2 or its associated factors eaf1 and eaf2. ELL2 shows the highest level of induction in plasma cells among this and all groups. B, pTEFb (cdk9 and cyclin T) and associated factors. The highly abundant 7SK RNA was primed by random oligonucleotides for the RT reaction and compared with an hypoxanthine-guanine phosphoribosyltransferase random-primed reaction; the amount of 7SK was divided by 1,000 to plot it on the same scale as the other pTEFb factors. Many of these factors are significantly increased in plasma cells. C, components of the super elongation complexes recently described (see “Results”). D, the mRNA abundance of the histone methylases (MLL, Dot1L, and Nsd1), cyclin C (CCNC), and its associated cdk8 and the polyadenylation cleavage factor CstF-64 (pA cstF2). Significant differences in plasma cells versus B cells were determined in the Bonferroni post test following two-way analysis of variance as indicated with *, p ≤ 0.05, **, p ≤ 0.01, ***, p ≤ 0.001, ****, p ≤ 0.0001; the error bars represent S.E. in this and all subsequent graphs derived from at least three different biological replicates.
A significant almost 3-fold increase is seen in the mRNA for Aff1 in the plasma cells (Fig. 1C). AFF1/af4 has been shown to influence epigenetic signatures and pTEFb kinase activation in the context of the MLL-af4 fusion (19). Among the remaining factors we surveyed, Dot1L, an H3K79 histone methyl transferase, and CstF2 (poly(A) CstF2), a polyadenylation factor, were increased slightly in plasma cells (Fig. 1D). Thus many factors involved in the transcriptional super elongation complex (8), Dot-com complex for H3K79 methylation (17), and the Tat-tar-associated factors (9, 10) are increased in plasma cells; ELL2 shows the largest increase of all of those tested.
IgH Secretory mRNA Expression Is Influenced by ELL2
To explore the mechanism of action of ELL2, B cells were transfected with an IgH reporter (Fig. 2A) capable of being processed to the secretory-specific and membrane-specific mRNA forms. In B cells or cells transfected by empty vector, the ratio of IgH sec to mb-specific form of mRNA produced is about 1.2:1 (Fig. 2B). As we showed previously (1), after transfection with ELL2, the amount of the secretory-specific mRNA increases to about five times that of the membrane form, whereas ELL1 and CstF2 have little or no effect on shifting mRNA production (Fig. 2B). Co-transfection with an shRNA encoding plasmid that produces an siell2 RNA against ELL2 reduced the sec:mb ratio about 5-fold more than empty vector-treated B cell. The sec:mb ratio observed for the same gene in plasma cells is 10:1, shown for comparison. Thus ELL2 alone is able to induce secretory-specific IgH mRNA production in B cells almost to the amount seen in plasma cells, which have increased expression of multiple elongation factors.
FIGURE 2.
ELL2 influences IgH secretory mRNA production, histone methylations, and factor associations with the IgH gene. A, The IgH and its mRNA products are diagrammed. Primers used for quantification of mRNA levels are indicated on the respective mRNAs and listed in Table 1. The regions indicated on the gene are: TATA, for transcription start site or TATA box; V:J2, for the V to J fusion region; J4 for the unused joining-region remaining in the intervening sequence; EH for the heavy chain enhancer region also known as Eμ; CH1, for constant heavy chain region 1; CH2, constant heavy chain region 2; sec pA for the secretory specific poly(A) site; sec + 200, the region 200 bp downstream of the secretory poly(A) site; IVS for the intervening sequence between the secretory and membrane specific regions; M1 for membrane-specific exon 1; pAmb3′ for a probe just 3′ of the membrane specific poly(A) site. B, the B cells were transfected with the intact IgH of γ2b on a plasmid and the indicated cDNAs for ELL2, ELL1, or CstF2 or siRNA as described previously. Cells were harvested 48 h after transfection, and the level of IgH secretory and membrane forms of mRNA was determined using quantitative RT-PCR with the probes diagramed in Panel A on the mRNAs. The value for the sec:mb mRNA ratio in plasma cells is shown for comparison. %IP, percentage immunoprecipitated. C and D, a plasma cell lacking its own IgH was co-transfected with an intact IgH gene and siRNA to ell2, a nonsense siRNA, or an ELL2 cDNA lacking the siell2 target sequence. After 48 h, the cells were subjected to chromatin immunoprecipitation with the indicated antibodies. The qPCR signals for the TATA and V-J region of the transfected IgH were averaged from at least three biological replicates. C, ChIP was performed with antibodies to the various H3K methylations. D, ChIP was performed with antibodies to the indicated initiation and elongation factors. ns, normal serum.
ELL2 Influences the Level of Histone H3 Methylations
As shown above, an increase in ELL2 in plasma cells appears causally linked to the alternative processing of IgH mRNAs. We asked whether ELL2 plays a role in changes in histone H3 methylations, some of which were associated with alternative mRNA processing (5). The density of histone methylation at the IgH transcription start site (TATA and V-J2) of the transfected IgH gene in plasma cells (location diagrammed in Fig. 2A) was determined by ChIP and qPCR. As shown in Fig. 2C, the presence of the siRNA that targets ELL2 substantially decreased H3K4 di- and trimethylation and H3K79 trimethylation. Adding an ELL2-specific cDNA (lacking the siell2 target site) back restores the methylations. Thus ELL2 affects both the H3K4 and the H3K79 methylase activities. The H3K36 methylation was unaffected by siell2.
ELL2 Influences Association of Many Factors and RNAP-II Modifications
To determine which steps in transcription elongation of the IgH gene are impacted after siell2 co-transfections, we performed ChIP experiments. As summarized in Fig. 2D, the amount of RNAP-II on the IgH transcription start site remained unchanged relative to a control actin promoter when siell2 was present. The amount of Ser-5 phosphorylation of the CTD of RNAP-II was elevated when siell2 was present, perhaps reflecting stalling of RNAP-II. The presence of siell2 decreased MLL association with the IgH transcription start site; the association of Dot1L with the complex was also diminished. The siell2 reduced the presence of ELL2, as expected, and also decreased pTEFb (cyclin T plus cdk9) and the Ser-2 phosphorylation of the CTD of RNAP-II and reduced association with CstF2, part of the mRNA cleavage/polyadenylation complex. Thus siell2 blocks the H3K4 and H3K79 methylation activities (Fig. 2C) by reducing the amount of the enzymes associated with the transcription complex (Fig. 2D). Adding back a cDNA with ELL2 lacking the siell2 target region restored the normal modifications on the gene.
The IgH Locus in Plasma Cells Displays High H3K Methylations beyond the Enhancer Region
We performed ChIP analyses of the identical IgH locus (upper panel) in the two different cell types to determine whether there were histone modification changes associated with the increase in ELL2 in plasma cells. In B cells, the histone H3K79 di- and trimethylations were clustered between the TATA box and the heavy chain enhancer and were significantly decreased downstream of that (Fig. 3B), a pattern similar to that seen for the Lys-79 methylations in LPS-activated splenic B cells (see supplemental Fig. 1). In contrast, in plasma cells (Fig. 3C), the initial levels of H3K79 di- and trimethylations at TATA were higher than in B cells by ∼2-fold. Those methylations of H3K79 extended into the region downstream of the enhancer and all the way to the 3′ membrane poly(A) site. H3K79 is within the histone core region, and modifications in this region could be significant in allowing DNA unwinding for transcription (22). The gastrin gene (Gast) transcription start site serves as a negative control as it is not expressed in B cells or plasma cells (data not shown). β-Actin (Actb) serves as a positive control in these studies. The H3K79 modifications on the actin promoter did not differ substantially between B cells and plasma cells. We conclude, based on the requirement for ELL2 to allow H3K79 trimethylation and Dot1L binding, that this pattern in plasma cells is the result of the action of this transcription elongation factor.
FIGURE 3.
The distribution of H3K79 di- and trimethylation is enhanced in the region downstream of the IgH enhancer in plasma cells. A, the 11-kb IgH γ2a gene in the two cell lines representing the B cell (membrane IgH > secretory) and plasma cell (sec IgH ≫ membrane) is identical and in the normal Ig locus context, as diagrammed in this panel. Probes used in the subsequent qPCRs are shown. The IgH γ2a-producing cells were fixed, and chromatin immunoprecipitation was performed as described using antibodies to the individual Lys-79 methylations. B and C, B cells (B) and plasma cells (C). %IP, percentage immunoprecipitated.
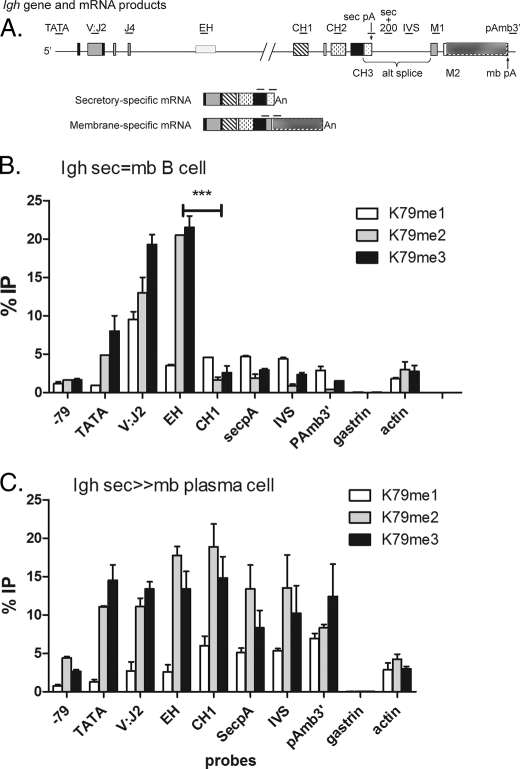
Because siell2 influenced MLL association with the IgH gene and H3K4 methylation (Fig. 2), we tested whether the distribution pattern of H3K4me across the IgH gene would differ between B cells and plasma cells. We saw a peak in both cell types of H3K4 me2 and me3 near the transcription start site at probes TATA and V-J2 (Fig. 4B). This is the pattern seen in many genes as reported in the literature A (14) and in splenic B cells (see data in supplemental Fig. 1). However, in plasma cells (Fig. 4C), the H3K4 me2 and me3 modifications continue downstream to the regions designated as CH1, sec poly(A), IVS, and poly(A) m3′ well beyond the IgH enhancer. The amount of RNAP-II on these genes in this region does not differ between cell types as shown below, but the chromatin modifications at H3K4 are indicative of more activation farther downstream in plasma cells through the action of elongation factors, especially ELL2.
FIGURE 4.
The distribution of H3K4 dimethylation differs between B cells versus plasma cells. A, the 11-kb IgH γ2a gene, is illustrated. The B cells and plasma cells were immunoprecipitated with antibodies to the H3K4 methylations as described above and subjected to qPCR analyses. B represents the data obtained with B cells, whereas C shows the data obtained in plasma cells. %IP, percentage immunoprecipitated.
Comparison of Associations of RNAP-II and Histone H3 on IgH between B Cells and Plasma Cells
The patterns of immunoprecipitations of fixed chromatin were performed using antibodies to the RNAP-II and histone H3 across the IgH gene (Fig. 5). There is a higher signal for polymerase in the plasma cells than in the B cells around V-J2, about 300 bp downstream of the transcription start site and about 1 kb upstream of the heavy chain enhancer region (Fig. 5B). The transcription start site region is open and thus depleted of histones in both cell types. The patterns obtained with total histone H3 and acetylated H3K9 and H3K14 (not shown) were similar to those we saw previously (20). There is no significant difference between the cell types in polymerase loading across the rest of the IgH gene despite the higher amount initially in the plasma cells. There is a small peak of RNAP-II in the region between the secretory-specific poly(A) site and the membrane exons. This region had been shown to be a polymerase pause site (23) that does not differ significantly in polymerase occupancy between B cells and plasma cells.
FIGURE 5.
The distribution of H3K36 trimethylation differs between B cells versus plasma cells. A, the 11-kb IgH γ2a gene in the two cell lines representing the B cell is shown. B, the cells were fixed, and chromatin immunoprecipitation was performed using antibodies to RNAP-II and histone H3 across the IgH gene in either B cells or plasma cells. The distribution in each cell type is indicated. %IP, percentage immunoprecipitated. C, the profile of H3K36 trimethylation across the IgH γ2a gene.
Histone H3K36 Methylation Is Increased beyond the IgH Enhancer in Plasma Cells
We performed ChIP assays of the H3K36 methylation patterns across the IgH γ2a gene in B cells versus plasma cells (Fig. 5C). We found that the amount of H3K36 trimethylation was significantly higher in plasma cells than in B cells, especially 3′ of the IgH enhancer region. Higher levels of modification are centered around the region (sec pA) where the secretory poly(A) site is located in both cell types. The higher level of H3K36 methylation in plasma cells in the region between CH1 and 200 bp beyond the sec poly(A) site cannot be attributed to a difference in the amount of the RNAP-II in that region based on the data shown in Fig. 5B.
DISCUSSION
We previously demonstrated that the addition of ELL2 can drive IgH secretory-specific mRNA production by facilitating exon skipping and proximal poly(A) site use (1). Higher Ser-2 phosphorylation of the CTD of RNAP-II and greater association of ELL2 and the polyadenylation factors with the start of IgH gene were seen in plasma cells versus B cells (20). Here we found that ELL2 influences factor associations and histone methylation at H3K79 and H3K4. These H3 modifications extend beyond the IgH enhancer in plasma cells in contrast to their promoter proximal clustering in B cells. Thus to process IgH mRNA to the secretory-specific form in plasma cells, several things occur as a result of the addition of ELL2: (a) pTEFb association and Ser-2 phosphorylation occur; (b) the polyadenylation capacity of the RNAP-II is increased, and the skipping of less optimal splice sites, like that of the one in the terminal secretory exon, is enhanced; and (c) changes occur in the methylation of the H3 histones, especially at H3K4 and H3K79, allowing the downstream chromatin to open more readily and be transcribed more efficiently.
Our experiments show that reducing ELL2 with siRNA decreases the association of pTEFb, the Ser-2 phosphorylation of RNAP-II, and the association of polyadenylation/cleavage factor CstF64 (CSTF2). Thus efficient elongation, Ser-2 phosphorylation, and the polyadenylation capacity of the polymerase are linked. This is similar to the paf1 subunit cdc73 where siRNA depletion experiments of cdc73 showed that mRNA abundance and first poly(A) site utilization were impacted negatively (24). The paf1 complex is necessary for association of the polyadenylation/cleavage factors with RNAP-II.
Histone Modifications
Methylation of the histone H3 tails has been closely linked through enzymes detecting the phosphorylation of the CTD of RNAP-II. H3K4 methylation near the promoter is associated with phosphorylation of the CTD of RNAP-II at Ser-5 and Ser-7 of the heptad repeats via TFIIH- cdk7 kinase activity (25) and the subsequent CTD interactions with MLL or SMYD2, the histone methylases (Ref. 15 and reviewed in Ref. 26). We found a reduction in MLL bound near the IgH promoter in the presence of siRNA to ELL2, although Ser-5 phosphorylation of CTD was not affected. This was surprising because it was known that MLL targets the 5′ end of genes and brings along the elongation factors, yet it was not previously known whether the absence of elongation factors such as ELL2 would influence MLL binding as we have seen here.
Because H3K79 resides within the histone core, its methylation may serve to help unwind the DNA from the histone more readily, leading to increased transcription readthrough. Point mutations in the yeast histones at a comparable position facilitate DNA unwinding, and the DNA is traversed by RNAP-II as if it were devoid of nucleosomes (22). H3K79 methylations are brought about by Dot1L (13). Previous studies showed that Dot1L preferentially occupies the proximal transcribed region of active genes, correlating with enrichment of H3K79 di- and trimethylation there (13). Here we show H3K79 methylation in downstream regions of the IgH gene when its RNA product is being processed to the secretory-specific form in plasma cells. Conversion of H3K79 that is monomethylated into di- and trimethylated forms is correlated with the transition from low to high level gene transcription, mostly likely because of a decrease in the histone-DNA interaction. Recent studies on leukemia have shown a requirement for Dot1L for leukemogenesis, whereas there is selective tumor killing by small molecule inhibitors of Dot1L (27, 28). Thus H3K79 methylation is emerging as an important step in gene dysregulation in cancer. The action of ELL2 on H3K79 methylation that we show here is notable, and the broad distribution pattern of H3K79 methylation in plasma cells may significantly impact the ability of the mRNA to be processed.
The multisubunit Dot1 complex (Dot-com) includes the MLL partner ENL, which may functionally link the super elongation complex (SEC) with H3K79 modifications. Some controversy in the literature exists; no stable complexes between SEC and Dot-com were isolated in one study (17), whereas in an analysis of MLL-AF4 fusions, large complexes involving not only pTEFb but also Dot1L have been observed (19). From in vitro studies, ELL proteins were shown to increase the catalytic rate of transcription elongation by RNA polymerase II (7, 29). That activity is consistent with what we see here, the enhancement of transcription by influencing the unwinding of the DNA from the histone core, thus bypassing transcription pause sites. Depleting ELL2 with siRNA led to decreased binding of Dot1L to the polymerase complex on the IgH gene and a consequential decrease in H3K79 trimethylation. Depleting ELL2 decreases H3K4 methylation and MLL binding. We cannot yet distinguish whether these are sequential events, whether ELL2 acts independently on both, or whether ELL2 is part of a larger complex, but this is an important question for future study.
Role of the IgH Enhancer
The IgH enhancer represents a unique barrier between high to low levels of H3K4 and H3K79 methylations in B cells, which is breached in plasma cells. The enhancer region is also an inflection point for H3K36 methylation, but that occurs in both cell types. In a recent review, the still enigmatic role of enhancers in maintenance and memory of transcriptional activation was explored (30). The enhancer (Eμ) between the Vh regions and Ch regions was the first transcriptional enhancer discovered (31), yet its mechanism of action remains obscure. The enhancer seems to be required for both establishment and maintenance of μ heavy chain gene expression (32). Loss of the IgH enhancer in germline DNA results in significant reduction in histone acetylation spanning several hundred kilobases (33). It is clear that there is cross-talk between 3′ end processing and the promoter (34) with proper polyadenylation and cleavage stimulating transcription. Exactly how the enhancer functions with the promoter and elongation factors in IgH to alter chromatin structure for efficient production of the IgH secretory mRNA remains an intriguing question.
The pattern of H3K4 and H3K79 methylations and pTEFb-ELL2 associations downstream of the IgH enhancer region in plasma cells may have implications not only for the production of the secretory-specific IgH mRNA but also for Ig somatic hypermutation and IgH internal enhancer function. The mRNA expression patterns of the Aicda (activation-induced cytidine deaminase) enzyme and Supt5h are reversed in B cells versus plasma cells; activation-induced cytidine deaminase is high in B cells, whereas supt5 is low. In B cells, activation-induced cytidine deaminase is associated with Spt5 on stalled polymerases and initiates Ig hypermutation and class switching (35). With high pTEFb in plasma cells, the threonine 4 residues in the C-terminal repeats of Spt5 are phosphorylated, thereby converting DSIF from a repressor of elongation to an activator (36). Thus the increased association of pTEFb-SEC-ELL2 in plasma cells with the RNAP-II, the consequent loss of stalling by higher levels of Spt5, coupled with a shutoff of the activation-induced cytidine deaminase enzyme, could effectively shut down somatic hypermutation. Therefore the increase of ELL2 in plasma cells may have many important consequences beyond alternative RNA processing.
Supplementary Material
Acknowledgments
We thank Lisa Borghesi for help with splenic B cell isolation and excellent discussions, Joshua Paul and Brandon Nelson for assistance with RT-qPCR, and Mark Nichols for editorial comments.
This work was supported by National Science Foundation Grant MCB-0842725 (to C. M.), National Institutes of Health Grant 5T32 CA82084-12 (to K. S. P.), and the Summer Undergraduate and Office of Research programs at the University of Pittsburgh, School of Medicine.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- ELL
- eleven-nineteen lysine-rich leukemia gene
- EAF
- ell-associated factor
- MLL
- multiple lineage leukemia gene
- sec
- secretory
- mb
- membrane
- SEC
- super elongation complex
- pTEFb
- positive transcription factor b
- CTD
- carboxyl-terminal domain
- RNAP-II
- RNA polymerase II
- qPCR
- quantitative PCR.
REFERENCES
- 1. Martincic K., Alkan S. A., Cheatle A., Borghesi L., Milcarek C. (2009) Nat. Immunol. 10, 1102–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Milcarek C., Suda-Hartman M., Croll S. C. (1996) Mol. Immunol. 33, 691–701 [DOI] [PubMed] [Google Scholar]
- 3. de la Mata M., Lafaille C., Kornblihtt A. R. (2010) RNA 16, 904–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Muñoz M. J., de la Mata M., Kornblihtt A. R. (2010) Trends Biochem. Sci. 35, 497–504 [DOI] [PubMed] [Google Scholar]
- 5. Luco R. F., Pan Q., Tominaga K., Blencowe B. J., Pereira-Smith O. M., Misteli T. (2010) Science 327, 996–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Luco R. F., Misteli T. (2011) Curr. Opin. Genet. Dev. 21, 366–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shilatifard A., Duan D. R., Haque D., Florence C., Schubach W. H., Conaway J. W., Conaway R. C. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 3639–3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lin C., Smith E. R., Takahashi H., Lai K. C., Martin-Brown S., Florens L., Washburn M. P., Conaway J. W., Conaway R. C., Shilatifard A. (2010) Mol. Cell 37, 429–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. He N., Liu M., Hsu J., Xue Y., Chou S., Burlingame A., Krogan N. J., Alber T., Zhou Q. (2010) Mol. Cell 38, 428–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sobhian B., Laguette N., Yatim A., Nakamura M., Levy Y., Kiernan R., Benkirane M. (2010) Mol. Cell 38, 439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sugiura N., Dadashev V., Corriveau R. A. (2004) Eur. J. Biochem. 271, 4629–4637 [DOI] [PubMed] [Google Scholar]
- 12. Gutiérrez N. C., Ocio E. M., de Las Rivas J., Maiso P., Delgado M., Fermiñán E., Arcos M. J., Sánchez M. L., Hernández J. M., San Miguel J. F. (2007) Leukemia 21, 541–549 [DOI] [PubMed] [Google Scholar]
- 13. Steger D. J., Lefterova M. I., Ying L., Stonestrom A. J., Schupp M., Zhuo D., Vakoc A. L., Kim J. E., Chen J., Lazar M. A., Blobel G. A., Vakoc C. R. (2008) Mol. Cell. Biol. 28, 2825–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Milcarek C. (2011) in RNA Processing (Grabowski P. ed) InTech, Rijeka, Croatia, in press [Google Scholar]
- 15. Wang P., Lin C., Smith E. R., Guo H., Sanderson B. W., Wu M., Gogol M., Alexander T., Seidel C., Wiedemann L. M., Ge K., Krumlauf R., Shilatifard A. (2009) Mol. Cell Biol. 29, 6074–6085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Spies N., Nielsen C. B., Padgett R. A., Burge C. B. (2009) Mol. Cell 36, 245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mohan M., Herz H. M., Takahashi Y. H., Lin C., Lai K. C., Zhang Y., Washburn M. P., Florens L., Shilatifard A. (2010) Genes Dev. 24, 574–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mueller D., Bach C., Zeisig D., Garcia-Cuellar M. P., Monroe S., Sreekumar A., Zhou R., Nesvizhskii A., Chinnaiyan A., Hess J. L., Slany R. K. (2007) Blood 110, 4445–4454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Benedikt A., Baltruschat S., Scholz B., Bursen A., Arrey T. N., Meyer B., Varagnolo L., Müller A. M., Karas M., Dingermann T., Marschalek R. (2011) Leukemia 25, 135–144 [DOI] [PubMed] [Google Scholar]
- 20. Shell S. A., Martincic K., Tran J., Milcarek C. (2007) J. Immunol. 179, 7663–7673 [DOI] [PubMed] [Google Scholar]
- 21. Krueger B. J., Varzavand K., Cooper J. J., Price D. H. (2010) PLoS ONE 5, e12335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Luse D. S., Spangler L. C., Újvári A. (2011) J. Biol. Chem. 286, 6040–6048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Peterson M. L., Bertolino S., Davis F. (2002) Mol. Cell Biol. 22, 5606–5615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rozenblatt-Rosen O., Nagaike T., Francis J. M., Kaneko S., Glatt K. A., Hughes C. M., LaFramboise T., Manley J. L., Meyerson M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 755–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Akhtar M. S., Heidemann M., Tietjen J. R., Zhang D. W., Chapman R. D., Eick D., Ansari A. Z. (2009) Mol. Cell 34, 387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Smith E., Lin C., Shilatifard A. (2011) Genes Dev. 25, 661–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nguyen A. T., Taranova O., He J., Zhang Y. (2011) Blood 117, 6912–6922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Daigle S. R., Olhava E. J., Therkelsen C. A., Majer C. R., Sneeringer C. J., Song J., Johnston L. D., Scott M. P., Smith J. J., Xiao Y., Jin L., Kuntz K. W., Chesworth R., Moyer M. P., Bernt K. M., Tseng J. C., Kung A. L., Armstrong S. A., Copeland R. A., Richon V. M., Pollock R. M. (2011) Cancer Cell 20, 53–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shilatifard A., Lane W. S., Jackson K. W., Conaway R. C., Conaway J. W. (1996) Science 271, 1873–1876 [DOI] [PubMed] [Google Scholar]
- 30. Sen R., Grosschedl R. (2010) Genes Dev. 24, 973–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Grosschedl R., Baltimore D. (1985) Cell 41, 885–897 [DOI] [PubMed] [Google Scholar]
- 32. Grosschedl R., Marx M. (1988) Cell 55, 645–654 [DOI] [PubMed] [Google Scholar]
- 33. Chakraborty T., Perlot T., Subrahmanyam R., Jani A., Goff P. H., Zhang Y., Ivanova I., Alt F. W., Sen R. (2009) J. Exp. Med. 206, 1019–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mapendano C. K., Lykke-Andersen S., Kjems J., Bertrand E., Jensen T. H. (2010) Mol. Cell 40, 410–422 [DOI] [PubMed] [Google Scholar]
- 35. Pavri R., Gazumyan A., Jankovic M., Di Virgilio M., Klein I., Ansarah-Sobrinho C., Resch W., Yamane A., Reina San-Martin B., Barreto V., Nieland T. J., Root D. E., Casellas R., Nussenzweig M. C. (2010) Cell 143, 122–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yamada T., Yamaguchi Y., Inukai N., Okamoto S., Mura T., Handa H. (2006) Mol. Cell 21, 227–237 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.